ABSTRACT
To exploit highly conserved and difficult drug targets, including multipass membrane proteins, monoclonal antibody discovery efforts increasingly rely on the advantages offered by divergent species such as rabbits, camelids, and chickens. Here, we provide an overview of antibody discovery technologies, analyze gaps in therapeutic antibodies that stem from the historic use of mice, and examine opportunities to exploit previously inaccessible targets through discovery now possible in alternate species. We summarize the clinical development of antibodies raised from divergent species, discussing how these animals enable robust immune responses against highly conserved binding sites and yield antibodies capable of penetrating functional pockets via long HCDR3 regions. We also discuss the value of pan-reactive molecules often produced by these hosts, and how these antibodies can be tested in accessible animal models, offering a faster path to clinical development.
Introduction
Therapeutic monoclonal antibodies (MAbs) now comprise the fastest growing class of drugs, and this market is expected to reach $300B in 2025.Citation1 There are currently over 100 approved MAbs, with over 800 in clinical trials as of 2020.Citation2,Citation3 Fierce competition abounds for those that bind the most tractable and best understood targets, such as CD20, epidermal growth factor receptor (EGFR), and programmed cell death ligand 1 (PDL1).Citation4 Despite the saturation of MAbs for the more easily druggable targets, i.e., the proverbial low-hanging fruit, substantial opportunity remains for treatments exploiting more difficult therapeutic targets.
The majority of approved MAb therapeutics has relied on mouse immunizations and hybridoma technologies originally developed in the 1970s.Citation1 While this strategy has been exceedingly successful, many targets remain untapped due to biological or technological MAb discovery hurdles, including high sequence identity between humans and mice, or specific MAb-targeting requirements for epitopes that are difficult to access. Valuable drug targets usually have essential functions and so are often highly conserved, especially among mammals.
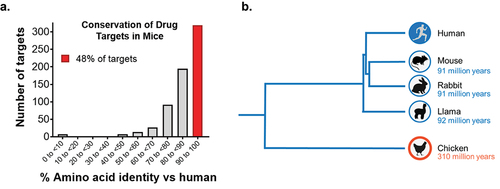
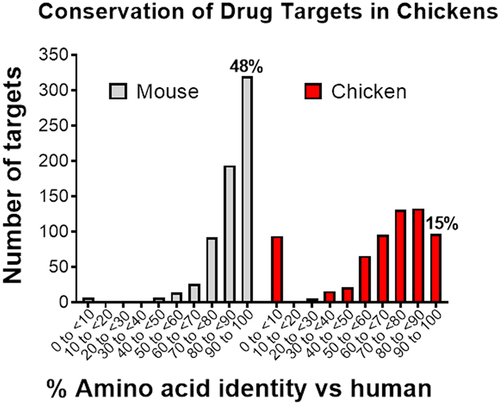
To understand the potential gaps in the therapeutic antibody space, we performed an analysis of 663 human therapeutic targets of large and small molecules using the ECOdrug database. We found that nearly half (48%) of these targets are highly conserved between humans and mice, with > 90% amino acid identity (). However, developing MAbs against these conserved targets is extremely difficult, as only 14% of approved and late-stage therapeutic MAb targets are conserved at the > 90% identity level.Citation5 This indicates that valuable conserved targets are underrepresented in the therapeutic MAb space. High target protein conservation is typically met with immune tolerance from the immunization host animal, substantially limiting the strength of the immune response and the epitope diversity of the generated MAbs.Citation6 By taking advantage of host species divergent from humans (), immune tolerance can be bypassed, enabling access to a plethora of yet untapped targets for MAb discovery.
Figure 1. Sequence conservation of drug targets. a) Human drug targets show high conservation with mouse orthologs. A collection of 663 small and large molecule human drug targets curated in the ECOdrug database7 were analyzed with respect to their sequence identity in mice. 48% of targets showed sequence identity of >90%, and 24% of targets showed sequence identity of >95% compared to their mouse orthologs. Seven targets were not predicted to have a murine ortholog. b) Phylogenetic tree of divergent species used as hosts for immunization, shown with respect to evolutionary distance from humans.8,9
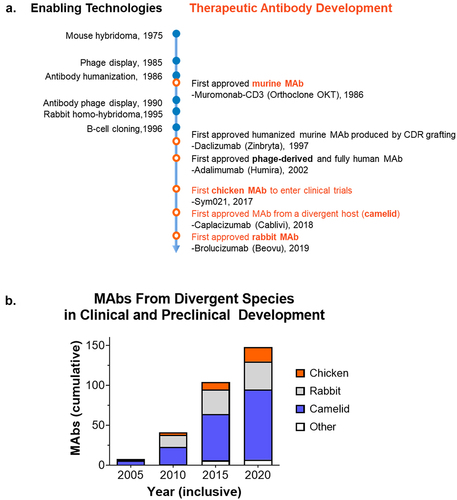
The emergence of technologies such as B-cell cloning and phage/yeast display has revolutionized antibody discovery, providing alternatives to traditional mouse hybridomasCitation10 and enabling the productive use of non-murine hosts to raise antibodies (). For the most complex targets such as membrane proteins, the most successful approach has been panning libraries derived from immunized divergent animal species.Citation11–13 The use of new DNA and mRNA immunization strategies, either alone, or in combination with protein, also contributes to the feasibility of successful antibody isolation campaigns using divergent animal hosts.Citation14,Citation15 This enables a robust immune response against native epitopes without requiring large quantities of difficult to obtain antigen.
Figure 2. Technologies enabling MAbs from divergent species. a) The development of antibody discovery technologies that provide an alternative to hybridomas have enabled the discovery and approval of therapeutic MAbs from divergent species, including camelid, rabbit, and chicken. b) Increasing use of divergent species for therapeutic MAb discovery and development. Analysis of the TABS therapeutic antibody database23 between 2005 and 2020 demonstrates the growing pipeline of MAbs from divergent species in clinical and preclinical development.
Phage and yeast panning can also be performed on naïve (non-immunized) animal libraries or synthetic libraries where protein conservation and immune tolerance or target toxicity do not present a barrier for antibody discovery.Citation4,Citation16 Panning a synthetic/naïve library resulted in the first phage-derived drug to be approved, the blockbuster drug adalimumab.Citation1,Citation17 While this approach has been successful, antibodies from synthetic sources often initially have lower affinity and developabilityCitation5,Citation18 and are difficult to isolate due to their low abundance in naïve libraries. Naïve or synthetic approaches are often not successful for the most difficult targets such as multipass membrane proteins, especially when the concentration of such antigens is limited. Even when isolated from synthetic/naïve libraries, such MAbs usually require substantial engineering to improve their affinity and create final candidates.
Here, our focus is animal-derived antibodies, as such approaches still represent the most successful campaigns for difficult and conserved targets. Animal-derived antibodies also benefit from natural immune selection mechanisms, including clonal selection, somatic hypermutation, and protein quality control that results in the isolated MAbs being both high affinity and developable.Citation19
With the introduction of newer antibody discovery technologies, the use of animals, such as llamas, rabbits, chickens, and even sharks, for antibody discovery is widespread. The first therapeutic antibodies derived from divergent species have now been approved for use, including the camel-derived caplacizumab (Cablivi®), approved by the European Medicines Agency in 2018 and by the US Food and Drug Administration FDA in 2019 (), followed by the first rabbit-derived MAb brolucizumab (Beovu®) in 2019. Antibodies derived from chickens are likely to follow since they have recently entered clinical trials. While there is excitement around the opportunity provided by antibody discovery in divergent species, there remain unanswered questions. For example, will there be unanticipated immunogenicity arising from antibody sequences derived from such animals, and will anti-drug antibodies (ADA) pose an issue in patients? This is difficult to predict as ADA can occur with any therapeutic protein, even those with 100% human origin. However, in the limited cases thus far of MAbs derived from divergent species (caplacizumab (camelid), brolucizumab (rabbit) and eptinezumab (rabbit)), ADA was not noted to cause decreased drug efficacy.Citation20–22 As of 2020, there were over 150 clinical and preclinical MAbs in development from divergent species (), and this growing trend is expected to continue.
In this review, we discuss the increasing value that divergent species bring to successful MAb discovery programs (). The use of rabbits, camelids, and chickens is emphasized, since MAbs from these species are the most prevalent among divergent species and are the most clinically advanced.
Table 1. MAb discovery challenges addressed by using divergent hosts.
Advantages of divergent species for MAb discovery
Obtaining a robust immune response and avoiding immune tolerance
The generation of large panels of MAbs with diversity in sequence, epitope, and function is key for discovering rare MAbs with desired therapeutic properties, such as conformational state sensitivity, functional inhibition of the target, or the ability to discriminate among closely related off-target proteins.Citation11,Citation14,Citation24 However, obtaining a robust immune response with broad epitope diversity is one of the most challenging aspects of MAb discovery, especially for highly conserved targets. Animal hosts generate antibodies when exposed to ‘non-self’ proteins. Due to immune tolerance, they generally do not mount effective immune responses to immunogens that are highly homologous to their own proteins. Our experience from thousands of immunization campaigns has shown that protein immunogens with 90% to 100% sequence identity to their host counterparts usually generate extremely poor immune responses and are challenging targets for antibody discovery. The difficulty in obtaining a robust immune response is exacerbated for functionally important epitopes, such as ligand binding sites that tend to be more highly conserved.Citation25,Citation26
Despite the historical use of mice (and more recently rats), their utility in mounting an effective immune response against human proteins is limited due to their evolutionary proximity to humans. In our analysis of human drug targets, nearly all (99%) have a murine ortholog, almost half (48%) have ≥ 90% sequence identity, and about a quarter (24%) are ≥ 95% identical (). Rat proteins are similarly highly conserved with their human counterparts. The median sequence identity for mouse and rat versus human orthologs was determined to be 88.0 and 88.3% by the Rat Genome Sequencing Project Consortium.Citation27 The issue of immune tolerance remains even if using transgenic humanized mice as host animals. Regardless of whether the elicited antibodies have mouse or human antibody frameworks, the host animal must recognize a protein target as foreign material to mount an effective antibody response.
Immune tolerance in antibody host species is a problem that extends across all mammals, since the class branched out in a relatively short period of time starting approximately 91 million years ago ().Citation8 Choosing a host species that provides a robust immune response requires a careful balance that provides enough evolutionary distance to avoid immune tolerance, a sufficiently advanced immune system that produces a diverse antibody repertoire, and antibody frameworks readily amenable to humanization and affinity maturation. For highly conserved targets, divergent non-mammalian species have proven to be more effective hosts. Chickens have gained popularity as a divergent species for immunization due to their long evolutionary distance from humans (310 million years), while retaining similar antibody structure, similar antibody frameworks, and the ability to generate high affinity antibodies.Citation28
Accessing Functional Epitopes
Endogenous ligands and small molecule drugs often interact with their target proteins in concave ‘pockets’ that antibodies cannot often access due to their large size. However, protein binding regions of an antibody, called paratopes, can exhibit different topologies, and some of these can better reach protein grooves such as ligand binding pockets, catalytic sites, and other functionally important structures.Citation29 The choice of immunization host can influence antibody characteristics such as paratope topology.
The paratope is largely made up of complementarity-determining regions (CDRs), 3 of which are contributed by the heavy chain, and 3 from the light chain of the antibody. HCDR3 (the third CDR on the antibody heavy chain) is the most important for interactions with the antigen, as it is the most diverse in sequence and length compared with the other CDR regions.Citation30 It is encoded by the junction of three gene sequences (V, D, and J), with additional diversity introduced by random insertions and deletions during gene recombination.Citation31 HCDR3 lengths vary among animals, and longer sequences have been shown to form protruding paratopes that can access functionally important recessed epitopes.Citation12
A survey of the crystal structures of 50 antibodies compared HCDR3 length and paratope conformation and found that protruding paratopes were generally formed by HCDR3 sequences of more than 14 amino acids and were not observed with HCDR3 sequences of fewer than 9 amino acids.Citation32 These findings are exemplified by the HCDR3 regions of co-crystalized therapeutic antibodies nivolumab (4 amino acids, concave paratope), cetuximab (11 amino acids, flat paratope), and erenumab (21 amino acids, protruding paratope) (). For example, the protruding paratope of erenumab is composed of a 21 amino acid HCDR3 region critical for binding a deep pocket of the G protein-coupled receptor (GPCR) calcitonin gene-related peptide receptor (CGRPR) with high affinity (low pM) and high specificity.Citation33 Another antibody with an unusually long HCDR3 region of 23 amino acids bound a cavity of the SARS-CoV-2 spike protein.Citation34 Cryo-electron microscopy revealed a unique neutralizing mechanism of this MAb that locked the virus into an inactive conformation. Another example of this is JN241–9, the first published antibody with agonist activity for a GPCR.Citation35 This single domain antibody against the apelin receptor has an especially long HCDR3 region of 22 amino acids (as per Kabat numbering) that can deeply bind into the ligand-binding region of the receptor, resulting in activation.
Figure 3. HCDR3 length and paratope shape. Antibodies co-crystalized with target proteins corroborate the influence of HCDR3 length with paratope shape. In these examples, MAbs with smaller HCDR3 regions have concave (4 amino acid) and flat (11 amino acid) paratopes (left and middle panels). However, erenumab (right panel) with an HCDR3 length of 21 amino acids forms a protruding convex paratope that reaches into a pocket of the CGRP receptor. Structures were obtained from the Protein Data Bank (nivolumab 5WT9, cetuximab 5SX5, and erenumab 6UMG). Cetuximab was derived from mouse immunization, while nivolumab and erenumab were derived from transgenic mice that produce human MAbs.
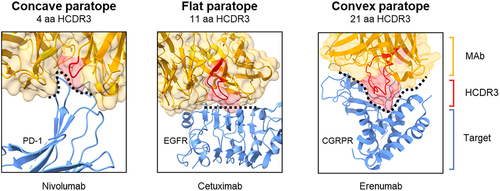
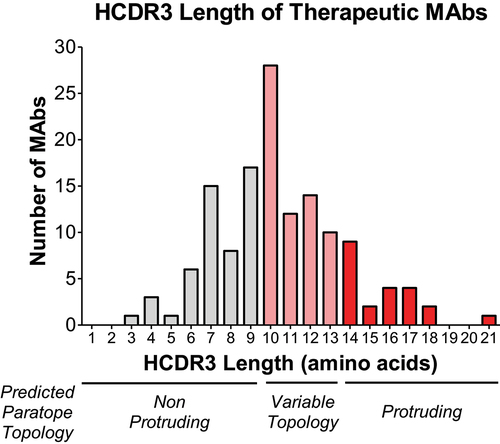
Despite the benefits of protruding paratopes, existing therapeutic MAbs overwhelmingly have short HCDR3 regions. Our analysis of 137 approved and late-stage therapeutic antibodiesCitation5 found that only 16% of these antibodies have HCDR3 sequences longer than 13 amino acids (). The lack of long HCDR3 regions in therapeutic MAbs corresponds to the routine use of mice in antibody discovery. HCDR3 lengths in mice are typically 9–13 amino acids and relatively short compared with many other animals.Citation36,Citation37 In a study by Wu et al., fewer than 7% of murine MAbs were found to have HCDR3 lengths greater than 12 amino acids.Citation38 Human antibodies have longer HCDR3 regions, with a mean length of approximately 13 amino acids (Kabat numbering).Citation36 Use of humanized transgenic mice in some cases has resulted in MAbs such as erenumab, which has an HCDR3 of 21 amino acids. Consistent with shorter HCDR3 regions, the paratopes of most native mouse MAbs are predicted to be flat and consequently bind flat epitopesCitation39 in a similar fashion to cetuximab which has an HCDR3 sequence of 11 amino acids.Citation40 Rather than penetrating a pocket, this MAb overlaps the ligand-binding region and sterically prevents dimerization ().
Figure 4. Few currently marketed therapeutic antibodies have long HCDR3. We analyzed the HCDR3 sequences of 137 FDA approved and late clinical stage MAbs.5 The average HCDR3 length of these MAbs was 10 amino acids. Only 16% of these MAbs have an HCDR3 length of 14 amino acids or more, the predicted threshold to form a protruding topology. The relationship between HCDR3 length and paratope shape noted here is based on the study by Ramsland et al.32
In theory, synthetic and engineered MAbs can also be designed with long HCDR3 regions. However, the dynamics associated with folding elongated amino acid structures are inherently unpredictable and make this difficult in practice. Paratope attributes encompass complexities beyond mere HCDR3 length, and include factors such as amino acid sequence, composition, and hydrophobicity. These can influence antibody developability due to factors such as aggregation and polyreactivity. For these reasons, antibody programs seeking protruding paratopes should consider leveraging animals with long HCDR3 regions where the host immune system’s natural selection provides architecture such as cysteine bridges for stabilization and has the ability to screen out antibodies with properties unsuitable for therapeutics. As antibody discovery parameters become more sophisticated, for example blocking functional sites or selectively binding conformational states, the use of alternate species with longer paratope regions becomes increasingly important.
Generating antibodies that can be used in preclinical animal testing
A practical advantage of using divergent species for immunization is the likelihood of obtaining cross-species reactive MAbs that can be used in easily accessible preclinical animal models. Therapeutic MAbs, like small molecules, must be tested in animals for efficacy and preclinical toxicology.Citation41,Citation42 Toxicology assessment typically requires two animal species, one rodent and one non-rodent;often rat and dog are used for small molecule testing. Toxicology studies for MAbs require testing in ‘relevant’ species that both express the target protein and retain the epitope sequence that is responsible for the binding interaction.Citation42,Citation43
Since antibody discovery has historically used mice, most late-stage MAbs have been non-reactive or poorly reactive in rodents and other mammals typically used in preclinical testing. As a result, non-human primates (NHPs) have been routinely used as relevant species for nonclinical safety and toxicology testing, and testing in such cases is permitted in a single species.Citation41 However, as antibody discovery pipelines continue to grow, so do ethical concerns about the increased use of NHPs in research.Citation44 Even chimpanzees, the most closely related animals to humans, have been used on occasion due to severely restricted MAb cross-reactivity. Efalizumab (anti-CD11a) and infliximab (anti-TNF) are both examples of antibodies that were tested in chimpanzees because they were not cross-reactive in any species other than chimpanzees and humans. Such studies would not be permitted now, and the use of chimpanzees for toxicology research has been discontinued by the NIH.Citation45 The FDA recently urged antibody developers to find alternatives to NHPs for MAb testing,Citation46 in part due to NHP shortages resulting from the COVID-19 pandemic.Citation47 In some cases, the FDA has indicated that toxicology studies may be conducted solely in rodents.Citation46 Such studies, however, still require an appropriately reactive MAb.
Surrogate antibodies are an indirect approach for toxicology testing used when a MAb’s narrow cross-reactivity makes it unfeasible to directly study in animals.Citation43 The binding characteristics of the surrogate MAb are supposed to resemble the clinical candidate as closely as possible. While some characteristics like affinity are relatively easy to reproduce, more relevant features such as the binding epitope are more difficult to replicate, but are crucial for assessing both on and off-target toxicities. In the case of efalizumab, some of the adverse effects seen in the clinic were not predicted by the safety profile of the surrogate, and efalizumab was later withdrawn from the market.Citation43 Surrogate MAbs involve a parallel antibody development program that is time-consuming, costly, and in the end only partially mirrors the actual drug, so this approach is generally used only when other options do not exist.
Divergent species as immunization hosts provide developers with a new pathway to avoid or reduce the use of surrogate MAbs and NHPs in toxicology studies, since this approach often yields pan-reactive antibodies that recognize multiple orthologous proteins in rodents and non-rodent mammals. Numerous examples of cross-reactive MAbs exist in the literature and in our own experience (). The first approved MAbs raised in divergent species were caplacizumab (camelid) and brolucizumab (rabbit), and both demonstrated cross-reactivity in rodents. These antibodies were tested in a combination of mice, rats, guinea pigs, and NHPs for their pharmacology and toxicology studies.Citation48,Citation49 The ability to obtain cross-reactive antibodies from chickens has been estimated at 80%,Citation6 which correlates with our own experience. The ability to use rodent animal models further increases the accuracy of toxicity evaluations, reduces dependence on non-human primates, and streamlines the process required to bring antibodies to the clinic.
Table 2. MAbs raised in divergent species frequently show cross-reactivity in rodents and other animals frequently used in preclinical studies.
The use of divergent species in MAb discovery
Rabbit
Key features for MAb discovery
Rabbits have a well-established role as immunological hosts due to their historical role in producing polyclonal antibodies. Rabbits also have a proven track record of producing high-titer responses to a diverse array of antigens, resulting in high-performance antibodies. Several characteristics of rabbits make them particularly attractive for MAb discovery and have been described in detail in several reviews (summarized across species in ).Citation59,Citation60 The long history of rabbits in research has resulted in highly accessible facilities to house rabbits.
Table 3. Comparative features and advantages of therapeutic MAbs generated in different animals.
Rabbits belong to the order Lagomorpha and are evolutionarily distinct from members of the order Rodentia that includes rats and mice.Citation59 The rabbit immune system can recognize unique epitopesCitation61,Citation62 and in some cases can yield human and rodent cross-reactive MAbs, as in the case of brolucizumab, which was approved by FDA in 2019.Citation49 The ability of the rabbit immune system to access unique epitopes is likely due to a combination of factors that include rabbits’ unusual gene conversion-based antibody diversification system. Here, numerous upstream pseudogenes provide donor sequences that mutate the antibody coding genes in multiple overlapping recombination events.Citation63 Access to epitopes is also facilitated by somewhat longer HCDR3, on average more than 3 amino acids longer than their mouse counterparts,Citation36 and greater sequence diversity in VL regions.Citation59 A subset of rabbit antibodies contains a disulfide bond connecting a variable and constant domain in the kappa light chain, which provides thermodynamic stability.Citation64
Rabbit MAbs essentially use a single VH (VH1) and VL (VK1) framework, making cloning, humanization, and engineering of their MAbs relatively straightforward. In addition, rabbits generally produce antibodies with high affinity and specificity. The rabbit immune response was originally thought to result in higher affinity MAbs than those elicited in mice, but such claims have been anecdotal and not supported by more quantitative studies. For instance, a detailed study of 1,410 rabbit MAbs found that while some rabbit MAbs showed high affinities close to 1 pM, the affinity range (20–200 pM) was not significantly different from the range for murine MAbs (30–300 pM).Citation65
Interest in rabbit monoclonal antibodies has developed along with a range of methods for their isolation, including B cell cloning, hybridomas, and phage display. B cell cloning technology for antibody discovery involves interrogating an animal’s immune cells individually and then selecting B cells of interest. For human and mouse immune responses, this process can be streamlined by first enriching for memory B cells or plasma cells to deplete irrelevant immune cell populations prior to screening. With human and mouse cells, this can be accomplished using flow cytometry and cell sorting with reagents that recognize surface markers such as CD38 and CD138. Well-established reagents are currently lacking to facilitate enrichment of rabbit B cells. Without enrichment, rabbit B cell cloning is still feasible, especially for targets that produce robust immune responses. However, for targets that yield low titer immune responses, this technique would be inefficient.
Given the tremendous success of hybridoma technology for MAb discovery, many attempts have been made to develop rabbit hybridomas. Such hybridomas, however, have been far less effective than mouse hybridomas because they have inferior fusion efficiency, are less stable, and secrete lower titers of antibodies.Citation59 Several of these drawbacks have been successfully addressed by RabMab hybridoma technology commercialized by Epitomics (now Abcam).Citation59 A transgenic rabbit with human antibody genes has also been developed, which enables the production of human antibodies that exploit rabbit characteristics such as gene conversion.Citation66
Clinical development
Rabbit full-sized IgG and scFv fragments are in clinical development by multiple companies (Supplementary Table S1). There are at least 13 humanized, rabbit derived MAbs in clinical trials and at least 23 in preclinical development.Citation23,Citation67 The first rabbit derived MAb to be FDA-approved was brolucizumab (ESBATech/Alcon/Novartis), approved in 2019 to treat wet age-related macular degeneration,Citation68 followed by eptinezumab in 2020 (Alder/Lundbeck) for migraine prevention. Anti-IL-6 MAb clazakizumab (Alder/BMS/Vitaeris/CSL Behring/Vitaeris) is presently in Phase 3 clinical trials for use in kidney transplant and end stage kidney disease (NCT03744910, NCT05485961), and crovalimab (Chugai/Roche) is also in Phase 3 trials for treatment of atypical hemolytic uremic syndrome and paroxysmal nocturnal hemoglobinuria (NCT04958265, NCT04861259; NCT04432584).
Several rabbit antibodies in advanced clinical stages, including eptinezumab,Citation69 crovalimab,Citation70 and clazakizumab,Citation69 were discovered using B cell cloning. Apexigen is a spin out from Epitomics, a company that developed some of the first rabbit monoclonal antibody technology. Apexigen’s sotigalimab (APX005M)Citation59 was discovered using rabbit hybridomas.
Camelid
Key features for MAb discovery
Camelids, including llamas, alpaca, and camels, belong to the order Cetartiodactyla, and their antibody system has been described in several recent reviews.Citation71–73 With approximately 92 million years of evolutionary distance from humans, camelids are nearly the same distance from humans as mice, which branched off from humans 91 million years ago.
Camelids produce conventional IgGs that are similar in structure to humans and can be humanized with 13 mutations to enable their use as therapeutics.Citation74 An abundantly produced camelid antibody structure is a small heavy-chain antibody (hCAb), which lacks a CH1 domain and does not pair with a light chain.Citation71 This unique structure has garnered much attention because it can be pared down to a more minimal ‘nanobody’ containing only the VHH antigen binding variable domain.Citation71
Nanobodies have numerous laboratory and therapeutic advantages. They are small (~15 kDa), stable under a relatively wide range of temperature and chemical conditions, easy to clone, and amenable to screening using phage display. They can be readily engineered into new therapeutic formats such as bi- and tri-specific MAbs and even targeted for delivery by gene therapy systems. Camelid VHH can recognize distinct epitopes from human antibodies, as was recently demonstrated with SARS-CoV-2 nanobodies that access a region of the virus inaccessible to human MAbs.Citation75 Their small size also simplifies humanization, manufacturing, and tissue penetration. However, their small size can also be associated with rapid clearance and poor pharmacokinetic properties.Citation73 For therapeutic use, nanobodies are generally engineered into conjugated or multivalent formats, which improve their avidity, potency, and half-life.Citation73
Camelid antibodies can also feature long HCDR3 regions that enable access to sterically hindered sites, such as ligand binding pockets and enzyme active sites. This has been demonstrated in many cases with inhibitory, antagonist, and agonist nanobodies to enzymes and GPCRs.Citation12,Citation29,Citation35,Citation76–78 Due to some evolutionary distance from rodents and humans, camelids can also produce some degree of rodent cross-reactive antibodies. This is the case for caplacizumab (Cablivi®).Citation48 Display technologies using phage and yeast are the predominant methods for antibody isolation from camelids. B cell cloning is technically possible for camelids, but the lack of reagents enabling enrichment of B cells from other immune cells prior to screening make this process inefficient. Hybridoma screening is not possible for camelid MAb discovery due to the lack of a fusion partner.
Although camelids provide attractive features for antibody discovery, numerous practical considerations can constrain their use as a routine host. The animal’s large size creates complex logistics in terms of animal maintenance and cost. However, given the upsurge of recent interest in camelid antibodies, a growing number of facilities around the world provide access to camelids. Due to their size and expensive maintenance, the accessible B cell repertoire is more limited because camelids are usually not sacrificed for their spleen or bone marrow, so only circulating B cells are harvested. Their physical size also often means that camelids are not as responsive to the same amount of immunogen as a mouse. Smaller animals are easier to handle and cheaper, which permits a wider array of immunization conditions and immunogens to be used. This is especially important for conserved targets and transmembrane proteins, which often require multiple immunizations and strategies.Citation79 Finally, while camelids are a divergent species, they are still mammalian. Since much of their genome is still highly homologous to human, this limits the diversity of their immune response against human proteins.
Clinical development
The first approved MAb obtained from a non-rodent animal source was the alpaca-derived bivalent nanobody caplacizumab developed by Ablynx, which targets von Willebrand factor for the treatment of thrombotic thrombocytopenic purpura and thrombosis. Ablynx (now part of Sanofi) is the major developer of engineered humanized camelid nanobodies, with at least four in clinical development and numerous others in their pipeline. ArgenX is another major developer of therapeutic camelid antibodies, and FairJourney Biologics is a major discovery engine for camelid nanobodies. There are at least 37 camelid antibodies in clinical trials, with numerous others in preclinical stages of development (Supplementary Table S1).Citation67 Approved and clinical stage antibodies caplacizumab,Citation80 envafolimab,Citation81 and ozoralizumabCitation82 were all derived using phage display.
Chicken
Key features for MAb discovery
When considering divergent animal species for MAb development, chickens provide a desirable balance of phylogenetic distance from humans, yet with an advanced immune system capable of producing high-affinity canonical antibodies closely related to humans. Chickens diverged from mammals 310 million years ago,Citation8,Citation9 making many conserved targets immunologically accessible in this species.Citation6,Citation9,Citation11
Only 15% of human drug targets are highly conserved (>90%) in chickens (), with the median sequence identity between chicken and human orthologs 75%.Citation9 For comparison, 48% of targets in mice have > 90% identity to their human homologs, with the median sequence identity 88%.Citation27 In our experience, sequence identities above the 90% threshold are predictive of a poor immune response, meaning that about half of human drug targets are not suitable for antibody discovery in rodents while most of these are suitable for discovery in chickens. To further capitalize on the divergence between human and chicken genomes, a transgenic chicken containing human antibody genes has been developed and is being used for antibody discovery.Citation83
Figure 5. Human drug targets are less conserved in chickens compared with mice. A collection of 663 human drug targets curated in the ECOdrug databaseCitation7 were analyzed with respect to their amino sequence homology. Almost half (48%) of the human drug targets have the highest degree of conservation (>90%) when compared with mice, but only 15% of targets are conserved at this high level with chickens.
The use of chickens as immunization hosts played an essential role in the isolation of rare, highly specific MAbs for the oncofetal target Claudin 6Citation14 that are being pursued as tumor-selective therapies for ovarian and endometrial cancers. The extracellular domain of CLND6 is highly conserved, being 95% identical in mice but only 82% in chickens. An immune response with high epitope diversity in chickens enabled the discovery of MAbs that bound the target, but not any of the other 23 widely expressed claudin family members, including CLDN9 that differs by only three extracellular residues. Detailed epitope mapping of these MAbs revealed that they access a very specific binding site unique to the structure of CLDN6 that differs from CLDN9 by only a single atomic contact point.Citation14 The lead MAb from these immunizations was further developed as a T-cell engaging bispecific (CTIM-76), which is now in preclinical testing by Context Therapeutics.
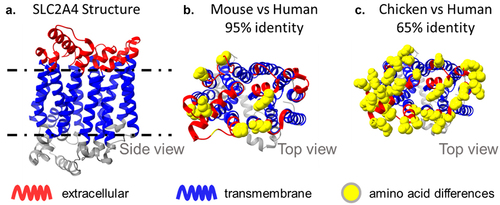
The discovery of MAbs targeting the glucose transporter SLC2A4 (GLUT4) also illustrates the advantages of chicken hosts. This transporter is highly conserved in mammals, reflecting its essential roles in glucose metabolism and insulin regulation, so MAbs targeting it had not been isolated previously. SLC2A4 shares 95% sequence identity with its mouse ortholog, but lacks an ortholog in chickens and shares only 65% identity with its SLC2A1 paralog (). Chicken immunization enabled the isolation of a diverse panel of conformationally sensitive MAbs able to discriminate functional states of the transporter.Citation11 These MAbs provide valuable molecular tools to monitor the transporter and for the discovery of novel diabetes therapeutics.
Figure 6. Sequence divergence of SLC2A4, a conserved 12-TM transporter. SLC2A4 is a complex immunogen with small extracellular loops and 95% sequence identity between mouse and human. The extracellular loops of human SLC2A4 have only 9 amino acid differences relative to mouse, but 20 amino acid differences (and only 65% identity) with the closest chicken paralog, SLC2A1. The use of a chicken host enabled the discovery of antibodies with diverse epitopes.11
Unlike the normally distributed HCDR3 repertoire of human antibodies, chicken sequences have a skewed distribution favoring longer sequences, with almost 90% of chicken MAbs having HCDR3 sequences 13 amino acids or longer, as per the Kabat numbering scheme.Citation37 This phenomenon was observed in our own antibody panel raised against SLC2A4 that included four highly characterized MAbs with HCDR3 lengths of 16 to 26 amino acids. Epitope mapping of one of these MAbs, LM048 with an HCDR3 length of 26 amino acids, indicated this antibody bound in a recessed cavity between two extracellular loops, providing the unique ability to selectively bind the outward-open state of the transporter.Citation11 In about half of chicken MAbs, the long protruding paratope structure is supported by disulfide bonding in the HCDR3 region that stabilizes the structure.
The affinity of chicken MAbs is similar to those of mice and rabbits. In comparative studies, MAbs elicited in all three species were able to bind in the picomolar range.Citation6,Citation57,Citation65 Cloning, humanization, and engineering chicken-derived MAbs is far easier than murine or human antibodies, since chickens have only a single germline gene encoding for a heavy and light chain, similarly to rabbits.Citation37 For comparison, mouse MAbs have more than 100 frameworks.Citation84 The IgG orthologous antibody type produced by chickens, IgY, is highly conserved with human IgG. As a result, chicken MAbs can be humanized by CDR grafting with minimal framework fine-tuning.Citation51
Phage display is the predominant method used for the isolation of chicken antibodies, and B cell cloning has been used for chickens with some success. A few antibody reagents that allow the enrichment of chicken B cells prior to B cell cloning are commercially available, but there are no fusion partners to enable chicken hybridomas. Although chickens are not as commonly used as small mammals in biomedical research, they are relatively easy and inexpensive to house and accessibility is not an issue for research.
Preclinical and clinical development
Sym021, targeting PD-1 and being developed by Symphogen, is the first chicken-derived MAb to enter clinical trials (NCT03311412, NCT04641871, NCT04672434). Numerous chicken antibodies are in preclinical development, and this trend is expected to grow as alternatives to traditional mice immunization continue to emerge. Many preclinical chicken antibodies described in the literature target exceedingly conserved proteins that would constitute poor immunogens in mice. This includes Pfizer’s BDNF chicken antibody (100% target sequence identity in mice), and the inhibitory chicken antibody developed by Ligand and Tetragenetics against Kv1.3 (96% target sequence identity in mice).Citation52,Citation85 Integral Molecular has a pipeline of chicken-derived antibodies targeting highly conserved human multipass membrane proteins, including SLC2A4 (95% identity with mouse), CLDN6 (88%), CLDN18.2 (90%), P2X3 (94%), and others. As described above, CTIM-76 is now in preclinical testing by Context Therapeutics. Bi- and trispecific molecules against GPRC5D, also derived from chicken immunizations, are in preclinical testing at Integral Molecular for multiple myeloma. Additional preclinical antibodies derived from chicken include MAbs against SIRP⍺ being developed by Ligand and ALX Oncology, antibodies targeting the GPCR GIPR from Ligand Pharmaceuticals and Boehringer Ingelheim,Citation56,Citation57 anti-MCT1 MAb from Immunext,Citation86 anti-MFSD2A antibody,Citation87 and an anti-GPCR antibody with agonist activities from Merck.Citation88 Symphogen employed B cell cloning in the discovery of Sym021,Citation51 and Integral Molecular uses phage display for chicken-derived antibodies in their pipeline.Citation11,Citation14
Other animals
Additional animals including sharks, hamsters, and cows are also being used for antibody discovery, and there is at least one hamster-derived antibody in clinical trials (Supplementary Table S1).Citation67 Sharks are the most evolutionary divergent animals from humans that still have an adaptive Ig-based immune system that is fundamentally similar to human. They have an IgNAR IgH isotype consisting of a heavy chain only, although these do not have a canonical IgG structure. While some groups have immunized sharks, others have made use of the IgNAR structure and inserted sequences from other sources. Ossianix and AdAlta are the major developers pursuing shark-based antibody structures for therapeutics, and AdAlta’s AD-214 is now in clinical development (NCT04415671).
Although not frequently used as an immunization host, cow antibodies have gained attention due to their unusually long HCDR3 regions, with HCDR3 sequences up to 60 amino acids reported.Citation89,Citation90 Their antigen-binding regions have a unique knob-and-stalk structure with the potential to reach into deep protein pockets.Citation91 Cow antibodies with long HCDR3 regions were shown to enable the development of broadly neutralizing HIV MAbs by using a 58aa HCDR3 to penetrate the dense glycan shield of HIV.Citation92
Conclusions
The therapeutic MAb discovery landscape has seen staggering advances since the first MAb approval over 30 years ago. While traditional mouse-derived therapeutic antibodies retain the lion’s share of therapeutic MAbs currently on the market, the opportunities for discovery using mouse hosts have been waning as more easily accessed targets have been plucked. Therapeutic programs in the crowded marketplace are increasingly reaching for the so-called “high-hanging fruit”, looking beyond simple binding activity, and reaching for molecules with demanding specifications for new therapeutics and therapeutic modalities. Whether this means the ability to contact a particular epitope, bind with a specific geometry or effect a given function, diversity in target proteins, epitope access and paratope topologies will be vital. Just as mouse hybridoma technology led to an explosive development of novel therapeutics, we anticipate that antibody discovery using divergent animals can drive the next phase of innovation in biotherapeutics.
Abbreviations
| aa | = | amino acid |
| ADA | = | anti-drug antibody |
| CDR | = | complementarity determining region |
| FDA | = | Food and Drug Administration |
| GPCR | = | G protein-coupled receptor |
| HCAb | = | heavy chain-only antibodies |
| HCDR3 | = | heavy-chain complementarity-determining region 3 |
| IgNAR | = | immunoglobulin new antigen receptor |
| MAb | = | monoclonal antibody |
| NHP | = | non-human primate |
| scFv | = | single-chain variable fragment |
| VHH | = | variable domain of the heavy chain of HCAb |
Supplemental Material
Download MS Excel (39.1 KB)Acknowledgments
The authors are grateful for helpful discussions and manuscript assistance from Joseph Rucker, Ginny Feltzin, and Edgar Davidson. We also thank Dr. Janice Reichert for helpful advice regarding the derivation of clinical-stage antibodies.
Disclosure statement
Integral Molecular is a biotech company that uses divergent species for antibody discovery. S.S.R.B., R.C., and B.J.D. are current employees and shareholders of Integral Molecular.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19420862.2023.2273018
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Lu RM, Hwang YC, Liu IJ, Lee CC, Tsai HZ, Li HJ, Wu HC. Development of therapeutic antibodies for the treatment of diseases. J Biomed Sci. 2020;27:1. doi:10.1186/s12929-019-0592-z. PMID: 31894001.
- Kaplon H, Reichert JM. Antibodies to watch in 2021. MAbs. 2021;13:1860476. doi:10.1080/19420862.2020.1860476. PMID: 33459118.
- Kaplon H, Chenoweth A, Crescioli S, Reichert JM. Antibodies to watch in 2022. MAbs. 2022;14:2014296. doi:10.1080/19420862.2021.2014296. PMID: 35030985.
- Carter PJ, Lazar GA. Next generation antibody drugs: pursuit of the ‘high-hanging fruit’. Nat Rev Drug Discov. 2018;17(3):197–14. PMID: 29192287. doi: 10.1038/nrd.2017.227.
- Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, Sharkey B, Bobrowicz B, Caffry I, Yu Y, et al. Biophysical properties of the clinical-stage antibody landscape. Proceedings of the National Academy of Sciences of the United States of America. 2017; 114:944–49. doi:10.1073/pnas.1616408114. PMID: 28096333.
- Abdiche YN, Harriman R, Deng X, Yeung YA, Miles A, Morishige W, Boustany L, Zhu L, Izquierdo SM, Harriman W. Assessing kinetic and epitopic diversity across orthogonal monoclonal antibody generation platforms. MAbs. 2016;8:264–77. doi:10.1080/19420862.2015.1118596. PMID: 26652308.
- Verbruggen B, Gunnarsson L, Kristiansson E, Osterlund T, Owen SF, Snape JR, Tyler CR. Ecodrug: a database connecting drugs and conservation of their targets across species. Nucleic Acids Res. 2018;46:D930–D36. doi:10.1093/nar/gkx1024. PMID: 29140522.
- Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–49. doi:10.1038/nrg929. PMID: 12415314.
- International Chicken Genome Sequencing C. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. PMID: 15592404. doi:10.1038/nature03154
- Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–97. doi:10.1038/256495a0. PMID: 1172191.
- Tucker DF, Sullivan JT, Mattia KA, Fisher CR, Barnes T, Mabila MN, Wilf R, Sulli C, Pitts M, Payne RJ, et al. Isolation of state-dependent monoclonal antibodies against the 12-transmembrane domain glucose transporter 4 using virus-like particles. Proceedings of the National Academy of Sciences of the United States of America. 2018; 115:E4990–E99. doi:10.1073/pnas.1716788115. PMID: 29769329.
- Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature. 2011;469(7329):175–80. PMID: 21228869. doi:10.1038/nature09648.
- Mujic-Delic A, RH DW, Verkaar F, MJ S. GPCR-targeting nanobodies: attractive research tools, diagnostics, and therapeutics. Trends Pharmacol Sci. 2014;35(5):247–55. PMID: 24690241. doi: 10.1016/j.tips.2014.03.003.
- Screnci B, Stafford LJ, Barnes T, Shema K, Gilman S, Wright R, Al Absi S, Phillips T, Azuelos C, Slovik K, et al. Antibody specificity against highly conserved membrane protein Claudin 6 driven by single atomic contact point. iScience. 2022;25:105665. doi:10.1016/j.isci.2022.105665. PMID: 36505931.
- Liu S, Wang S, Lu S. DNA immunization as a technology platform for monoclonal antibody induction. Emerg Microbes Infect. 2016;5:e33. doi:10.1038/emi.2016.27. PMID: 27048742.
- Beck A, Goetsch L, Dumontet C, Corvaia N. Strategies and challenges for the next generation of antibody–drug conjugates. Nat Rev Drug Discov. 2017;16(5):315–37. PMID: 28303026. doi: 10.1038/nrd.2016.268.
- Marks JD, Bradbury A. Selection of human antibodies from phage display libraries. Methods Mol Biol. 2004;248:161–76. doi:10.1385/1-59259-666-5:161. PMID: 14970495.
- Shehata L, Maurer DP, Wec AZ, Lilov A, Champney E, Sun T, Archambault K, Burnina I, Lynaugh H, Zhi X, et al. Affinity maturation enhances antibody specificity but compromises conformational stability. Cell Reports. 2019;28(13):3300–8 e3304. PMID: 31553901. doi:10.1016/j.celrep.2019.08.056.
- Prabakaran P, Rao SP, Wendt M. Animal immunization merges with innovative technologies: a new paradigm shift in antibody discovery. MAbs. 2021;13:1924347. doi:10.1080/19420862.2021.1924347. PMID: 33947305.
- Ablynx Ghent B. CABLIVI [package insert]. 2023.
- Novartis East Hanover NJ. BEOVU [package insert]. 2022.
- Lundbeck Seattle BioPharmaceuticals IB, WA. VYEPTI [prescribing information]. 2022.
- Tabs - therapeutic antibody database. https://tabs.craic.com/users. Date [accessed 2020-2023].
- Edwards BM, Barash SC, Main SH, Choi GH, Minter R, Ullrich S, Williams E, Du Fou L, Wilton J, Albert VR, et al. The remarkable flexibility of the human antibody repertoire; isolation of over one thousand different antibodies to a single protein, BLyS. J Mol Biol. 2003;334:103–18. doi:10.1016/j.jmb.2003.09.054. PMID: 14596803.
- Madabushi S, Gross AK, Philippi A, Meng EC, Wensel TG, Lichtarge O. Evolutionary trace of G protein-coupled receptors reveals clusters of residues that determine global and class-specific functions. J Biol Chem. 2004;279:8126–32. doi:10.1074/jbc.M312671200. PMID: 14660595.
- Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257:342–58. doi:10.1006/jmbi.1996.0167. PMID: 8609628.
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Genome sequence of the brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi:10.1038/nature02426. PMID: 15057822.
- Finlay WJ, Bloom L, Varghese S, Autin B, Cunningham O. Optimized generation of high-affinity, high-specificity single-chain fv antibodies from multi-antigen immunized chickens. Methods Mol Biol. 2017;1485:319–38. doi:10.1007/978-1-4939-6412-3_16. PMID: 27730560.
- De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S, Wyns L Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2006; 103:4586–91. doi:10.1073/pnas.0505379103. PMID: 16537393.
- Stave JW, Lindpaintner K. Antibody and antigen contact residues define epitope and paratope size and structure. J Immunol. 2013;191(3):1428–35. PMID: 23797669. doi: 10.4049/jimmunol.1203198.
- JA F, Koehler LJ, JR W, Cisneros A, JE C, Meiler J. Improving loop modeling of the antibody complementarity-determining region 3 using knowledge-based restraints. PloS One. 2016;11(5):e0154811. PMID: 27182833. doi: 10.1371/journal.pone.0154811.
- Ramsland PA, Kaushik A, Marchalonis JJ, Edmundson AB. Incorporation of long CDR3s into V domains: implications for the structural evolution of the antibody-combining site. Exp Clin Immunogenet. 2001;18:176–98. doi:10.1159/000049197. PMID: 11872949.
- Garces F, Mohr C, Zhang L, Huang CS, Chen Q, King C, Xu C, Wang Z. Molecular insight into recognition of the CGRPR complex by migraine prevention therapy aimovig (erenumab). Cell Reports. 2020;30(6):1714–23 e1716. PMID: 32049005. doi: 10.1016/j.celrep.2020.01.029.
- Barnes CO, Jette CA, Abernathy ME, Dam KA, Esswein SR, Gristick HB, Malyutin AG, Sharaf NG, Huey-Tubman KE, Lee YE, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682–87. PMID: 33045718. doi:10.1038/s41586-020-2852-1.
- Ma Y, Ding Y, Song X, Ma X, Li X, Zhang N, Song Y, Sun Y, Shen Y, Zhong W, et al. Structure-guided discovery of a single-domain antibody agonist against human apelin receptor. Sci Adv. 2020;6(3):eaax7379. PMID: 31998837. doi:10.1126/sciadv.aax7379.
- Lavinder JJ, Hoi KH, Reddy ST, Wine Y, Georgiou G. Systematic characterization and comparative analysis of the rabbit immunoglobulin repertoire. PloS One. 2014;9(6):e101322. PMID: 24978027. doi: 10.1371/journal.pone.0101322.
- Wu L, Oficjalska K, Lambert M, Fennell BJ, Darmanin-Sheehan A, Ni Shuilleabhain D, Autin B, Cummins E, Tchistiakova L, Bloom L, et al. Fundamental characteristics of the immunoglobulin VH repertoire of chickens in comparison with those of humans, mice, and camelids. J Immunol. 2012;188(1):322–33. PMID: 22131336. doi:10.4049/jimmunol.1102466.
- Wu TT, Johnson G, Kabat EA. Length distribution of CDRH3 in antibodies. Proteins. 1993;16:1–7. doi:10.1002/prot.340160102. PMID: 8497480.
- Muyldermans S, Smider VV. Distinct antibody species: structural differences creating therapeutic opportunities. Curr Opin Immunol. 2016;40:7–13. doi:10.1016/j.coi.2016.02.003. PMID: 26922135.
- Li S, Schmitz KR, Jeffrey PD, Wiltzius JJ, Kussie P, Ferguson KM. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005;7:301–11. doi:10.1016/j.ccr.2005.03.003. PMID: 15837620.
- ICH S6. Preclinical safety evaluation of biotechnology-derived pharmaceuticals. https://www.ich.org/. Date [accessed May 4, 2022].
- ICH harmonisation for better health: safety guidelines, biotechnology products. https://www.ich.org/page/safety-guidelines. Date [accessed May 42022].
- Bussiere JL, Martin P, Horner M, Couch J, Flaherty M, Andrews L, Beyer J, Horvath C. Alternative strategies for toxicity testing of species-specific biopharmaceuticals. Int J Toxicol. 2009;28:230–53. doi:10.1177/1091581809337262. PMID: 19546261.
- Sewell F, Chapman K, Couch J, Dempster M, Heidel S, Loberg L, Maier C, Maclachlan TK, Todd M, van der Laan JW. Challenges and opportunities for the future of monoclonal antibody development: improving safety assessment and reducing animal use. MAbs. 2017;9:742–55. doi:10.1080/19420862.2017.1324376. PMID: 28475417.
- Collins FS NIH will no longer support biomedical research on chimpanzees. https://www.nih.gov/about-nih/who-we-are/nih-director/statements/nih-will-no-longer-support-biomedical-research-chimpanzees. 2015 Date [accessed November 22, 2021].
- GUIDANCE DOCUMENT: nonclinical considerations for mitigating nonhuman primate supply constraints arising from the COVID-19 pandemic. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/nonclinical-considerations-mitigating-nonhuman-primate-supply-constraints-arising-covid-19-pandemic 2022 Date [accessed May 4, 2022].
- Zhang S. America is running low on a crucial resource for COVID-19 vaccines. The Atlantic. 2020 2020; August 13. https://www.theatlantic.com/science/archive/2020/08/america-facing-monkey-shortage/615799/.
- Food and Drug Administration. Caplacizumab. Center for drug evaluation and research application number: 761112Orig1s000 multi-discipline Review. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2019/761112Orig1s000MultiR.pdf. Date [accessed April 11, 2022].
- Food and Drug Administration. Brolucizumab. Center for drug evaluation and research application number 761125Orig1s000. Non-Clinical Review. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2019/761125Orig1s000PharmR.pdf. 2019 Date [accessed April 11, 2022].
- Food and Drug Administration. Eptinezumab. Center for drug evaluation and research application number: 761119Orig1s000 non-clinical Review. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2020/761119Orig1s000PharmR.pdf. Date [accessed April 11, 2022].
- Gjetting T, Gad M, Frohlich C, Lindsted T, Melander MC, Bhatia VK, Grandal MM, Dietrich N, Uhlenbrock F, Galler GR, et al. Sym021, a promising anti-PD1 clinical candidate antibody derived from a new chicken antibody discovery platform. MAbs. 2019;11:666–80. doi:10.1080/19420862.2019.1596514. PMID: 31046547.
- Stack E, McMurray S, McMurray G, Wade J, Clark M, Young G, Marquette K, Jain S, Kelleher K, Chen T, et al. In vitro affinity optimization of an anti-BDNF monoclonal antibody translates to improved potency in targeting chronic pain states in vivo. MAbs. 2020;12:1755000. doi:10.1080/19420862.2020.1755000. PMID: 32329655.
- Shih HH, Tu C, Cao W, Klein A, Ramsey R, Fennell BJ, Lambert M, Ni Shuilleabhain D, Autin B, Kouranova E, et al. An ultra-specific avian antibody to phosphorylated tau protein reveals a unique mechanism for phosphoepitope recognition. Journal Of Biological Chemistry. 2012;287(53):44425–34. PMID: 23148212. doi:10.1074/jbc.M112.415935.
- Chockalingam K, Kumar A, Song J, Chen Z. Chicken-derived CD20 antibodies with potent B-cell depletion activity. Br J Haematol. 2022;199:560–71. doi:10.1111/bjh.18438. PMID: 36039695.
- Yokosaki Y, Nishimichi N. New therapeutic targets for hepatic fibrosis in the integrin family, α8β1 and α11β1, induced specifically on activated stellate cells. Int J Mol Sci. 2021;22(23):12794. PMID: 34884600. doi: 10.3390/ijms222312794.
- Sim J, Sockolosky JT, Sangalang E, Izquierdo S, Pedersen D, Harriman W, Wibowo AS, Carter J, Madan A, Doyle L, et al. Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRPα. MAbs. 2019;11(6):1036–52. PMID: 31257988. doi:10.1080/19420862.2019.1624123.
- Konitzer JD, Pramanick S, Pan Q, Augustin R, Bandholtz S, Harriman W, Izquierdo S. Generation of a highly diverse panel of antagonistic chicken monoclonal antibodies against the GIP receptor. MAbs. 2017;9:536–49. doi:10.1080/19420862.2016.1276683. PMID: 28055305.
- Xiang Y, Sang Z, Bitton L, Xu J, Liu Y, Schneidman-Duhovny D, Shi Y. Integrative proteomics identifies thousands of distinct, multi-epitope, and high-affinity nanobodies. Cell Syst. 2021;12(3):220–34 e229. PMID: 33592195. doi: 10.1016/j.cels.2021.01.003.
- Weber J, Peng H, Rader C. From rabbit antibody repertoires to rabbit monoclonal antibodies. Experimental & Molecular Medicine. 2017;49:e305. doi:10.1038/emm.2017.23. PMID: 28336958.
- Zhang Z, Liu H, Guan Q, Wang L, Yuan H. Advances in the isolation of specific monoclonal rabbit antibodies. Front Immunol. 2017;8:494. doi:10.3389/fimmu.2017.00494. PMID: 28529510.
- Yu Y, Lee P, Ke Y, Zhang Y, Yu Q, Lee J, Li M, Song J, Chen J, Dai J, et al. A humanized anti-VEGF rabbit monoclonal antibody inhibits angiogenesis and blocks tumor growth in xenograft models. PloS One. 2010;5:e9072. doi:10.1371/journal.pone.0009072. PMID: 20140208.
- Popkov M, RG M, CB A, Thundivalappil S, CF B, Rader C. Rabbit immune repertoires as sources for therapeutic monoclonal antibodies: the impact of kappa allotype-correlated variation in cysteine content on antibody libraries selected by phage display. J Mol Biol. 2003;325(2):325–35. PMID: 12488098. doi: 10.1016/s0022-2836(02)01232-9.
- Leighton PA, Morales J, Harriman WD, Ching KH. V(D)J rearrangement is dispensable for producing CDR-H3 sequence diversity in a gene converting species. Front Immunol. 2018;9:1317. doi:10.3389/fimmu.2018.01317. PMID: 29951062.
- Kawade R, Akiba H, Entzminger K, Maruyama T, Okumura CJ, Tsumoto K. Roles of the disulfide bond between the variable and the constant domains of rabbit immunoglobulin kappa chains in thermal stability and affinity. Protein Eng Des Sel. 2018;31:243–47. doi:10.1093/protein/gzy008. PMID: 29850878.
- Landry JP, Ke Y, Yu GL, Zhu XD. Measuring affinity constants of 1450 monoclonal antibodies to peptide targets with a microarray-based label-free assay platform. J Immunol Methods. 2015;417:86–96. doi:10.1016/j.jim.2014.12.011. PMID: 25536073.
- Ros F, Offner S, Klostermann S, Thorey I, Niersbach H, Breuer S, Zarnt G, Lorenz S, Puels J, Siewe B, et al. Rabbits transgenic for human IgG genes recapitulating rabbit B-cell biology to generate human antibodies of high specificity and affinity. MAbs. 2020;12:1846900. doi:10.1080/19420862.2020.1846900. PMID: 33228444.
- The antibody society. https://www.antibodysociety.org/. Date [accessed October 12, 2023].
- Markham A. Brolucizumab: first approval. Drugs. 2019;79:1997–2000. doi:10.1007/s40265-019-01231-9. PMID: 31768932.
- Alder BioPharmaceuticals I Annual report on form 10-K. In: Annual Report on Form 10-K. https://www.sec.gov/Archives/edgar/data/1423824/000156459019004084/aldr-10k_20181231.htm. 2019.
- Fukuzawa T, Sampei Z, Haraya K, Ruike Y, Shida-Kawazoe M, Shimizu Y, Gan SW, Irie M, Tsuboi Y, Tai H, et al. Long lasting neutralization of C5 by SKY59, a novel recycling antibody, is a potential therapy for complement-mediated diseases. Sci Rep. 2017;7:1080. doi:10.1038/s41598-017-01087-7. PMID: 28439081.
- de Los Rios M, Criscitiello MF, Smider VV. Structural and genetic diversity in antibody repertoires from diverse species. Curr Opin Struct Biol. 2015;33:27–41. doi:10.1016/j.sbi.2015.06.002. PMID: 26188469.
- Arbabi-Ghahroudi M. Camelid single-domain antibodies: historical perspective and future outlook. Front Immunol. 2017;8:1589. doi:10.3389/fimmu.2017.01589. PMID: 29209322.
- Jovcevska I, Muyldermans S. 2020. The therapeutic potential of nanobodies. BioDrugs: clinical immunotherapeutics, biopharmaceuticals and gene therapy. PMID: 31686399 34(1):11–26. doi:10.1007/s40259-019-00392-z
- Vincke C, Loris R, Saerens D, Martinez-Rodriguez S, Muyldermans S, Conrath K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem. 2009;284:3273–84. doi:10.1074/jbc.M806889200. PMID: 19010777.
- Xu J, Xu K, Jung S, Conte A, Lieberman J, Muecksch F, Lorenzi JCC, Park S, Schmidt F, Wang Z, et al. Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants. Nature. 2021;595:278–82. doi:10.1038/s41586-021-03676-z. PMID: 34098567.
- Lauwereys M, Arbabi Ghahroudi M, Desmyter A, Kinne J, Holzer W, De Genst E, Wyns L, Muyldermans S. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 1998;17(13):3512–20. PMID: 9649422. doi: 10.1093/emboj/17.13.3512.
- Desmyter A, Transue TR, Ghahroudi MA, Thi MH, Poortmans F, Hamers R, Muyldermans S, Wyns L. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol. 1996;3:803–11. doi:10.1038/nsb0996-803. PMID: 8784355.
- Heukers R, De Groof TWM, Smit MJ. Nanobodies detecting and modulating GPCRs outside in and inside out. Curr Opin Cell Biol. 2019;57:115–22. doi:10.1016/j.ceb.2019.01.003. PMID: 30849632.
- Belanger K, Tanha J. High-efficacy, high-manufacturability human VH domain antibody therapeutics from transgenic sources. Protein engineering, design & selection: PEDS. 2021. 34. 10.1093/protein/gzab012. PMID: 33991089
- Alfaleh MA, Alsaab HO, Mahmoud AB, Alkayyal AA, Jones ML, Mahler SM, Hashem AM. Phage display derived monoclonal antibodies: from bench to bedside. Front Immunol. 2020;11:1986. doi:10.3389/fimmu.2020.01986. PMID: 32983137.
- Zhang F, Wei H, Wang X, Bai Y, Wang P, Wu J, Jiang X, Wang Y, Cai H, Xu T, et al. Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade. Cell Discov. 2017;3:17004. doi:10.1038/celldisc.2017.4. PMID: 28280600.
- Tanaka Y. Ozoralizumab: first nanobody® therapeutic for rheumatoid arthritis. Expert Opin Biol Ther. 2023;23(7):579–87. PMID: 37431762. doi: 10.1080/14712598.2023.2231344.
- Ching KH, Collarini EJ, Abdiche YN, Bedinger D, Pedersen D, Izquierdo S, Harriman R, Zhu L, Etches RJ, van de Lavoir MC, et al. Chickens with humanized immunoglobulin genes generate antibodies with high affinity and broad epitope coverage to conserved targets. MAbs. 2018;10:71–80. doi:10.1080/19420862.2017.1386825. PMID: 29035625.
- IMGT®, The international ImMunoGeneTics information system®. 2022 Date [accessed May 20, 2023].
- Bednenko J, Harriman R, Marien L, Nguyen HM, Agrawal A, Papoyan A, Bisharyan Y, Cardarelli J, Cassidy-Hanley D, Clark T, et al. A multiplatform strategy for the discovery of conventional monoclonal antibodies that inhibit the voltage-gated potassium channel Kv1.3. MAbs. 2018;10:636–50. doi:10.1080/19420862.2018.1445451. PMID: 29494279.
- Anti-MCT1 antibodies and uses thereof. WO2019136300A2;
- Wood CAP, Zhang J, Aydin D, Xu Y, Andreone BJ, Langen UH, Dror RO, Gu C, Feng L. Structure and mechanism of blood–brain-barrier lipid transporter MFSD2A. Nature. 2021;596(7872):444–48. PMID: 34349262. doi: 10.1038/s41586-021-03782-y.
- Handa M Designing discovery strategies to Maximize Molecular recognition to find agonist antibodies. In: Proceedings of the Discovery on Target Conference 2022; Boston: 2022.
- Pasman Y, Soliman C, Ramsland PA, Kaushik AK. Exceptionally long CDR3H of bovine scFv antigenized with BoHV-1 B-epitope generates specific immune response against the targeted epitope. Mol Immunol. 2016;77:113–25. doi:10.1016/j.molimm.2016.07.014. PMID: 27497190.
- Saini SS, Allore B, Jacobs RM, Kaushik A. Exceptionally long CDR3H region with multiple cysteine residues in functional bovine IgM antibodies. Eur J Immunol. 1999;29:2420–26. doi:10.1002/(SICI)1521-4141(199908)29:08<2420:AID-IMMU2420>3.0.CO;2-A. PMID: 10458755.
- Hawkins A, Joyce C, Brady K, Hold A, Smith A, Knight M, Howard C, van den Elsen J, Lawson ADG, Macpherson A. The proximity of the N- and C- termini of bovine knob domains enable engineering of target specificity into polypeptide chains. MAbs. 2022;14:2076295. doi:10.1080/19420862.2022.2076295. PMID: 35634719.
- Stanfield RL, Berndsen ZT, Huang R, Sok D, Warner G, Torres JL, Burton DR, Ward AB, Wilson IA, Smider VV. Structural basis of broad HIV neutralization by a vaccine-induced cow antibody. Sci Adv. 2020;6(22):eaba0468. PMID: 32518821. doi: 10.1126/sciadv.aba0468.
