ABSTRACT
Since their initial observation, contact sites formed between different organelles have transitioned from ignored curiosities to recognized centers for the exchange of metabolites and lipids. Contact formed between the ER and endomembrane system (eg. the plasma membrane, endosomes, and lysosomes) is of particular biomedical interest, as it governs aspects of lipid metabolism, organelle identity, and cell signaling. Here, we review the field of ER-endolysosomal communication from the perspective of three model systems: budding yeast, the fruit fly D. melanogaster, and mammals. From this broad perspective, inter-organelle communication displays a consistent role in metabolic regulation that was differentially tuned during the development of complex metazoan life. We also examine the current state of understanding of lipid exchange between organelles, and discuss molecular mechanisms by which this occurs.
Introduction: Membrane contact sites in cell biology and disease
Since their initial observation over 60 years ago, contact sites formed between different organelles have transitioned from ignored curiosities to recognized centers for the exchange of metabolites and lipids.Citation1 Recent studies have identified several conserved protein families that function as “tethers” at inter-organelle membrane contact sites (MCSs), as well as soluble proteins that dock at these sites to mediate the non-vesicular trafficking of lipids. Many of these proteins have important roles in human physiology, and their loss contributes to serious genetic conditions, underscoring the need to understand their roles in cell biology, and ultimately, human physiology and pathophysiology ().
Table 1. Location, proposed functions, and associated diseases of ER-organelle MCS proteins.
The purpose of this short review is two-fold: 1) to examine the growing list of proteins that function at sites of inter-organelle contact, and 2) to discuss how perturbing their functions impact metabolism and disease across evolution from yeast and fruit flies to mammals. As this is a broad topic, most attention will be paid to proteins involved in contact between the Endoplasmic Reticulum (ER) and the endomembrane trafficking pathway (the plasma membrane, endosomes, and lysosomes). Since several MCS-localizing proteins play important roles in lipid metabolism, we will also discuss how lipid metabolism integrates into general physiology, and how this relationship governs organismal health and aging. Finally, we will briefly examine current models describing how lipids are exchanged at MCSs, and how perturbing this affects metabolism and organelle identity. For additional information regarding other inter-organelle contact sites, as well as discussions of inter-organelle communication, endosome movement, lipid transport, and signaling, the reader is directed to other excellent reviews.Citation2-5
Yeast ER-endolysosomal contact sites in cell metabolism and disease
Although ER-endolysosomal contact sites are currently the focus of intense study in mammalian systems, proteins that localize to sites of contact between the ER and vacuole (the yeast lysosome) have been well known for over a decade.Citation6-8 Many of these act as inter-organelle “tethers” and were originally described in the budding yeast Saccharomyces cervisiae. Here the site of contact between the ER and vacuole is known as the Nuclear ER (nER) Vacuolar Junction (NVJ), and comprises a ˜200–500 nm long patch-like strip juxtaposed between the outer nuclear envelope (continuous with the peripheral ER) and vacuole surface. The NVJ is principally formed by the direct interaction of the integral ER protein Nvj1, which directly binds to the vacuolar-resident protein Vac8.Citation6 Notably, Vac8 is not an integral membrane protein, but binds tightly to the vacuole surface including numerous post-translational palmitoylations on its N-terminus.Citation9 Both Nvj1 and Vac8 have no obvious mammalian orthologs. However, a second NVJ-resident ER integral membrane protein, Nvj2, has a clear ortholog in humans: Tex2/HT008,Citation7 although its function is unclear. Nvj2 contains two lipid-binding domains: a predicted PH domain, and a lipid-binding synaptotagmin-like-mitochondrial-lipid binding protein (SMP) domain that is present in numerous MCS-localizing proteins, including the Extended-Synaptotagmins (E-Syts).Citation10-12 E-Syts and their yeast homologs, the Tricalbins (Tcbs), act as tethers at ER-plasma membrane (PM) contact sites, and also interact with VAP proteins, which have been implicated in ALS disease.Citation13,14
Numerous lipid-modifying and transport proteins also show enrichment at yeast NVJs. The enoyl reductase Tsc13, which catalyzes the final step of very long chain fatty acid (VLCFA) elongation, exhibits clear human homologs in the TECR protein family,Citation15,16 which are linked to intellectual disability. Loss of Tsc13 function affects sphingolipid synthesis, leading to decreases in mature sphingolipids and C24 ceramide. Notably, loss of Tsc13 can be rescued by reintroduction of TECR, indicating a conservation of function between yeast and man. Osh1, a member of the Oxysterol binding OSBP/ORP protein family, also enriches at NVJs in response to stationary growth stress.Citation8 Osh/ORP family proteins play key roles in lipid metabolism, and are implicated in the non-vesicular transport of sterols and phospholipids between organelles. Their loss may contribute to numerous familial hypercholesteraemias and lysosomal cholesterol-sphingolipid storage diseases.Citation17
More recently, other proteins with clear functions in lipid metabolism have been found to localize at NVJs. Yeast Mdm1/Snx13 is an ER-resident protein with clear orthologs in all metazoans (). It contains a PI3P-binding Phox homology (PX) domain that binds the vacuole surface, and is thus capable of tethering the nER and vacuole at NVJs.Citation18 Yeast also express a soluble paralog of Mdm1, named Nvj3, which also localizes to NVJs in an Mdm1-dependent manner. Notably, Sorting Nexin 14 (Snx14), one of four human orthologs of Mdm1, was recently implicated in pediatric cerebellar ataxia and intellectual disability.Citation19,20 The precise function of Mdm1 and its homologs is still unknown, but intriguingly over-expression of disease-analogous Mdm1 alleles that mimic the Snx14 alleles found in SNX14-deficient patients perturbs yeast sphingolipid metabolism, suggesting Mdm1 may function in lipid metabolism at NVJs.Citation18 A role for Mdm1 family proteins in lipid metabolism is also supported by the fact that Snazurus (Snz), the Drosophila homolog of Mdm1, is highly expressed in the fly fat body, the lipid metabolic center of the insect.Citation21
Figure 1. Domain architecture of proteins implicated in ER-endolysosomal membrane contact sites. Protein orthologs among Saccharomyces cerevisiae (Sc top), Drosophila melanogaster (Dm, middle) and Homo sapiens (Hs, bottom) are depicted. Abbreviations: PHg, Pleckstrin homology gram; MENTAL, MLN64 N-terminal alignment; StART, StAR-related Lipid Transfer; PH, Pleckstrin homology; OSBP, Oxysterol binding domain; PXA, PX-Associated; RGS, Regulator of G-protein Signaling; PX, Phox Homology; MSP, Major Sperm Protein; SMP, Synaptotagmin-like mitochondrial lipid-binding protein; DUF, domain of unknown function.
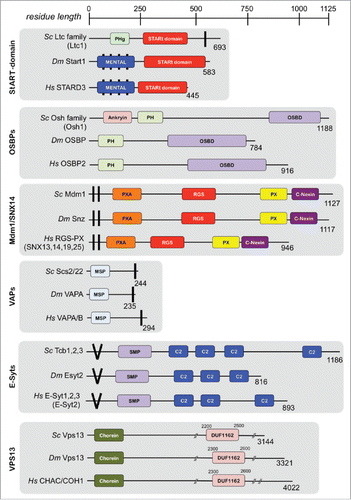
Recently, other proteins have been shown to dynamically localize to NVJs, and several of these are implicated in lipid metabolism and human disease. The Lipid Transfer Anchored at Membrane contact site (Lam) protein Lam6/Ltc1 localizes to both ER-mitochondrial and ER-vacuole MCSs, and is a member of the STAR-related lipid-transfer (StART) domain-containing protein family.Citation22-24 Notably, StART domain-containing proteins bind sterols, and several are implicated in diseases of cholesterol metabolism, including diabetes. Members of this protein family such as STARD3 may also function with Niemann-Pick proteins NPC1 and NPC2 to traffic cholesterol out of lysosomes, the failure of which causes the pathological accumulation of sterols and other lipids in lysosomal storage diseases.
Another protein, Vps13, was recently found to dynamically localize to both ER-mitochondrial and ER-vacuole MCSs.Citation25 When yeast are grown in glycerol-containing media, known to suppress formation of vacuole-mitochondria contact sites, Vps13 concentrates at NVJs. The reason for this localization is currently unclear, but it is tempting to speculate that Vps13 functions in lipid-exchange at these different MCSs. At least two Vps13 homologs exist in humans, VPS13A/CHAC and VPS13B/COH1, which have been implicated in chorea-acanthocytosis and Cohen Syndrome, respectively.Citation26 Intriguingly, both these disorders exhibit significant neurological and neuromuscular defects, as well as alterations in lipid metabolism in patients. The molecular mechanisms governing this remain obscure, however in yeast Vps13 appears to function on a parallel pathway to the ERMES complex, which controls ER-mitochondrial phospholipid flux. Vps13 alleles have been identified, which suppress the toxicity associated with growing ERMES-deficient strains on non-fermentable media, indicating that establishing alternative inter-organelle contact sites may compensate for the loss of ER-mitochondrial contact.Citation25 Further studies will be needed to understand the function of Vps13 and its homologs, as well as the roles of ER-endolysosomal contact sites in cellular lipid metabolism.
Thus, many yeast MCSs display clear orthologs in metazoans with roles in metabolism. Understanding their metabolic roles in metazoans is a clear priority in order to understand their connections to human disease. This review now turns to studies in the fruit fly Drosophila melanogaster as a potential model system to understand metazoan lipid metabolism and inter-organelle communication.
Organelle contact site proteins in Drosophila metabolism and aging
Numerous parallels exist between Drosophila and vertebrate metabolism, especially with respect to nutrient sensing and lipid metabolic pathways. The fly genome is also remarkably well shared between both yeasts and mammals, making it an ideal model system linking unicellular and multicellular organisms. Additionally, neuropeptides, enzymes, organs, and even disease phenotypes are highly conserved from flies to humans.Citation27-29
Numerous proteins involved in ER-endolysosomal MCSs in yeast are also conserved in Drosophila, and a few show clear disease pathologies. VAP-B/ALS8, the metazoan homolog of yeast Scs2/22 that promotes ER-PM contact sites, is linked to ALS disease in humans. Consistent with this, a fly model for ALS using mutant VAPP58S manifests protein aggregation and defects in synaptic morphology.Citation30 Ist2, which in yeast also participates in ER-PM contact sites, is homologous to Drosophila Axs (Abnormal X segregation), which is ER-localized and linked to defects in chromosomal segregation during meiosis.Citation31 It is unclear whether this phenotype is linked to inter-organelle communication. Stim1, which couples ER-PM Ca2+ flux in mammals, is also highly conserved in flies and is essential for proper larval development and mechano-sensory bristle differentiation.Citation32,33 The yeast enoyl-reductase Tsc13 that localizes to ER-vacuole NVJs in yeast is conserved in flies as Sc2, whose loss is embryonic lethal. Mutant Sc2 alleles have also been shown to exacerbate fly models of spinocerebellar ataxia.Citation34
Previous studies have identified Snazarus (Snz), the fly ortholog of yeast NVJ tether Mdm1, as a novel lifespan-associated gene by executing a fat body enhancer trap screen followed by longevity testing of the enhancer trap fly collection.Citation21 Snz is highly expressed in the fly fat body, the central organ for insect lipid metabolism, implying a role in fat metabolism. Consistent with this, Snz-deficient flies exhibit remarkable longevity, living approximately twice as long as their wildtype brethren. As lifespan is traditionally associated with alterations in metabolism and caloric intake/expenditure, it begs the question: what is the role for Snz in fly metabolism? Furthermore, how does alteration of Snz promote changes in aging and extend fly lifespan?
Metabolism and lifespan in Drosophila
Classic life-span studies in Caenorhabditis elegans implicate roles for insulin signaling in aging.Citation35 As in mammals, fruit fly insulin metabolism is regulated through neuroendocrine signaling. Drosophila secrete 8 insulin-like peptides (DILPS) that control growth, lipid and carbohydrate homeostasis, stress response, and aging.Citation36-39 Flies also produce a glucagon homolog, adipokinetic hormone (AKH), which regulates sugar levels in the hemolymph.Citation40,41 Recent studies have highlighted the significance of the minor glucose fraction in fly metabolism and its relevance to mammalian physiology.Citation42 Moreover, a high degree of functional conservation is observed among organ systems involved in metabolic processes, for example, the fat body is analogous to the mammalian adipose tissue and liver, storing excess energy as lipids and glycogen.Citation27,28,40,43
Like vertebrates, flies mobilize their fat reserves to provide energy during conditions of starvation or stress, and regulate sugar levels in response to environmental changes.Citation27,40 The processes of lipolysis and lipid storage are highly conserved between mammals and flies; flies express numerous cytoplasmic lipases including adipose triglyceride lipase (ATGL) homolog, brummer lipase, and a perilipin-like protein, LSD2 (lipid storage droplet 2), which localize to the outer membrane of lipid droplets.Citation44-46 Lysosomal lipases are less defined, but include Lip4 which are implicated in the turnover of lipid droplets during lipophagy.Citation47 In addition to maintaining lipid and carbohydrate homeostasis, the fat body also secretes lipoproteins and hormones to maintain metabolic balance; for example, the fat body produces DIPL6, the closest homolog to IGF, on induction by dFOXO and starvation. Interestingly, increased DILP6 levels have been linked to increased lifespan in flies, possibly by inhibiting secretion of DILP2 from brain insulin producing cells (IPCs).Citation37,39,48
Numerous studies have also established a relationship between energy homeostasis and aging in yeast, worms, flies, and mice. For example, attenuated insulin and TOR signaling, AMPK activation, and caloric restriction prolong lifespan.Citation28,37,49,50 Nutritional deprivation, short of malnutrition, is a potent stimulator of cellular autophagy: a process of self-degradation of cytoplasmic components by lysosomes and recycling of the catabolized products back to the cytosol to meet the nutritional demands of the cell, and to use as building blocks for new cellular components.Citation51,52 Defects in autophagy have been associated with accelerated aging, as aging is often characterized by accumulation of proteotoxic aggregates and dysfunctional organelles that contribute to cellular damage.Citation53-55 Autophagy is also involved in the breakdown of lipid droplets and triglycerides by a process called lipophagy, thereby playing a vital role in maintaining lipid homeostasis.Citation56 Studies in C. elegans have shown that up-regulation of lipid hydrolysis increases lifespan; intestinal induction of lysosomal acid lipase LIPL-4 triggers a lipid chaperone-mediated lysosome-to-nucleus longevity pathway.Citation57 However, the observed longevity may not be a consequence of reduced “adiposity,” as paradoxically, many long-lived mutants have increased fat storage.Citation58,59
Consistent with the relationship between autophagy and aging is the observation that several lifespan-extending mutations affect fat storing tissues. In mice, targeted loss of the murine insulin receptor in adipose tissue extends lifespan.Citation60 Murine Sir2 may confer anti-aging effects by stimulating fat hydrolysis via repression of the master regulator of adipogenesis (PPAR-g)Citation61; in the fruit fly, fat body-specific transgenic expression of Sir2 produces a similar result.Citation62 Drosophila is especially suitable to aging and longevity studies as it is possible to perform loss-of-function and gain-of-function screening, in a tissue-specific manner.Citation63
Mammalian ER-endosome contact sites in health and disease
As in Drosophila, mammals must also organize and regulate sophisticated metabolic pathways, and numerous studies are beginning to highlight inter-organelle MCSs as hubs for this metabolic regulation. Collectively, these studies indicate that zones of ER-endolysosomal contact regulate metabolism.
Unlike the singular and stable nuclear-vacuole junction of yeast, live-cell imaging and electron microscopy indicate that mammalian ER-endosome/lysosome MCSs are highly dynamic and prevalent within cells. ER-endosome MCSs are currently thought to be relatively tight connections between the 2 membranes (10–30 nm).Citation64 Intriguingly, this contact begins early in endosome biogenesis, and changes throughout its maturation. As endosomes mature, the number of ER-endosome contact sites increase and it has been reported that 99% of late endosomes (LEs) form contacts with the ER.Citation64-66 However, the dynamics and regulation of ER–endosome contact sites still remain poorly understood.
How do mammalian cells utilize ER-endolysosomal MCSs? Numerous studies indicate that these sites play roles in endosomal fission, receptor dephosphorylation, cholesterol transfer and Ca2+ exchange.Citation67 Specific proteins have also been identified which localize to ER-endosomal MCSs. Protrudin, an ER-localized protein that regulates protrusion and neurite outgrowth in mammals (although there is no obvious fly or yeast homolog), was recently found to localize to ER-LE contact sites, where it governs endosome mobility.Citation68 Protrudin facilitates LE translocation to the cell periphery through recruiting kinesin-1. Notably, mutations in the Protrudin gene ZFYVE27 are associated with an inherited neurological disorder and hereditary spastic paraplegia (HSP).Citation69 Thus, Protrudin is also named spastic paraplegia (SPG33). Consistent with this, previous studies indicate that Protrudin interacts with numerous HSP-associated proteins. A subset of HSP patients exhibit the ProtrudinG191V mutant, which promotes protein aggregation and exacerbates ER stress response and abnormal ER morphology. Another ER-endosome MCS protein is PTP1B, an ER-localized protein tyrosine phosphatase that interacts with the epidermal growth factor receptor (EGFR) at ER-endosome MCSs,Citation70 has been reported as both an oncogene and tumor suppressor in different cancer types,Citation71 and has also been implicated in metabolic disease.Citation72
Numerous sterol-binding proteins have also been implicated in ER-endolysosomal lipid metabolism and trafficking. The STARD3 StAR (steroidogenic acute regulatory protein) related lipid transfer (START) domain-3, previously known as MLN64 (metastatic lymph node 64),Citation73 and STARD3NL (STARD3 N-terminal like), previously known as MENTHOCitation74 proteins, are components of ER-LE MCS bridging complexes.Citation75 Tichauer et al. reported that overexpression of STARD3 in mice promotes liver damage,Citation76 and studies also found that high-grade prostate cancer and HER2-positive breast cancer are associated with STARD3.Citation77,78 The Drosophila STARD3 homolog, Start1, is linked to insect egg development. Research also revealed that bone mineral density patients have single-nucleotide polymorphisms of STARD3NL.Citation79
Lipid flux at inter-organelle contact sites in human health and disease
Several studies collectively implicate inter-organelle MCSs in non-vesicular lipid transport.Citation80,81 An increasing number of proteins are being identified as “tethers” that bring together membranes of opposing organelles into close proximity (<30 nm).Citation82,83 In addition to sustaining the integrity of the MCS, these “tethers” may function in shuffling lipids between nearby membranes. Major efforts in the field have been aimed at distinguishing between these two functions.Citation81 Alternatively, MCS “tethers” may recruit soluble effector molecules known as lipid transfer proteins (LTPs) that mediate lipid transfer between organelles.Citation84,85 Therefore, the closely juxtaposed MCSs provide a convenient center for both spontaneous and LTP-mediated non-vesicular lipid transport.Citation86,87
Based on their specificity to different lipids, well-defined LTPs can be grouped into three main categories: (1) sphingolipid-, (2) sterol- and (3) phospholipid-transfer proteins.Citation86 The crystal structures of several LTPs have been resolved both in the presence or absence of their lipid-binding ligands, which aid in understanding their mechanism of action ().Citation85,86 In general, LTPs bind individual lipids in a hydrophobic pocket or tunnel formed within the tertiary fold of the protein. In addition to the core hydrophobic pocket, most LTPs also contain protein-binding and/or lipid-binding domains that allow them to directly interact with membranes that donate and/or accept lipids. Additionally, the interaction of some LTPs with membranes is thought to induce conformational changes which expose the lipid-binding pocket within the LTP. The physical and chemical properties of this hydrophobic pocket dictate the lipid-binding specificity and affinity of different LTPs.Citation86 Typically, a lid-like domain shields the transferred lipids and acts as a gate during lipid exchange.Citation88,89
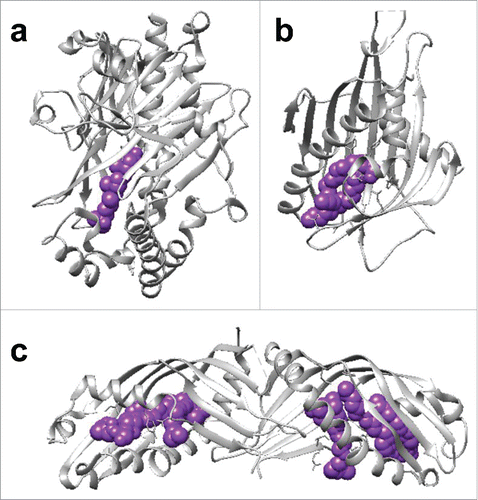
Figure 2. Three-dimensional structure of LTPs showing the protein backbone (gray ribbon representation) and the bound lipid (purple sphere representation) (a) The crystal structure of the yeast oxysterol-related domain ORD of oxysterol binding protein (OSBP) homolog 4 (Osh4) in complex with 25-hydroxycholesterol (PDB: 1ZHX). (b) The crystal structure of CERT (STAR)-related transfer (STARt) domain in complex with C16-ceramide (PDB: 2E3P). (c) The crystal structure of extended-synaptotagmin 2 (E-syt2) SMP domain in complex with TritonX-100 and DOPE (PDB: 4P42).
Several LTPs have been directly implicated in lipid metabolism at the MCSs (). LTPs are classically defined by their ability to facilitate lipid transfer between membranes in vitro.Citation90,91 However, in many cases, it remains unclear whether LTPs can also shuttle lipids between membranes in vivo.Citation85 One alternative hypothesis is that LTPs may function as lipid sensors or even lipid chaperones that localize to MCSs to escort or “present” specific lipids to metabolic enzymes to be processed.Citation85 So far, only a few studies have demonstrated in vivo lipid transfer activities by LTPs.Citation85,92 One example is the ceramide-transfer protein CERT which mediates the transport of various molecular species of ceramides between the ER and Golgi.Citation93-95 CERTs are cytosolic proteins that consist of an N-terminal pleckstrin homology (PH) domain, an FFAT motif (two Phe residues in an acidic tract),Citation96 and a C-terminal START domain (). The START domain is sufficient to mediate ceramide transfer in vivo.Citation97 The crystal structure of CERT START domain has been solved both in the apo-form and in complex with different ceramides providing structural insights into the mechanism by which it recognizes different ceramide species.Citation98
CERT. CERTs have been reported to function at the ER-Golgi MCSs,Citation93 and mediate the transfer of ceramides from their site of synthesis in the ER to the Golgi complex where they are converted to sphingomyelin by sphingomyelin synthase (SMS).Citation97,99 The interaction with the ER is mediated by the CERTs FFAT motif, which binds to the VAMP-associated proteins (VAPA and VAPB), while the PH domain of CERT mediates the interaction with the Golgi complex by binding PtdIns4P.Citation100,101 Mutations in either the FFAT motif or the PH domain not only abrogate the ER-Golgi MCS localization of CERTs, but also inhibit ceramide transport and consequently sphingolipid synthesis.Citation97,100 This suggests that the spatial restriction of CERT-mediated ceramide transfer at the ER-Golgi MCSs is required for sphingolipid metabolism.Citation86
FAPP2. Another example of LTPs is FAPP2, which was shown to transfer glycosylated ceramides (GlcCer) in vivo.Citation102 FAPP2 have similar domain organization as CERT and contains an N-terminal PH domain and a C-terminal glycolipid transfer protein or GLTP-homology domain.Citation102 The crystal structure of GLTP has been solved both in the presence and absence of the bound ligand, which provides molecular insights into the specificity of glycolipid binding and the mechanism of transfer.Citation103 The amino acid residues that are required for GLTP-mediated glycolipid transfer are highly conserved in FAPP2.Citation102,103 FAPP2 has been demonstrated to promote non-vesicular transfer of GlcCer between the Golgi compartmentsCitation102 as well as from the Golgi to the ER.Citation104 GlcCer transport to the ER lumen was reported to be significantly reduced due to the knockdown of FAPP2, which also severely impaired glycosphingolipids (GSL) metabolism.Citation104
Nir2. Nir2 is an example of a phosphoinositol transfer protein (PITP). PITPs were shown to transfer phosphatidylcholine (PtdCho) and phosphatidylioinositol (PtdIns) in vitro.Citation105 In cells, lipid transfer activity of PITPs was demonstrated using the Drosophila PITP DrdgBα. The levels of PtdIns(4,5)P2 was found to be significantly reduced in the DrdgBα mutants cells indicating that DrdgBα may play a role in moving PtnIns from their site of synthesis in the ER to the PM for local conversion to PtdIns(4,5)P2.Citation106,107 Similar to CERT, Nir2 contains an FFAT motif, which mediates its interaction with the ER. Nir2 also contains an N-terminal PITP domain, which binds lipids. Structural analysis of two human PITPs (PITPα and β)Citation108,109 and the yeast PITP Sec14pCitation110,111 and its closest homolog Sfh1Citation112 provided insights into the mechanism of lipid exchange by PITPs.
Oxysterol binding proteins (OSBP/ORP). Oxysterol binding proteins (OSBPs) bind to phospholipids and sterols, and often contain a lipid binding PH domain and a FFAT motif, which mediate dual interactions with lipid and protein, respectively. Additionally, mammalian OSBP has a C-terminal oxysterol-related domain (ORD), which binds 25-hydroxycholesterol (25-OH). OSBP is a member of a bigger family of sterol-binding proteins which includesCitation11 OSBP-related proteins (ORPs). The yeast OSBP ortholog Osh1 localizes to the nuclear vacuole junction (NVJ) by binding to Nvj1.Citation8 The crystal structure of another yeast ortholog Osh4 has been solved,Citation89 and the mechanism of sterol transfer has been demonstrated in vivo.Citation113 In addition, more recent work has demonstrated that Osh/ORP proteins can exchange phospholipids including phosphatidylserine (PS) and PtdIns4P between the ER and plasma membrane, and do so at ER-PM contact sites.Citation89
SMP domain-containing proteins. The synaptotagmin-like mitochondrial-lipid-binding protein (SMP) domain is found in several proteins that localize to inter-organelle contact sites such as the ERMES complex (ER-mitochondria) and the extended synaptotagmins (E-Syts), known in yeast as tricalbins (ER-PM).Citation10-12 SMP domains are found to bind several different lipids,Citation12 and they are proposed to transfer lipids between organelles at membrane contact sites, although the molecular mechanisms governing this require more study.
Lipid exchange at MCSs may plays an important role in lipid metabolism. Abnormal lipid metabolism can lead to several life-threatening conditions including obesity, diabetes, and heart disease. Moreover, genetic defects in several LTPs were found to have severe consequences on lipid metabolic pathways, which underlines the significance of these proteins in human health and development (). One of the well-studied examples of diseases associated with lipid metabolism is the Neimann-Pick Disease.
As mentioned in the previous section, two recent studied have implicated Snx14, the human homolog of the yeast MCS protein Mdm1, in a neurological disease with hallmarks of lipid storage diseases.Citation19,20 Cells derived from patients showed enlarged lysosomes and accumulation of cellular debrisCitation20 which may implicate Snx14 in the turnover of lysosomal lipids. Finally, few diseases have been directly linked to defects in MCS “tethering” proteins suggesting that these contact sites are important locations for inter-organelle communication. A mutation in VAPB, which may reduce the level of ER anchoring of lipid-binding proteins, has been linked to motor-neuron degeneration in amyotrophic lateral sclerosis (ALS) ().Citation114
Conclusions and perspectives
In conclusion, the study of inter-organelle contact sites has transformed from a curiosity of electron microscopy to a source of new understanding in the fields of lipid metabolism, nutrient exchange, signaling, and organelle identity. The discovery of key proteins and protein complexes which mediate inter-organelle tethering, or localize to MCSs for their functions, has been instrumental in the understanding of membrane contact sites in general cell biology. Most notably, it is now appreciated that key proteins with essential roles in human physiology appear to utilize MCSs for their functions. Loss of these proteins contributes to major genetic or acquired diseases, thus prompting the need to further understand these proteins, as well as how sites of inter-organelle contact are maintained, regulated, and integrate into general cellular physiology and pathophysiology. Future studies will no doubt continue to identify new and exciting proteins and pathways that utilize inter-organelle membrane contact sites.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of the Henne lab, in particular Ryan Feathers, for helpful discussions during the preparation of this manuscript.
Funding
This work is supported by a Welch Grant I-1873, and the American Federation for Aging Research (AFAR). WMH is a W.W. Caruth, Jr. Endowed Scholar at UT Southwestern.
References
- Porter KR, Palade GE. Studies on the endoplasmic reticulum. J Biophys Biochem Cytol 1957; 3:269-300; PMID:13438910; http://dx.doi.org/10.1083/jcb.3.2.269
- Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol advance online publication 2015; 17(2):69-82; PMID:26627931
- Raiborg C, Wenzel EM, Stenmark H. ER–endosome contact sites: molecular compositions and functions. EMBO J 2015; 34:1848-58; PMID:26041457; http://dx.doi.org/10.15252/embj.201591481
- Wijdeven RH, Jongsma MLM, Neefjes J, Berlin I, ER contact sites direct late endosome transport. Bioessays 2015; 37:1298-302; PMID:26440125; http://dx.doi.org/10.1002/bies.201500095
- Stefan CJ, Manford AG, Emr SD. ER-PM connections: sites of information transfer and inter-organelle communication. Curr Opin Cell Biol 2013; 25:434-42; PMID:23522446; http://dx.doi.org/10.1016/j.ceb.2013.02.020
- Pan X, Roberts P, Chen Y, Kvam E, Shulga N, Huang K, Lemmon S, Goldfarb DS. Nucleus–vacuole junctions in saccharomyces cerevisiae Are formed through the direct interaction of Vac8p with Nvj1p, Mol. Biol. Cell. 2000; 11:2445-57; PMID:10888680; http://dx.doi.org/10.1091/mbc.11.7.2445
- Toulmay A, Prinz WA. A conserved membrane-binding domain targets proteins to organelle contact sites. J Cell Sci 2012; 125:49-58; PMID:22250200; http://dx.doi.org/10.1242/jcs.085118
- Kvam E, Goldfarb DS. Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J Cell Sci 2004; 117:4959-68; PMID:15367582; http://dx.doi.org/10.1242/jcs.01372
- Peng Y, Tang F, Weisman LS. Palmitoylation plays a role in targeting Vac8p to specific membrane subdomains. Traffic 2006; 7:1378-87; PMID:16978392; http://dx.doi.org/10.1111/j.1600-0854.2006.00472.x
- Schauder CM, Wu X, Saheki Y, Narayanaswamy P, Torta F, Wenk MR, De Camilli P, Reinisch KM. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature 2014; 510:552-5; PMID:24847877; http://dx.doi.org/10.1038/nature13269
- Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology Dev Cell 2012; 23:1129-40; PMID:23237950; http://dx.doi.org/10.1016/j.devcel.2012.11.004
- Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 2013; 153:1494-509; PMID:23791178; http://dx.doi.org/10.1016/j.cell.2013.05.026
- Tavassoli S, Chao JT, Young BP, Cox RC, Prinz WA, de Kroon AI, Loewen CJ. Plasma membrane–endoplasmic reticulum contact sites regulate phosphatidylcholine synthesis. EMBO Rep 2013; 14:434-40; PMID:23519169; http://dx.doi.org/10.1038/embor.2013.36
- Nishimura AL, Mitne-Neto M, Silva HCA, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet 2004; 75:822-31; PMID:15372378; http://dx.doi.org/10.1086/425287
- Abe K, Ohno Y, Sassa T, Taguchi R, Çalışkan M, Ober C, Kihara A. Mutation for nonsyndromic mental retardation in the trans-2-enoyl-CoA reductase TER gene involved in fatty acid elongation impairs the enzyme activity and stability, leading to change in sphingolipid profile. J Biol Chem 2013; 288:36741-9; PMID:24220030; http://dx.doi.org/10.1074/jbc.M113.493221
- Kvam E, Gable K, Dunn TM, Goldfarb DS. Targeting of Tsc13p to nucleus-vacuole junctions: a role for very-long-chain fatty acids in the biogenesis of microautophagic vesicles Mol Biol Cell 2005; 16:3987-98; PMID:15958487; http://dx.doi.org/10.1091/mbc.E05-04-0290
- Olkkonen VM, Béaslas O, Nissilä E, Oxysterols and their cellular effectors. Biomolecules 2012; 2:76-103; PMID:24970128; http://dx.doi.org/10.3390/biom2010076
- Henne WM, Zhu L, Balogi Z, Stefan C, Pleiss JA, Emr SD. Mdm1/Snx13 is a novel ER–endolysosomal interorganelle tethering protein, J Cell Biol 2015; 210:541-51; PMID:26283797; http://dx.doi.org/10.1083/jcb.201503088
- Akizu N, Cantagrel V, Zaki MS, Al-Gazali L, Wang X, Rosti RO, Dikoglu E, Gelot AB, Rosti B, Vaux KK, et al., Biallelic mutations in SNX14 cause a syndromic form of cerebellar atrophy and lysosome-autophagosome dysfunction. Nat Genet 2015; 47:528-34; PMID:25848753; http://dx.doi.org/10.1038/ng.3256
- Thomas AC, Williams H, Setó-Salvia N, Bacchelli C, Jenkins D, O'Sullivan M, Mengrelis K, Ishida M, Ocaka L, Chanudet E, et al., Mutations in SNX14 cause a distinctive autosomal-recessive cerebellar ataxia and intellectual disability syndrome. Am J Hum Genet 2014; 95:611-21; PMID:25439728; http://dx.doi.org/10.1016/j.ajhg.2014.10.007
- Suh JM, Stenesen D, Peters JM, Inoue A, Cade A, Graff JM. An RGS-Containing sorting nexin controls drosophila lifespan. PLoS One 2008; 3:e2152; PMID:18478054; http://dx.doi.org/10.1371/journal.pone.0002152
- Murley A, Sarsam RD, Toulmay A, Yamada J, Prinz WA, Nunnari J. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts, J Cell Biol 2015; 209:539-48; PMID:25987606; http://dx.doi.org/10.1083/jcb.201502033
- Gatta AT, Wong LH, Sere YY, Calderón-Noreña DM, Cockcroft S, Menon AK, Levine TP. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport Elife 2015; 4; PMID:26001273; http://dx.doi.org/10.7554/eLife.07253
- Elbaz-Alon Y, Eisenberg-Bord M, Shinder V, Stiller SB, Shimoni E, Wiedemann N, Geiger T, Schuldiner M. Lam6 Regulates the Extent of Contacts between Organelles. Cell Rep 2015; 12:7-14; PMID:26119743; http://dx.doi.org/10.1016/j.celrep.2015.06.022
- Lang AB, Peter ATJ, Walter P, Kornmann B, ER–mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J Cell Biol 2015; 210:883-90; PMID:26370498; http://dx.doi.org/10.1083/jcb.201502105
- Velayos-Baeza A, Vettori A, Copley RR, Dobson-Stone C, Monaco AP. Analysis of the human VPS13 gene family. Genomics 2004; 84:536-49; PMID:15498460; http://dx.doi.org/10.1016/j.ygeno.2004.04.012
- Bharucha KN. The epicurean fly: using Drosophila melanogaster to study metabolism. Pediatr Res 2009; 65:132-7; PMID:18978647; http://dx.doi.org/10.1203/PDR.0b013e318191fc68
- Owusu-Ansah E, Perrimon N. Modeling metabolic homeostasis and nutrient sensing in Drosophila: implications for aging and metabolic diseases. Dis Model Mech 2014; 7:343-50; PMID:24609035; http://dx.doi.org/10.1242/dmm.012989
- Schlegel A, Stainier DYR. Lessons from “Lower” Organisms: What Worms, Flies, and Zebrafish Can Teach Us about Human Energy Metabolism. PLoS Genet 2007; 3:e199; PMID:18081423; http://dx.doi.org/10.1371/journal.pgen.0030199
- Ratnaparkhi A, Lawless GM, Schweizer FE, Golshani P, Jackson GR. A Drosophila model of ALS: human ALS-associated mutation in VAP33A suggests a dominant negative mechanism. PLoS One 2008; 3:e2334; PMID:18523548; http://dx.doi.org/10.1371/journal.pone.0002334
- Kramer J, Hawley RS. The spindle-associated transmembrane protein Axs identifies a membranous structure ensheathing the meiotic spindle, Nat. Cell Biol 2003; 5:261-3; PMID:12646877; http://dx.doi.org/10.1038/ncb944
- Eid JP, Arias A, Robertson H, Hime GR, Dziadek M. The Drosophila STIM1 orthologue, dSTIM, has roles in cell fate specification and tissue patterning. BMC Dev Biol 2008; 8:104; PMID:18950512; http://dx.doi.org/10.1186/1471-213X-8-104
- Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci USA 2007; 104:9301-6; PMID:17517596; http://dx.doi.org/10.1073/pnas.0702866104
- Fernandez-Funez P, Nino-Rosales ML, de Gouyon B, She WC, Luchak JM, Martinez P, Turiegano E, Benito J, Capovilla M, Skinner PJ, et al. Identification of genes that modify ataxin-1-induced neurodegeneration. Nature 2000; 408:101-6; PMID:11081516; http://dx.doi.org/10.1038/35040584
- Dorman JB, Albinder B, Shroyer T, Kenyon C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics 1995; 141:1399-406; PMID:8601482
- Broughton S, Alic N, Slack C, Bass T, Ikeya T, Vinti G, Tommasi AM, Driege Y, Hafen E, Partridge L. Reduction of DILP2 in Drosophila triages a metabolic phenotype from lifespan revealing redundancy and compensation among DILPs. PLoS One 2008; 3:e3721; PMID:19005568; http://dx.doi.org/10.1371/journal.pone.0003721
- Kannan K, Fridell YWC. Functional implications of Drosophila insulin-like peptides in metabolism, aging, and dietary restriction Front Physiol 2013; 4:288; PMID:24137131; http://dx.doi.org/10.3389/fphys.2013.00288
- Géminard C, Rulifson EJ, Léopold P. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab 2009; 10:199-207; http://dx.doi.org/10.1016/j.cmet.2009.08.002
- Grönke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular Evolution and Functional Characterization of Drosophila Insulin-Like Peptides. PLoS Genet 2010; 6:e1000857; PMID:20195512; http://dx.doi.org/10.1371/journal.pgen.1000857
- Baker KD, Thummel CS. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila Cell Metab 2007; 6:257-66; PMID:17908555; http://dx.doi.org/10.1016/j.cmet.2007.09.002
- Kim SK, Rulifson EJ. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature 2004; 431 316-20; PMID:15372035; http://dx.doi.org/10.1038/nature02897
- Ugrankar R, Berglund E, Akdemir F, Tran C, Kim MS, Noh J, Schneider R, Ebert B, Graff JM. Drosophila glucome screening identifies Ck1alpha as a regulator of mammalian glucose metabolism. Nat Commun 2015; 6:7102; PMID:25994086; http://dx.doi.org/10.1038/ncomms8102
- Leopold P, Perrimon N. Drosophila and the genetics of the internal milieu. Nature 2007; 450:186-8; PMID:17994083; http://dx.doi.org/10.1038/nature06286
- Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, Müller G, Jäckle H, Kühnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab 2005; 1:323-30; PMID:16054079; http://dx.doi.org/10.1016/j.cmet.2005.04.003
- Teixeira L, Rabouille C, Rørth P, Ephrussi A, Vanzo NF. Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech Dev 2003; 120:1071-81; PMID:14550535; http://dx.doi.org/10.1016/S0925-4773(03)00158-8
- Kühnlein RP. Lipid droplet-based storage fat metabolism in Drosophila. J Lipid Res 2012; 53:1430-6; PMID:22566574; http://dx.doi.org/10.1194/jlr.R024299
- Singh R, Cuervo AM, Singh R, Cuervo AM, Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol 2012; 2012:e282041
- Tatar M, Post S, Yu K. Nutrient control of Drosophila longevity. Trends Endocrinol Metab 2014; 25:509-17; PMID:24685228; http://dx.doi.org/10.1016/j.tem.2014.02.006
- Stenesen D, Suh JM, Seo J, Yu K, Lee KS, Kim JS, Min KJ, Graff JM. Adenosine nucleotide biosynthesis and AMPK regulate adult life span and mediate the longevity benefit of caloric restriction in flies. Cell Metab 2013; 17:101-12; PMID:23312286; http://dx.doi.org/10.1016/j.cmet.2012.12.006
- Barzilai N, Huffman DM, Muzumdar RH, Bartke A. The critical role of metabolic pathways in aging. Diabetes 2012; 61:1315-22; PMID:22618766; http://dx.doi.org/10.2337/db11-1300
- Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 2010; 90:1383-435; PMID:20959619; http://dx.doi.org/10.1152/physrev.00030.2009
- Rubinsztein DC, Mariño G, Kroemer G, Autophagy and aging. Cell 2011; 146:682-95; PMID:21884931; http://dx.doi.org/10.1016/j.cell.2011.07.030
- Bergmann A. Autophagy and Cell Death: No Longer at Odds. Cell 2007; 131:1032-4; PMID:18083090; http://dx.doi.org/10.1016/j.cell.2007.11.027
- Markaki M, Tavernarakis N. Metabolic control by target of rapamycin and autophagy during ageing - a mini-review. Gerontology 2013; 59:340-8; PMID:23594965; http://dx.doi.org/10.1159/000348599
- Madeo F, Zimmermann A, Maiuri MC, Kroemer G. Essential role for autophagy in life span extension. J Clin Invest 2015; 125:85-93; PMID:25654554; http://dx.doi.org/10.1172/JCI73946
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature 2009; 458:1131-5; PMID:19339967; http://dx.doi.org/10.1038/nature07976
- Folick A, Oakley HD, Yu Y, Armstrong EH, Kumari M, Sanor L, Moore DD, Ortlund EA, Zechner R, Wang MC. Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science 2015; 347:83-6; PMID:25554789; http://dx.doi.org/10.1126/science.1258857
- Gems D, Partridge L. Genetics of longevity in model organisms: debates and paradigm shifts. Annu Rev Physiol 2013; 75:621-44; PMID:23190075; http://dx.doi.org/10.1146/annurev-physiol-030212-183712
- Hansen M, Flatt T, Aguilaniu H. Reproduction, Fat Metabolism, and Lifespan – What Is the Connection?. Cell Metab 2013; 17:10-9; PMID:23312280; http://dx.doi.org/10.1016/j.cmet.2012.12.003
- Selman C, Lingard S, Choudhury AI, Batterham RL, Claret M, Clements M, Ramadani F, Okkenhaug K, Schuster E, Blanc E, et al. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J 2008; 22:807-18; PMID:17928362; http://dx.doi.org/10.1096/fj.07-9261com
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 2004; 429:771-6; PMID:15175761; http://dx.doi.org/10.1038/nature02583
- Whitaker R, Faulkner S, Miyokawa R, Burhenn L, Henriksen M, Wood JG, Helfand SL. Increased expression of Drosophila Sir2 extends life span in a dose-dependent manner. Aging (Albany NY) 2013; 5:682-91; PMID:24036492
- Proshkina EN, Shaposhnikov MV, Sadritdinova AF, Kudryavtseva AV, Moskalev AA. Basic mechanisms of longevity: A case study of Drosophila pro-longevity genes. Ageing Res Rev 2015; 24:218-31; PMID:26318059; http://dx.doi.org/10.1016/j.arr.2015.08.005
- Friedman JR, Dibenedetto JR, West M, Rowland AA, Voeltz GK. Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol Biol Cell 2013; 24:1030-40; PMID:23389631; http://dx.doi.org/10.1091/mbc.E12-10-0733
- Hönscher C, Ungermann C. A close-up view of membrane contact sites between the endoplasmic reticulum and the endolysosomal system: from yeast to man. Crit Rev Biochem Mol Biol 2014; 49:262-8; PMID:24382115; http://dx.doi.org/10.3109/10409238.2013.875512
- van der Kant R, Neefjes J. Small regulators, major consequences – Ca2+ and cholesterol at the endosome–ER interface. J Cell Sci 2014; 127:929-38; PMID:24554437; http://dx.doi.org/10.1242/jcs.137539
- Rowland AA, Chitwood PJ, Phillips MJ, Voeltz GK. ER Contact Sites Define the Position and Timing of Endosome Fission. Cell 2014; 159:1027-41; PMID:25416943; http://dx.doi.org/10.1016/j.cell.2014.10.023
- Raiborg C, Wenzel EM, Pedersen NM, Olsvik H, Schink KO, Schultz SW, Vietri M, Nisi V, Bucci C, Brech A, et al. Repeated ER–endosome contacts promote endosome translocation and neurite outgrowth. Nature 2015; 520:234-8; PMID:25855459; http://dx.doi.org/10.1038/nature14359
- Hashimoto Y, Shirane M, Matsuzaki F, Saita S, Ohnishi T, Nakayama KI. Protrudin regulates endoplasmic reticulum morphology and function associated with the pathogenesis of hereditary spastic paraplegia. J Biol Chem 2014; 289:12946-61; PMID:24668814; http://dx.doi.org/10.1074/jbc.M113.528687
- Eden ER, White IJ, Tsapara A, Futter CE, Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol 2010; 12:267-72; PMID:20118922
- Liu H, Wu Y, Zhu S, Liang W, Wang Z, Wang Y, Liu H, Wu Y, Zhu S, Liang W, Wang Z, Wang Y. PTP1B promotes cell proliferation and metastasis through activating src and ERK1/2 in non-small cell lung cancer. Cancer Lett 2015; 359:218-25; PMID:25617799; http://dx.doi.org/10.1016/j.canlet.2015.01.020
- Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL, Normandin D, Cheng A, Himms-Hagen J, Chan CC, et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 1999; 283:1544-8; PMID:10066179; http://dx.doi.org/10.1126/science.283.5407.1544
- Alpy F, Stoeckel ME, Dierich A, Escola JM, Wendling C, Chenard MP, Vanier MT, Gruenberg J, Tomasetto C, Rio MC. The steroidogenic acute regulatory protein homolog MLN64, a late endosomal cholesterol-binding protein. J Biol Chem 2001; 276:4261-9; PMID:11053434; http://dx.doi.org/10.1074/jbc.M006279200
- Alpy F, Wendling C, Rio MC, Tomasetto C, MENTHO, a MLN64 homologue devoid of the START domain J Biol Chem 2002; 277:50780-7; PMID:12393907; http://dx.doi.org/10.1074/jbc.M208290200
- Alpy F, Rousseau A, Schwab Y, Legueux F, Stoll I, Wendling C, Spiegelhalter C, Kessler P, Mathelin C, Rio MC, et al. STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J Cell Sci 2013; 126:5500-12; PMID:24105263; http://dx.doi.org/10.1242/jcs.139295
- Tichauer JE, Morales MG, Amigo L, Galdames L, Klein A, Quinones V, Ferrada C, Alvarez AR, Rio MC, Miquel JF, et al. Overexpression of the cholesterol-binding protein MLN64 induces liver damage in the mouse. World J Gastroenterol 2007; 13:3071-9; PMID:17589922
- Vinatzer U, Dampier B, Streubel B., Pacher M, Seewald MJ, Stratowa C, Kaserer K, Schreiber M. Expression of HER2 and the coamplified genes GRB7 and MLN64 in human breast cancer: quantitative real-time reverse transcription-PCR as a diagnostic alternative to immunohistochemistry and fluorescence in situ hybridization. Clin Cancer Res 2005; 11:8348-57; PMID:16322295; http://dx.doi.org/10.1158/1078-0432.CCR-05-0841
- Stigliano A, Gandini O, Cerquetti L, Gazzaniga P, Misiti S, Monti S, Gradilone A, Falasca P, Poggi M, Brunetti E, et al. Increased metastatic lymph node 64 and CYP17 expression are associated with high stage prostate cancer. J Endocrinol 2007; 194:55-61; PMID:17592021; http://dx.doi.org/10.1677/JOE-07-0131
- Rivadeneira F, Styrkársdottir U, Estrada K, Halldórsson BV, Hsu YH, Richards JB, Zillikens MC, Kavvoura FK, Amin N, Aulchenko YS, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies Nat Genet 2009; 41:1199-206; PMID:19801982; http://dx.doi.org/10.1038/ng.446
- Toulmay A, Prinz WA. Lipid transfer and signaling at organelle contact sites: the tip of the iceberg. Curr Opin Cell Biol 2011; 23:458-63; PMID:21555211; http://dx.doi.org/10.1016/j.ceb.2011.04.006
- Prinz WA. Bridging the gap: Membrane contact sites in signaling, metabolism, and organelle dynamics J Cell Biol 2014; 205:759-69; PMID:24958771; http://dx.doi.org/10.1083/jcb.201401126
- Helle SCJ, Kanfer G, Kolar K, Lang A, Michel AH, Kornmann B. Organization and function of membrane contact sites. Biochimica Et Biophysica Acta (BBA) - Mol Cell Res 2013; 1833:2526-41; http://dx.doi.org/10.1016/j.bbamcr.2013.01.028
- English AR, Voeltz GK. Endoplasmic Reticulum Structure and Interconnections with Other Organelles. Cold Spring Harb Perspect Biol 2013; 5:a013227; PMID:23545422; http://dx.doi.org/10.1101/cshperspect.a013227
- Lev S. Nonvesicular Lipid Transfer from the Endoplasmic Reticulum. Cold Spring Harb Perspect Biol 2012; 4:a013300; PMID:23028121; http://dx.doi.org/10.1101/cshperspect.a013300
- D'Angelo G, Vicinanza M, De Matteis MA, Lipid-transfer proteins in biosynthetic pathways. Curr Opin Cell Biol 2008; 20:360-70; http://dx.doi.org/10.1016/j.ceb.2008.03.013
- Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol 2010; 11:739-50; PMID:20823909; http://dx.doi.org/10.1038/nrm2971
- Holthuis JCM, Levine TP. Lipid traffic: floppy drives and a superhighway. Nat Rev Mol Cell Biol 2005; 6:209-20; PMID:15738987; http://dx.doi.org/10.1038/nrm1591
- Wirtz KWA, Schouten A, Gros P. Phosphatidylinositol transfer proteins: from closed for transport to open for exchange. Adv Enzyme Regul 2006; 46:301-11; PMID:16854452; http://dx.doi.org/10.1016/j.advenzreg.2006.01.020
- Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature 2005; 437:154-8; PMID:16136145; http://dx.doi.org/10.1038/nature03923
- Wirtz KW, Zilversmit DB. Exchange of phospholipids between liver mitochondria and microsomes in vitro. J Biol Chem 1968; 243:3596-602; PMID:4968799
- Zilversmit DB. Lipid transfer proteins: overview and applications. Meth Enzymol 1983; 98:565-73; PMID:6669064; http://dx.doi.org/10.1016/0076-6879(83)98183-1
- Levine TP. A lipid transfer protein that transfers lipid. J Cell Biol 2007; 179:11-3; PMID:17923527; http://dx.doi.org/10.1083/jcb.200709055
- Kawano M, Kumagai K, Nishijima M, Hanada K. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J Biol Chem 2006; 281:30279-88; PMID:16895911; http://dx.doi.org/10.1074/jbc.M605032200
- Hanada K, Kumagai K, Tomishige N, Kawano M. CERT and intracellular trafficking of ceramide. Biochim Biophys Acta 2007; 1771:644-53; PMID:17314061; http://dx.doi.org/10.1016/j.bbalip.2007.01.009
- Kumagai K, Yasuda S, Okemoto K, Nishijima M, Kobayashi S, Hanada K. CERT mediates intermembrane transfer of various molecular species of ceramides J Biol Chem 2005; 280:6488-95; PMID:15596449; http://dx.doi.org/10.1074/jbc.M409290200
- Loewen CJR, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J 2003; 22:2025-35; PMID:12727870; http://dx.doi.org/10.1093/emboj/cdg201
- Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide Nature 2003; 426:803-9; PMID:14685229; http://dx.doi.org/10.1038/nature02188
- Kudo N, Kumagai K, Tomishige N, Yamaji T, Wakatsuki S, Nishijima M, Hanada K, Kato R. Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide. Proc Natl Acad Sci USA 2008; 105:488-93; PMID:18184806; http://dx.doi.org/10.1073/pnas.0709191105
- Huitema K, van den Dikkenberg J, Brouwers J.F.HM, Holthuis JCM. Identification of a family of animal sphingomyelin synthases. EMBO J 2004; 23:33-44; PMID:14685263; http://dx.doi.org/10.1038/sj.emboj.7600034
- Hanada K. Discovery of the molecular machinery CERT for endoplasmic reticulum-to-Golgi trafficking of ceramide. Mol Cell Biochem 2006; 286:23-31; PMID:16601923; http://dx.doi.org/10.1007/s11010-005-9044-z
- Lev S, Halevy DB, Peretti D, Dahan N. The VAP protein family: from cellular functions to motor neuron disease. Trends Cell Biol 2008; 18:282-90; PMID:18468439; http://dx.doi.org/10.1016/j.tcb.2008.03.006
- D'Angelo G, Polishchuk E, Di Tullio G, Santoro M, Di Campli A, Godi A, West G, Bielawski J, Chuang CC, van der Spoel AC, et al. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature 2007; 449:62-7; http://dx.doi.org/10.1038/nature06097
- Malinina L, Malakhova ML, Teplov A, Brown RE, Patel DJ. Structural basis for glycosphingolipid transfer specificity, Nature 2004; 430:1048-53; PMID:15329726; http://dx.doi.org/10.1038/nature02856
- Halter D, Neumann S, van Dijk SM, Wolthoorn J, de Mazière AM, Vieira OV, Mattjus P, Klumperman J, van Meer G, Sprong H. Pre- and post-Golgi translocation of glucosylceramide in glycosphingolipid synthesis. J Cell Biol 2007; 179:101-15; PMID:17923531; http://dx.doi.org/10.1083/jcb.200704091
- Helmkamp GM. Phospholipid transfer proteins: mechanism of action J Bioenerg Biomembr 1986; 18:71-91; PMID:3522562; http://dx.doi.org/10.1007/BF00743477
- Hardie RC, Raghu P, Moore S, Juusola M, Baines RA, Sweeney ST. Calcium influx via TRP channels is required to maintain PtdIns(4,5)P2 levels in Drosophila photoreceptors. Neuron 2001; 30:149-59; PMID:11343651; http://dx.doi.org/10.1016/S0896-6273(01)00269-0
- Milligan SC, Alb JG, Elagina RB, Bankaitis VA, Hyde DR. The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J Cell Biol 1997; 139:351-63; PMID:9334340; http://dx.doi.org/10.1083/jcb.139.2.351
- Tilley SJ, Skippen A, Murray-Rust J, Swigart PM, Stewart A, Morgan CP, Cockcroft S, McDonald NQ. Structure-Function Analysis of Phosphatidylinositol Transfer Protein Alpha Bound to Human Phosphatidylinositol. Structure 2004; 12:317-26; PMID:14962392; http://dx.doi.org/10.1016/j.str.2004.01.013
- Vordtriede PB, Doan CN, Tremblay JM, George HM, Yoder MD. Structure of PITPβ in Complex with Phosphatidylcholine: Comparison of Structure and Lipid Transfer to Other PITP Isoforms†,‡. Biochem 2005; 44:14760-71; http://dx.doi.org/10.1021/bi051191r
- Sha B, Phillips SE, Bankaitis VA, Luo M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol- transfer protein. Nature 1998; 391:506-10; PMID:9461221; http://dx.doi.org/10.1038/35179
- Ryan MM, Temple BRS, Phillips SE, Bankaitis VA. Conformational Dynamics of the Major Yeast Phosphatidylinositol Transfer Protein Sec14p: Insight into the Mechanisms of Phospholipid Exchange and Diseases of Sec14p-Like Protein Deficiencies. Mol Biol Cell 2007; 18:1928-42; PMID:17344474; http://dx.doi.org/10.1091/mbc.E06-11-1024
- Schaaf G, Ortlund EA, Tyeryar KR, Mousley CJ, Ile KE, Garrett TA, Ren J, Woolls MJ, Raetz CR, Redinbo MR, et al. Functional Anatomy of Phospholipid Binding and Regulation of Phosphoinositide Homeostasis by Proteins of the Sec14 Superfamily. Mol Cell 2008; 29:191-206; PMID:18243114; http://dx.doi.org/10.1016/j.molcel.2007.11.026
- Raychaudhuri S, Im YJ, Hurley JH, Prinz W. A. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J Cell Biol 2006; 173:107-19; PMID:16585271; http://dx.doi.org/10.1083/jcb.200510084
- Teuling E, Ahmed S, Haasdijk E, Demmers J, Steinmetz MO, Akhmanova A, Jaarsma D, Hoogenraad CC. Motor neuron disease-associated mutant vesicle-associated membrane protein-associated protein (VAP) B recruits wild-type VAPs into endoplasmic reticulum-derived tubular aggregates. J Neurosci 2007; 27:9801-15; PMID:17804640; http://dx.doi.org/10.1523/JNEUROSCI.2661-07.2007
