Abstract
European plaice (Pleuronectes platessa) is quite frequently substituted with other flatfish, especially with Yellow fin sole (Limanda aspera), which differs not only in meat quality but also particularly in its origin and manipulation chain. We propose an integrated approach of laboratory and in silico mtDNA PCR-RFLP procedures generating a set of restriction patterns easily resolvable in agarose gel, able to discriminate with certainty P. platessa from other 20 flatfish species. The herein proposed procedure is an economical and valid tool in detecting mislabelled seafoods.
Frecuentemente, la solla europea (Pleuronectes platessa) es sustituida por otros peces planos, utilizándose especialmente la limanda (Limanda aspera). Ésta se distingue de la primera no sólo por la calidad de su carne, sino también por su origen y por su cadena de manejo. En el presente artículo, los autores proponen realizar un enfoque integrado de procedimientos de laboratorio y de in silico mtDNA PCR-RFLP, a partir de los cuales se genera un conjunto de patrones de restricción fácilmente resolubles en gel de agarosa. Dicho enfoque puede discriminar con seguridad entre P. platessa y otras veinte especies de peces planos. El procedimiento que aquí se propone representa un método económico y válido para la detección de pescados mal etiquetados.
Introduction
The EC law No.2065/2001 requests the appropriate species traceability and labelling of fishery and seafood products, with a particular care for the scientific name of the species. Valid species identification is often lacking and no traceability procedure exists to ensure the exact nature and origin of the product, particularly those that are indistinguishable after processing and freezing (Filonzi, Chiesa, Vaghi, & Nonnis Marzano, Citation2010).
European plaice Pleuronectes platessa Linnaeus 1785 is the most commercially important flatfish in European waters, especially considering the frozen fillets market. Indeed, its fillets can be easily confused with other species belonging to the same family, Pleuronectidae, for example, with the genus Limanda (L. aspera, L. limanda, L. ferruginea) less valuable as to both economical and nutritional values (Tepedino et al., Citation2001), or with species belonging to the family Soleidae, for example, Common sole (Solea solea), Egyptian sole (Solea aegyptiaca) or Senegalese sole (Solea senegalensis). In 2008, Espiñeira, González-Lavín, Vieites, and Santaclara reported that the species with the highest rate of mislabelling was the European plaice, followed by the Common sole (2008). Specifically they found out that 17% of the European plaice labels were incorrect and that the species present in the commercial product concerned Platichthys flesus in the case of whole fish, and Hippoglossoides spp. in the case of frozen fillets.
Differences concern not only the meat quality of each species but also the FAO Area of origin and the manipulation chain. So, food mystification can lead to involuntary ingestion of cryptic species from contaminated areas (van Leeuwen et al., Citation2009); for example, the European plaice belonging to FAO Area 27, corresponding to the Northeast of the Atlantic Ocean, on the contrary the Yellow fin sole, L. aspera belonging to FAO Area 61, subjected to observation after the Tohoku earthquake and tsunami of 11 March 2011 that caused extensive damage to the Fukushima Daiichi nuclear power plant. Furthermore, the European plaice is killed in water and ice, eviscerated on board, put in ice and frozen without further manipulations. On the contrary, Yellow fin sole, L. aspera, is eviscerated and frozen on board, thereafter defrosted, filleted, frozen again and slightly glazed to prevent premature oxidation; the high fat content of this animal increases its tendency to become rancid if the cold chain is not strictly observed.
Food mystification can also lead to involuntary ingestion of fish species that, in highly allergic individuals, can lead to serious or even fatal allergic reactions (Le Fresne, Popova, Le Vacon, & Carton, Citation2011). Even if cross-reactivity among fish and seafood in general is largely reported, different IgE levels were observed for various fish species, including flatfishes, and a correlation between IgE levels and expression of symptoms after fish ingestion was demonstrated (Koyama et al., Citation2006). An accurate identification of fish products could shed light on possible allergenic responses due to adulterations. It is known that parvalbumins are the major fish allergens and that their contents vary in different fish species (Kuehn, Scheuermann, Hilger, & Hentges, Citation2010). For this reason, it would be important to consider the exclusion of L. aspera from children’s diets, especially of those individuals who showed IgE-mediated reactions during weaning and who require special care in food introductions.
It is almost impossible to distinguish some flatfishes, these species in particular, only on the basis of their frozen, breaded, filleted aspects. As well synthetised by Filonzi et al. (Citation2010), molecular genetics is gaining increasing attention and several papers were published with the aim to describe methods distinguishing fish species, especially flatfishes (see Asensio, Citation2007; Teletchea, Citation2009 and Filonzi et al., Citation2010 for a review). Even though some molecular markers might be considered highly reproducible and informative on flatfishes distinction, as sequencing of target fragment (Espiñeira et al., Citation2008; Filonzi et al., Citation2010), they are often time- and money-consuming and cannot be applied in a routine food control chain. To date, DNA barcoding, particularly if improved by the simultaneous use of Cytochrome b (Cytb) and Cytochrome Oxidase I (COI), seems the most promising approach to solve taxonomic attribution in foiling food mystification (Filonzi et al., Citation2010; Kochzius et al., Citation2010; Lucentini et al., Citation2011a). Although the sequencing of Cytb and COI fragments is considered the best choice, the need may arise for more rapid and economical approaches. PCR-RFLP fulfils the requirements of accuracy, repeatability, fast procedure and is also less expensive to employ (Gigliarelli et al., Citation2013). This method has been used widely in fishes discrimination (Teletchea, Citation2009), providing a valid tool for monitoring the marketing of fish fillets. Here, we first analysed Cytb and COI in a classical DNA-barcode approach for three flatfish species; then, on the basis of DNA-barcoding data, we developed both Cytb- and COI- specific PCR-RFLP procedure and in silico RFLP analysis that distinguishes P. platessa from the other 17 species of the family Pleuronectidae and 3 species of the family Soleidae.
Materials and methods
Samples collection and DNA extraction
Thirteen specimens of P. platessa, 11 specimens of L. aspera and 4 of S. solea were sampled. Specimens, both fresh and frozen, were purchased in different markets, including pre-fried breaded fillets that were subjected to heat processing. To analyse breaded fillets, immediately after the removal of bread layers they were put in absolute ethanol and stored at −20°C until DNA extraction. DNA was extracted following a modified protocol (Lucentini, Caporali, Palomba, Lancioni, & Panara, Citation2006).
Amplification and sequencing
A fragment of 327 base pairs (bp) of Cytb gene was amplified using primers L14841(5′ AAAAAGCTTCCATCCAACATCTCAGCATGATGAAA 3′)/H15149 (5′AAACTGCAGCCCCTCAGAATGATATTTGTCCTCA 3′) (Kocher et al., Citation1989) and the amplification programme, 3 min, 94°C; 35 cycles of 45 s, 94°C, 30 s, 44°C, 2 min, 72°C; 10 min,72°C. Furthermore, a fragment of 558 bp of COI was amplified using primers ff2d-fish (5′TTCTCCACCAACCACAA(AG)GA(CT)AT(CT)GG 3′)/fr1d-fish (5′CACCTCAGGGTGTCCGAA(AG)AA(CT)CA(AG)AA 3′) (Ivanova, Dewaard, & Hebert, Citation2007) and the programme, 5 min, 94°C; 35 cycles of 30 s, 94°C, 45 s, 52°C, 1 min, 72°C; 10 min, 72°C.
Amplifications were performed as reported in Filonzi et al. (Citation2010), in a Biometra TPersonal thermal cycler. PCR products were purified using ExoSAP-IT® for PCR Product Clean-Up (USB, Cleveland, USA)® following the manufacturer’s instructions. They were sequenced in forward directions using ABI PRISM® BigDyeTM Terminator Cycle Sequencing Ready Reaction Kit v. 3.1 (Applera, Life Technologies Limited, Paisley, Scotland) as reported by Lucentini et al. (Citation2011b). The labelling programme consisted of 96°C, 1 min; 26 cycles: 96°C, 10 s, 50°C, 5 s, 60°C, 4 min. Furthermore, since COI-ff2d-fish has three degenerations, to sequence COI amplicons, a nested forward primer PplCOInestF (5′GGTGCCTGAGCCGGAATAGT3′) was designed on the basis of P. platessa GenBank EU513681 and L. aspera GenBank EU513663 sequences (Espiñeira et al., Citation2008).
The entire procedure (DNA extraction, PCR amplifications and sequencing reactions) was performed for each of the collected samples and replicated three times by different operators.
Sequence analysis
Obtained sequences of Cytb and COI were compared through blasting procedure (http://www.ncbi.nlm.nih.gov/BLAST) and aligned using MEGA 5.1 software (Tamura et al., Citation2011) together with the sequences of the following species, for which Cytb and COI GenBank codes were reported: L. aspera (Cytb: EU513800; COI: JQ354173), L. limanda (GU168913; EU513669), L. ferruginea (EU513804; KC015555), Pleuronichthys cornutus (JN204296; JN204309), P. flesus (EU492120; JQ775095), Reinhardtius hippoglossoides (EU513822; KC015875), Hippoglossus hippoglossus (HQ283277; KC015481), Hippoglossus stenolepis (EU513794; HQ712487), Hippoglossoides platessoides (EU513786; KC015475), Hippoglossoides elassodon (EU513782J; JQ354126), Hippoglossoides robustus (DQ464121; HM421780), Verasper moseri (AB326991; DQ242491), Microstomus pacificus (EU513812; JQ354230), Microstomus kitt (EU513810; EU513671), Glyptocephalus cynoglossus (EU513778; KC015419), Glyptocephalus stelleri (AB114910; JF952743), Lepidopsetta bilineata (EU513798; JQ354157), S. solea (JF969246; JQ774922), S. aegyptiaca (JN225430; EU513743) and S. senegalensis (EF439590; JQ775127).
Selective restriction sites were analysed through NEBcutterV2.0 software (http://tools.neb.com/NEBcutter/index.php3), allowing the identification of three enzymes for Cytb and one for COI which are able to distinguish these species, as detailed in the ‘Results and conclusions’ section.
Restriction
To test the restriction profile, PCR amplifications were performed as already reported, 10 μl of PCR product were digested with 1.5 μl of buffer, 10–15 units of enzyme and sdH20 to a final volume of 15 μl and incubating samples for 3 h at 37°C; restriction products were run in 2.5% EtBr-agarose gel and photographed.
Results and conclusions
The entire procedure was successfully carried out both on fresh and on frozen/breaded fillets and obtained sequences were registered in GenBank (COI:JN858067-JN977582, Cytb:JN858068-JN977583).
The number of mutations and the variation percentages identified by the alignments through MEGA 5.1, between P. platessa and all the other investigated species, ranged from 25, 4.48% (L. ferruginea), to 111, 19.90% (S. senegalensis) for COI and from 18, 5.50% (P. flesus), to 77, 23.55% (S. solea) for Cytb ().
Table 1. Number of mutations and percentages of genetic variation between Pleuronectes platessa and all the other investigated species.
Tabla 1. Número de mutaciones y porcentajes de variación genética entre Pleuronectes platessa y las restantes especies investigadas.
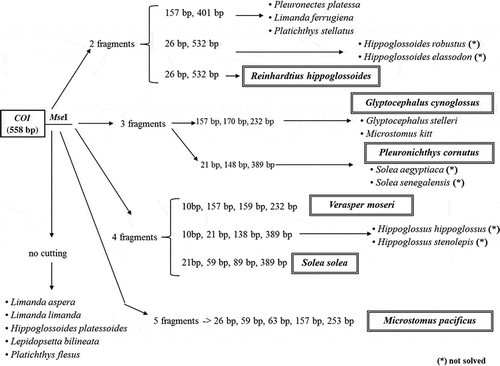
The in silico digestion of Cytb allowed the identification of three enzymes: NlaIII, BslI and MboI. Their sequential application permitted the unambiguous distinction of P. platessa from all other species (); furthermore, these enzymes allowed a unique species attribution to P. platessa, M. kitt, L. aspera, L. ferruginea, G. stelleri, P. cornutus, P. flesus and L. bilineata (). The digestion of COI with the enzyme MseI allowed to distinguish six species, using only COI or in conjunction with Cytb. In fact, R. hippoglossoides, V. moseri, S. solea and M. pacificus were clearly identified by the COI digestion with MseI (). Furthermore, G. cynoglossus and P. cornutus were correctly attributed by the combined use of COI and Cytb restriction profiles, because Cytb allowed the exclusion of M. kitt, S. aegyptiaca, S. senegalensis and G. stelleri ().
Figure 1. Cytb digestion workflow. Fragments produced and species identification through the digestion of the 327bp-Cytb fragment with NlaIII and BslI and, in some cases, MboI.
Figura 1. Flujo digestivo Cytb. Fragmentos producidos y especies identificadas a través de la digestión del fragmento 327bp-Cytb con NlaIII y BslI y, en algunos casos, con MboI.
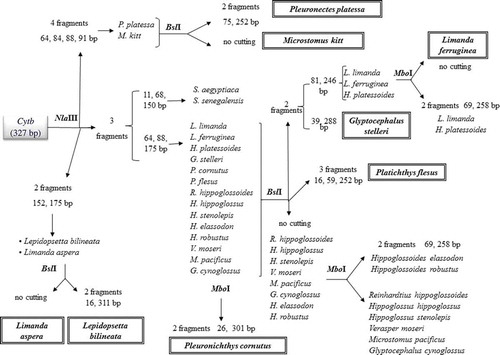
Figure 2. COI digestion workflow. Fragments produced and species identification through the digestion of the 558bp-COI fragment with MseI. *The species is not solved with both Cytb and COI digestion.
Figura 2. Flujo digestivo coi. Fragmentos producido y especias identificadas a través de la digestión del fragmento 558bp-coi con MseI. El asterisco (*) indica que la especie no es detectada con la digestión tanto Cytb como COI.
No mislabelled samples were found through the application of such methodology, so the fraud ratio was zero within this sample set.
Electrophoretic courses confirmed the in silico predicted patterns, allowing a rapid and easy identification of high commercial value species. Although the confusion in species attribution might be alimented also by the existence of hybrids of some of flatfish species, for example, P. flesus × P. platessa (Kijewska, Burzynski, & Wenne, Citation2009), this article did not focus on these aspects in view of the existence of papers specifically dedicated to this delicate biological aspect (Kijewska et al., Citation2009).
This PCR-RFLP combined methodology presents many advantages with respect to classical sequencing-based species attribution procedures already performed for these species. In fact, the method developed in the present study is easy to execute, it is less time-consuming, cheaper in comparison with classical barcoding procedures and can also be successfully employed for the identification of fish species on heat-processed products such as pre-fried fillets. At the same time, the possibility of digesting two genes using multiple restriction enzymes provides a multiple internal control of the species attribution. So it could be recommended as a valid and economical tool for the rapid identification of commercial plaice samples and the detection of mislabelling in routine analysis.
Acknowledgements
The authors are very grateful to Dr Gloria María González Fortes, University of York, UK, for Spanish translation and to Dr Anna Rita Vignati for English corrections. The authors are also grateful to the anonymous reviewers for their precious suggestions.
References
- Asensio, L. G. (2007). PCR-based methods for fish and fishery products authentication. Trends in Food Science & Technology, 18, 558–566. doi:10.1016/j.tifs.2007.04.016
- Espiñeira, M., González-Lavín, N., Vieites, J. M., & Santaclara, F. J. (2008). Development of a method for the genetic identification of flatfish species on the basis of mitochondrial DNA sequences. Journal of Agricultural and Food Chemistry, 56, 8954–8961. doi:10.1021/jf800570r
- Filonzi, L., Chiesa, S., Vaghi, M., & Nonnis Marzano, F. (2010). Molecular barcoding reveals mislabelling of commercial fish products in Italy. Food Research International, 43, 1383–1388. doi:10.1016/j.foodres.2010.04.016
- Gigliarelli, L., Caldelli, A., Morozzi, G., Giannetto, D., Panara, F., Lorenzoni, M., & Lucentini, L. (2013). Nuclear PCR-RFLP detects the brook chub, Squalius lucumonis (Leuciscinae: Cyprinidae), and related hybrids with other cyprinid species. Italian Journal of Zoology, 80, 462–465. doi:10.1080/11250003.2013.787462
- Ivanova, N., Zemlak, T. S., Hanner, R. H., & Hebert, P. D. N. (2007). Universal primer cocktails for fish DNA barcoding. Molecular Ecology Notes, 7, 544–548. doi:10.1111/j.1471-8286.2007.01748.x
- Kijewska, A., Burzynski, A., & Wenne, R. (2009). Molecular identification of European flounder (Platichthys flesus) and its hybrids with European plaice (Pleuronectes platessa). ICES Journal of Marine Science, 66, 902–906. doi:10.1093/icesjms/fsp110
- Kocher, T. D., Thomas, W. K., Meyer, A., Edwards, S. V., Paabo, S., Villablanca, F. X., & Wilson, A. C. (1989). Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proceedings of the National Academy of Sciences, 86, 6196–6200. doi:10.1073/pnas.86.16.6196
- Kochzius, M., Seidel, C., Antoniou, A., Botla, S. K., Campo, D., Cariani, A., … Blohm, D. (2010). Identifying fishes through DNA barcodes and microarrays. PLoS One, 5, e12620. doi:10.1371/journal.pone.0012620
- Koyama, H., Kakami, M., Kawamura, M., Tokuda, R., Kondo, Y., Tsuge, I., … Urisu, A. (2006). Grades of 43 fish species in Japan based on IgE-binding activity. Allergology International, 55, 311–316. doi:10.2332/allergolint.55.311
- Kuehn, A., Scheuermann, T., Hilger, C., & Hentges, F. (2010). Important variations in parvalbumin content in common fish species: A factor possibly contributing to variable allergenicity. International Archives of Allergy and Immunology, 153, 359–366. doi:10.1159/000316346
- Le Fresne, S., Popova, M., Le Vacon, F., & Carton, T. (2011). Application of Denaturing High-Performance Liquid Chromatography (DHPLC) for the identification of fish: A new way to determine the composition of processed food containing multiple species. Journal of Agricultural and Food Chemistry, 59, 12302–12308. doi:10.1021/jf2030242
- Lucentini, L., Caporali, S., Palomba, A., Lancioni, H., & Panara, F. (2006). A comparison of conservative DNA extraction methods from fins and scales of freshwater fish: A useful tool for conservation genetics. Conservation Genetics, 7, 1009–1012. doi:10.1007/s10592-006-9137-6
- Lucentini, L., Puletti, M. E., Ricciolini, C., Gigliarelli, L., Fontaneto, D., Lanfaloni, L., … Panara, F. (2011a). Molecular and phenotypic evidence of a ew species of genus Esox (Esocidae, Esociformes, Actinopterygii): The southern pike, Esox flaviae. PLoSOne, 6, e25218. doi:10.1371/journal.pone.0025218
- Lucentini, L., Rebora, M., Puletti, M. E., Gigliarelli, L., Fontaneto, D., Gaino, E., & Panara, F. (2011b). Geographical and seasonal evidence of cryptic diversity in the Baetis rhodani complex (Ephemeroptera, Baetidae) revealed by means of DNA taxonomy. Hydrobiologia, 673, 215–228. doi:10.1007/s10750-011-0778-1
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution, 28, 2731–2739.
- Teletchea, F. (2009). Molecular identification methods of fish species: Reassessment and possible applications. Reviews in Fish Biology and Fisheries, 19, 265–293. doi:10.1007/s11160-009-9107-4
- Tepedino, V., Berrini, A., Borromeo, V., Gaggioli, D., Cantoni, C., Manzoni, P., & Secchi, C. (2001). Identification of commercial fish species belonging to the orders pleuronectiformes and gadiformes: Library of isoelectric focusing patterns. Journal of AOAC International, 84, 1600.
- van Leeuwen, S. P. J., van Velzen, M. J. M., Swart, C. P., van der Veen, I., Traag, W. A., & de Boer, J. (2009). Halogenated contaminants in farmed salmon, trout, tilapia, pangasius, and shrimp. Environmental Science and Technology, 43, 4009–4015. doi:10.1021/es803558r
