ABSTRACT
DNA plays an important role in molecular authentication of dairy products, both heat treatment and storage may impair the detection limit of the DNA-based method. In this study, DNA was extracted from commercial ultra-high temperature treated (UHT) milkfrom the different storage stages for monitoring DNA yield, purity, integrity, as well as by the specific detection of UHT milk composition in goat milk. Results showed that DNA yield and purity remained stable during the whole storage stage (P > .05). In addition, all the extracted DNA from UHT milk was degraded into short DNA fragments with 100–2000 bp, but these fragments can be still used for the amplification of long target sequence (>1000 bp). Furthermore, it was found that PCR could readily detect UHT milk composition in goat milk mixture, and gave a good sensitivity threshold (0.1%) during the storage of UHT milk, but the detection efficiency was gradually decreased.
RESUMEN
El ADN desempeña una función importante en la autenticación molecular de los productos lácteos. Tanto el tratamiento térmico como el almacenamiento pueden alterar el límite de detección del método basado en el ADN. Para el presente estudio se extrajo ADN de leche comercial tratada a temperatura ultra alta (UHT) durante las diferentes etapas de almacenamiento, con el objetivo de controlar su rendimiento, su pureza y su integridad. Asimismo, se hizo una detección específica de la composición de la leche UHT en una mezcla de leche de cabra. Los resultados permiten constatar que el rendimiento y la pureza del ADN permanecen estables durante toda la etapa de almacenamiento (P > .05). Además, todo el ADN extraído de la leche UHT se degradó en fragmentos cortos con 100-2000 bp, los cuales pueden seguir utilizándose para amplificar la secuencia diana larga (>1000 bp). Al mismo tiempo, si bien se comprobó que la PCR puede detectar fácilmente la composición de la leche UHT en la mezcla de leche de cabra y da un buen umbral de sensibilidad (0.1%) durante el almacenamiento de la leche UHT, la eficacia de la detección fue disminuyendo gradualmente.
KEYWORDS:
PALABRAS CLAVE:
1. Introduction
Mislabels and ingredient substitutions of food are major issues in the food industry (Cawthorn et al., Citation2013; Everstine et al., Citation2013; Wong & Hanner, Citation2008). To ensure food quality, the DNA-based method has been a reliable, accurate, and speedy tool to correct mislabeling (Liao et al., Citation2017a, Citation2018a). However, the PCR-based method largely depends on the purity and intactness of DNA extracted from food (Spaniolas et al., Citation2008). As a complex matrix, food most often requires physical, chemical, and biochemical processing (Mano et al., Citation2017). It has been demonstrated that physical force, heat, pH, and enzymes can cause DNA fragmentation (Chen et al., Citation2005; Klein et al., Citation1998; Quirasco et al., Citation2004; Vijayakumar et al., Citation2009), which affects PCR analysis results (Chen et al., Citation2007; Yoshimura et al., Citation2005). Therefore, detecting DNA from highly processed food is of great significance for molecular traceability.
Milk is often pasteurized or ultra-high temperature sterilized to prolong its shelf-life. Pasteurized milk is only heated to a relatively low temperature of 72°C for a short period of time (15 s), thereby retaining more nutrients. Although the pasteurized milk is preferred by the public, it can only be preserved at 4°C for a short time, which limits its transportation and sale. By comparison, UHT sterilization involves the heating of milk to a high temperature (135°C–150°C) for just a few seconds (4 s) followed by aseptic packaging in order to obtain a product with a long shelf-life at room temperature (Valero et al., Citation2001). Until now, UHT milk has been one of the most consumed dairy products worldwide, especially in China, where 60% of milk consumption is UHT milk (Liem et al., Citation2016).
As a commonly used milk heat processing method, the UHT treatment can disturb some of the physicochemical and nutritional properties of milk. The previous study has shown that the DNA in milk can be affected by different heat treatments, such as pasteurization, boiling, and autoclaving (Liao et al., Citation2018b). So the UHT treatment may also result in DNA degradation and subsequently increase the difficulty of DNA detection. On the other hand, it was suggested that storage shows a significant influence on the DNA extracted from raw milk (Liao et al., Citation2018c), so there might be DNA degradation during the storage of UHT milk. Considering that monitoring the changes of DNA in UHT milk during storage not only contributes to optimizing DNA-based analysis methods but also provides a way to distinguish UHT milk composition in other dairy products. Therefore, this study aims to study the changes in DNA in UHT milk during the 6-month storage period. In addition, the influence of storage on the PCR detection sensitivity of UHT milk composition in goat milk was also investigated.
2. Materials and methods
2.1. Reagents and materials
Commercial UHT milk samples were provided by Yinqiao Dairy Co., Ltd. (Xi’an, Shaanxi province, China). Proteinase K, sodium dodecyl sulfonate, and Tris-base was purchased from Sigma–Aldrich. The molecular biology reagents were obtained from CWBIO Co., Ltd. (Beijing, China). All other chemical reagents used in the experiment were of analytical grade and purchased from Jingbo Chemical Reagent Co., Ltd. (Xi’an, Shaanxi province, China).
2.2. Extraction of DNA
A total of 42 UHT milk samples were prepared and stored at 25°C, and 6 UHT milk samples were used for DNA extraction at 1-month intervals during storage periods of 0 to 6 months. DNA extraction method was according to previous reported work with slight modifications (Liao & Liu, Citation2018). In detail, 10 mL of milk was centrifuged at 4150 × g for 10 min at room temperature. After removing the top fat layer and middle whey layer, the bottom sediment was suspended with 600 μL phosphate-buffered saline solution (pH 7.4); then, the mixture was centrifuged at 10625 × g for 10 min at room temperature. The obtained sediment was mixed with 350 μL DNA extraction buffer (pH 8.0, 100 mM Tris Cl, 100 mM NaCl, and 5 mM EDTA), 50 μL sodium dodecyl sulfate (200 mg/mL) and 10 µL proteinase K (20 mg/mL), and incubated at 56°C for 4 h. After incubation, the mixture was mixed with an equal volume of Tris-phenol before being centrifuged at 10625 × g for 10 min at room temperature. The obtained supernate was mixed with an equal volume of phenol: chloroform: isoamyl alcohol (volume ratio of 25:24:1) and then being centrifuged at 10625 × g for 10 min at room temperature. After that, the obtained supernate was mixed with an equal volume of chloroform: isoamyl alcohol (volume ratio of 24:1) and recentrifuged at 10625 × g for 10 min at room temperature. The obtained supernate was mixed with ice-cold absolute ethanol and then centrifuged at 10625 × g for 10 min at room temperature. After discarding the supernate, 25 μL of Tris-EDTA (pH = 8.0, 1 mM Tris-Cl and 0.5 mM EDTA) was added to dissolve DNA.
2.3. DNA yield and purity determination
The DNA yield was expressed as the mass (ng) of DNA per mL of milk and determined by a UV/VIS spectrophotometer (Nanodrop ND-1000, Thermo Fisher Scientific Inc., USA). The absorption ratio of A260 and A280 was used as DNA purity.
2.4. DNA integrity analysis
The integrity of DNA was evaluated by agarose gel electrophoresis. Specifically, 4 μL DNA solution extracted from UHT milk at different storage stages was mixed with 1 μL 6 × DNA loading buffer before analyzed in 1% (w/v) agarose gel electrophoresis at 100 V for 40 min. After electrophoresis, the gel was photographed under UV light imaging analyzer (BIO-BEST200E, SIM, USA). The DNA from raw milk and distilled water was used as a positive and negative control, respectively.
For further evaluating the integrity of the extracted DNA, the bovine B2 microglobulin (B2M) gene was randomly selected as representing a bovine-specific functional gene. The corresponding sequences for primer were from our previous work (Liu et al., Citation2014): 5ʹ-CAT CTG TCT TTC CCT GCC GC-3ʹ and 5ʹ-CTA CAG CCT TCC TCA TCT CCC CT-3ʹ (amplifying a 1019 bp sequence). The PCR amplification reaction was performed in a mixture (10 μL), which comprises 3.4 μL of TaqMan Universal PCR Master Mixture, 0.3 μL of each primer, 1 μL of the template DNA, and 5 μL of DNase-free water. The PCR protocol was 95°C for 10 min followed by 30 cycles of 94°C for 30 s, 63°C annealing for 30 s, and 72°C for 30 s, and a final extension at 72°C for 10 min, which was reacted in a universal PCR instrument (Aritik, Thermo Fisher Scientific Inc., USA). The amplification products were also examined by 1% (w/v) agarose gel electrophoresis.
2.5. PCR detection of UHT milk composition in goat milk
To evaluate the effect of storage on the detection sensitivity of UHT milk in other dairy products, different content of UHT milk was added into raw goat milk to make UHT milk proportions were 0.1%, 0.5%, 1%, 10%, and 30%, respectively. These mixtures were used for DNA extraction and subsequent PCR analysis. The goat milk samples were collected from a local dairy farm and their species were authenticated before the test. For PCR detection, cow-specific gene 12SBT-REV was selected for detecting UHT milk composition in goat milk. The sequences of the primer were adopted from López-Calleja et al. (Citation2005): 5ʹ-CTA GAG GAG CCT GTT CTA TAA TCG ATA A-3ʹ and 5ʹ-AAA TAG GGT TAG ATG CAC TGA ATC CAT-3ʹ. The PCR reaction system consisted of 3.4 μL of TaqMan Universal PCR Master Mix, 0.3 μL of assay-specific forward and reverse primers (0.3 μM), 1 μL of the template DNA, and 5 μL DNase-free water. The PCR protocol was 95°C for 10 min followed by 30 cycles of 94°C for 30 s, 60°C annealing for 30 s and 72°C for 30 s, and a final extension at 72°C for 10 min using a universal PCR System (Aritik, Thermo Fisher Scientific Inc., USA). The reaction products were analyzed by agarose gel electrophoresis according to the method as described in section 2.4. The DNA extracted from UHT milk and distilled water are used as a positive and negative control, respectively.
2.6. Statistical analysis
Data obtained in this study were subjected to statistical analysis using SPSS 21.0 (SPSS Inc., USA) and expressed as means ± standard deviation (SD). Six samples were prepared for each trial, and each measurement was repeated at least 3 times. The mean values were compared using Duncan’s multiple range tests and independent sample t-test. Results with a P-value <0.05 were considered significant.
3. Results and discussion
3.1. DNA yield and purity
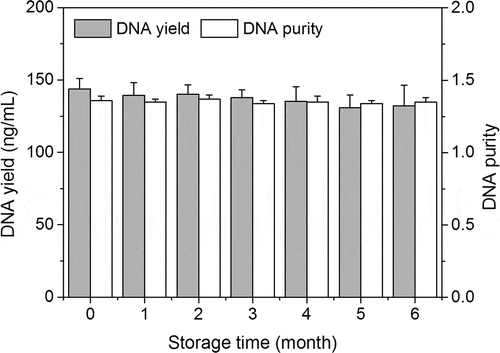
DNA yield and purity are crucial parameters for downstream applications such as PCR analysis. shows the changes in DNA yield and purity during UHT milk storage, DNA yield was in the range of 130.7–143.6 ng/mL during storage of UHT milk, and it was not affected by storage (P> .05). Interestingly, the DNA yield was found to be higher than previous-reported experiment with unheated milk samples (97 ng/mL) (Liao & Liu, Citation2018). The increased DNA yield that was observed in UHT milk probably attributes to the DNA degradation after UHT treatment. Several studies also suggested that the DNA will degrade when food products were subjected to heat treatment (Musto et al., Citation2014; Novak et al., Citation2007; Şakalar et al., Citation2012). On the other hand, the DNA purity was also not influenced by storage (P > .05), and it was ranged from 1.34 to 1.37, which was lower than pure DNA that generally possesses the purity of 1.8 (Sambrook et al., Citation1989). Although the DNA purity for all tested samples was ranged from 1.3 to 1.4 in the present work, it might be acceptable for PCR analysis according to the literature (Liao et al., Citation2017b).
3.2. DNA integrity
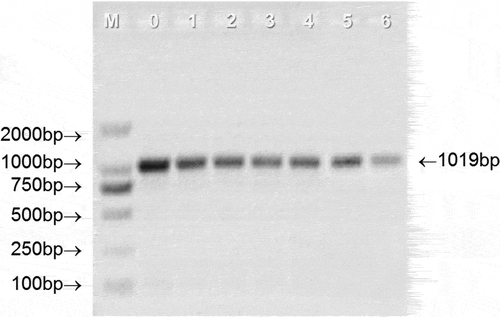
DNA integrity is also an important parameter for DNA quality evaluation, and it has been applied to evaluate the DNA quality of raw milk (Liao & Liu, Citation2018; Liu et al., Citation2014; Pokorska et al., Citation2016). DNA integrity could directly reflect the length of the extracted DNA fragment, a longer DNA fragment is helpful for PCR amplification as it contains more desired amplification regions than a short DNA fragments (Liao & Liu, Citation2018). shows the integrity of the DNA extracted from UHT milk during the whole storage period. Compared with the positive control, all the extracted DNA samples showed serious smearing and the fragment length is between 100 bp and 2000 bp, which is shorter than the positive control, indicating that UHT treatment severely degraded the DNA in the milk. This result is similar to the previous report, which suggested that the DNA extracted from raw milk will gradually degrade into small fragments when the raw milk was treated by autoclaving from 0 min to 15 min (Liao et al., Citation2018a). In addition, there were no obvious differences in DNA degradation at different storage stages of UHT milk. Based on this fact, a 1019 bp target fragment was randomly selected for PCR amplification to further elucidate the DNA integrity. As depicted in , all the tested samples gave the positive results at 1019 bp, suggesting the desired amplification regions of the extracted DNA still existed and could be recovered by PCR amplification during the whole storage of UHT milk. However, it is worth noting that the intensity of PCR product bands decreases with the storage time. Therefore, from the PCR results, it can be speculated that storage will affect the integrity of DNA in UHT milk.
Figure 2. Representative gel electrophoresis of total DNA extracted from UHT milk at different storage stage. M = DM 2000 marker; 0–6 represents the storage time (month) of UHT milk; NC = negative control; PC = positive control.
Figura 2. Electroforesis en gel representativa del ADN total extraído de la leche UHT en diferentes etapas de almacenamiento. M = marcador DM 2000; 0–6 representa el tiempo (mes) de almacenamiento de la leche UHT; NC = control negativo; PC = control positivo
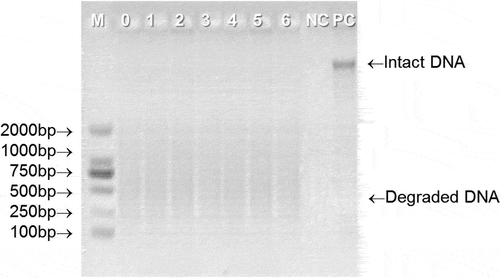
Figure 3. Representative PCR results from agarose gel electrophoresis analysis. 0–6 represents the storage time (month) of UHT milk.
Figura 3. Resultados representativos de la PCR a partir del análisis de electroforesis en gel de agarosa. 0–6 representa el tiempo (mes) de almacenamiento de la leche UHT
3.3. PCR detection of UHT milk composition in goat milk
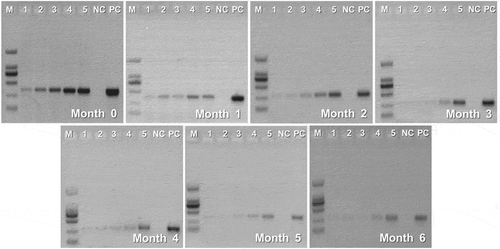
Conventional PCR is considered as one of the most important technique for adulteration detection and can be used in most laboratories as its low limit of detection (LOD), strong specificity, and lower cost (Liao et al., Citation2017b). Overall, many efforts have been made for species identification in dairy products by PCR techniques (I. Lopez-Calleja et al., Citation2004; I. M. Lopez-Calleja et al., Citation2007; Mafra et al., Citation2007). Those studies have focussed to validate the reliability and accuracy of the PCR-based method, while other factors that can influence the PCR results were usually neglected. In this study, a laboratory adulteration model was set up for evaluating the effect of storage time on the detection of UHT milk composition in goat milk by PCR. As shown in , the detected intensity of cow-specific gene bands at 346 bp showed an enhanced trend with an increasing proportion of adulteration for all tested samples, indicating favorable detection sensitivity. In addition, during 6-month storage, UHT milk composition in the goat milk mixture can be detected to 0.1% (wt/wt) (). However, we also found that the intensity of the target bands gradually decreased with the storage of UHT milk. Especially when the UHT milk was stored over 4 months, the brightness of the produced bands is too weak to be observed when the adulteration level is below 1%, suggesting that the storage time can interfere with the PCR detection efficiency of UHT milk composition.
Figure 4. Representative gel electrophoresis of 12SBT- REV PCR products (346bp) obtained from different-stored UHT milk in goat milk mixtures. Lanes 1, 2, 3, 4, and 5 = mixture of UHT milk and goat milk containing 0.1 %, 0.5 %, 1 %, 10 %, and 30 % UHT milk composition, respectively; NC = negative control; PC = positive control; M = molecular weight marker (2,000-bp DNA ladder).
Figura 4. Electroforesis en gel representativa de los productos de PCR 12SBT- REV (346bp) obtenidos a partir de diferentes leches UHT almacenadas en mezclas de leche de cabra. Columnas 1, 2, 3, 4 y 5 = mezcla de leche UHT y leche de cabra que contiene 0.1, 0.5, 1, 10 y 30% de composición de leche UHT, respectivamente; NC = control negativo; PC = control positivo; M = marcador de peso molecular (escalera de ADN de 2,000 bp)
4. Conclusions
In conclusion, sufficient DNA with acceptable purity can be extracted from UHT milk. However, the DNA integrity will gradually deteriorate with the storage of UHT milk, which was reflected by the less amplified long-chain PCR products with the storage of UHT milk. During 6-month storage, the detection of UHT milk composition in goat milk is still highly specific and reliable in the range from 0.1% to 30% content. However, the detection efficiency of the method is gradually falling down with the storage of UHT milk. To improve the method sensitivity, it needs a reliable estimation of the storage length to get the reasonable LOD. Therefore, our results provide the potential insights for food inspection services to trace the species origin of highly processed dairy products.
Disclosure statement
The authors declare that they have no conflict of interest.
Additional information
Funding
References
- Cawthorn, D. M., Steinman, H. A., & Hoffman, L. C. (2013). A high incidence of species substitution and mislabelling detected in meat products sold in South Africa. Food Control, 32(2), 440–449. https://doi.org/10.1016/j.foodcont.2013.01.008
- Chen, Y., Ge, Y., & Wang, Y. (2007). Effect of critical processing procedures on transgenic components in quality and quantity level during soymilk processing of roundup ready soybean. European Food Research and Technology, 225(1), 119–126. https://doi.org/10.1007/s00217-006-0389-7
- Chen, Y., Wang, Y., Ge, Y., & Xu, B. (2005). Degradation of endogenous and exogenous genes of Roundup-Ready soybean during food processing. Journal of Agricultural and Food Chemistry, 53(26), 10239–10243. https://doi.org/10.1021/jf0519820
- Everstine, K., Spink, J., & Kennedy, S. (2013). Economically motivated adulteration (EMA) of food: Common characteristics of EMA incidents. Journal of Food Protection, 76(4), 723–735. https://doi.org/10.4315/0362-028X.JFP-12-399
- Klein, J., Altenbuchner, J., & Mattes, R. (1998). Nucleic acid and protein elimination during the sugar manufacturing process of conventional and transgenic sugar beets. Journal of Biotechnology, 60(3), 145–153. https://doi.org/10.1016/S0168-1656(98)00006-6
- Liao, J., Gao, J., Ku, T., & Liu, Y. (2018c). Assessment of milk quality during storage based on DNA extracted from milk. CyTA-Journal of Food, 16(1), 786–792. https://doi.org/10.1080/19476337.2018.1474265
- Liao, J., & Liu, Y. (2018). Purification procedures meaningfully influence DNA quantification in milk. LWT, 94(8), 8–12. https://doi.org/10.1016/j.lwt.2018.04.031
- Liao, J., Liu, Y., & Ku, T. (2018b). Changes in physicochemical properties and DNA quality of milk as affected by different heat treatments. International Journal of Dairy Technology, 71(2), 333–339. https://doi.org/10.1111/1471-0307.12446
- Liao, J., Liu, Y. F., Ku, T., Liu, M. H., & Huang, Y. (2017b). Qualitative and quantitative identification of adulteration of milk powder using DNA extracted with a novel method. Journal of Dairy Science, 100(3), 1657–1663. https://doi.org/10.3168/jds.2016-11900
- Liao, J., Liu, Y. F., Yang, L., Li, F. P., & Sheppard, A. M. (2017a). Development of a rapid mitochondrial DNA extraction method for species identification in milk and milk products. Journal of Dairy Science, 100(9), 7035–7040. https://doi.org/10.3168/jds.2017-12653
- Liao, J., Yang, L., Sheppard, A. M., & Liu, Y. F. (2018a). Comparison of DNA quality in raw and reconstituted milk during sterilization. Journal of Dairy Science, 101(1), 147–153. https://doi.org/10.3168/jds.2017-13461
- Liem, D. G., Bolhuis, D. P., Hu, X., & Keast, R. S. J. (2016). Influence of labeling on Australian and Chinese consumers’ liking of milk with short (pasteurized) and long (UHT) shelf life. Journal of Dairy Science, 99(3), 1747–1754. https://doi.org/10.3168/jds.2015-10516
- Liu, Y. F., Gao, J. L., Yang, Y. F., Ku, T., & Zan, L. S. (2014). Novel extraction method of genomic DNA suitable for long-fragment amplification from small amounts of milk. Journal of Dairy Science, 97(11), 6804–6809. https://doi.org/10.3168/jds.2014-8066
- López-Calleja, I., Alonso, I. G., Fajardo, V., Rodríguez, M. A., Hernández, P. E., García, T., & Martín, R. (2005). PCR detection of cows’ milk in water buffalo milk and mozzarella cheese. International Dairy Journal, 15(11), 1122–1129. https://doi.org/10.1016/j.idairyj.2004.12.003
- Lopez-Calleja, I., Gonzalez, I., Fajardo, V., Rodriguez, M. A., Hernandez, P. E., Garcia, T., & Martin, R. (2004). Rapid detection of cows’ milk in sheeps’ and goats’ milk by a species-specific polymerase chain reaction technique. Journal of Dairy Science, 87(9), 2839–2845. https://doi.org/10.3168/jds.S0022-0302(04)73412-8
- Lopez-Calleja, I. M., Gonzalez, I., Fajardo, V., Hernandez, P. E., Garcia, T., & Martin, R. (2007). Application of an indirect ELISA and a PCR technique for detection of cows’ milk in sheep’s and goats’ milk cheeses. International Dairy Journal, 17(1), 87–93. https://doi.org/10.1016/j.idairyj.2006.01.006
- Mafra, I., Roxo, Á., Ferreira, I. M., & Oliveira, M. B. P. (2007). A duplex polymerase chain reaction for the quantitative detection of cows’ milk in goats’ milk cheese. International Dairy Journal, 17(9), 1132–1138. https://doi.org/10.1016/j.idairyj.2007.01.009
- Mano, J., Nishitsuji, Y., Kikuchi, Y., Fukudome, S. I., Hayashida, T., Kawakami, H., Kurimoto, Y., Noguchi, A., Kondo, K., Teshima, R., Takabatake, R., & Kitta, K. (2017). Quantification of DNA fragmentation in processed foods using real-time PCR. Food Chemistry, 226(9), 149–155. https://doi.org/10.1016/j.foodchem.2017.01.064
- Musto, M., Faraone, D., Cellini, F., & Musto, E. (2014). Changes of DNA quality and meat physicochemical properties in bovine supraspinatus muscle during microwave heating. Journal of the Science of Food and Agriculture, 94(4), 785–791. https://doi.org/10.1002/jsfa.6441
- Novak, J., Grausgruber-Gröger, S., & Lukas, B. (2007). DNA-based authentication of plant extracts. Food Research International, 40(3), 388–392. https://doi.org/10.1016/j.foodres.2006.10.015
- Pokorska, J., Kułaj, D., Dusza, M., Żychlińska-Buczek, J., & Makulska, J. (2016). New rapid method of DNA isolation from milk somatic cells. Animal Biotechnology, 27(2), 113–117. https://doi.org/10.1080/10495398.2015.1116446
- Quirasco, M., Schoel, B., Plasencia, J., Fagan, J., & Galvez, A. (2004). Suitability of real-time quantitative polymerase chain reaction and enzyme-linked immunosorbent assay for cry9C detection in Mexican corn tortillas: Fate of DNA and protein after alkaline cooking. Journal of AOAC International, 87(3), 639–646. https://doi.org/10.1093/jaoac/87.3.639
- Şakalar, E., Abasiyanik, M. F., Bektik, E., & Tayyrov, A. (2012). Effect of heat processing on DNA quantification of meat species. Journal of Food Science, 77(9), N40–N44. https://doi.org/10.1111/j.1750-3841.2012.02853.x
- Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular cloning: A laboratory manual (No. Ed. 2). Cold spring harbor laboratory press.
- Spaniolas, S., Bazakos, C., Ntourou, T., Bihmidine, S., Georgousakis, A., & Kalaitzis, P. (2008). Use of lambda DNA as a marker to assess DNA stability in olive oil during storage. European Food Research and Technology, 227(1), 175–179. https://doi.org/10.1007/s00217-007-0707-8
- Valero, E., Villamiel, M., Miralles, B., Sanz, J., & Martınez-Castro, I. (2001). Changes in flavour and volatile components during storage of whole and skimmed UHT milk. Food Chemistry, 72(1), 51–58. https://doi.org/10.1016/S0308-8146(00)00203-X
- Vijayakumar, K. R., Martin, A., Gowda, L. R., & Prakash, V. (2009). Detection of genetically modified soya and maize: Impact of heat processing. Food Chemistry, 117(3), 514–521. https://doi.org/10.1016/j.foodchem.2009.04.028
- Wong, E. H. K., & Hanner, R. H. (2008). DNA barcoding detects market substitution in North American seafood. Food Research International, 41(8), 828–837. https://doi.org/10.1016/j.foodres.2008.07.005
- Yoshimura, T., Kuribara, H., Kodama, T., Yamata, S., Futo, S., Watanabe, S., Aoki, N., Iizuka, T., Akiyama, H., Maitani, T., Naito, S., & Hino, A. (2005). Comparative studies of the quantification of genetically modified organisms in foods processed from maize and soy using trial producing. Journal of Agricultural and Food Chemistry, 53(6), 2060–2069. https://doi.org/10.1021/jf0483265