ABSTRACT
Since seafood can be a reservoir for diverse pathogenic bacteria, the risk to human health associated with seafood merits evaluation. The potentially pathogenic bacteria were isolated from seafood in Thailand. The 40 obtained strains were distinguished by ERIC-PCR fingerprints and identified to the species level by the VITEK 2 system. Their 16S rRNA gene had 97.74–99.79% nucleotide sequence identities to those of bacteria in 11 genera including Aeromonas, Citrobacter, Enterobacter, Escherichia, Hafnia, Klebsiella, Morganella, Salmonella, Proteus, Providencia, and Vibrio. Among the eight detected virulence genes, the tlh gene was expressed under all conditions, which, contrary to the cnf2, LTI, ssaO, and vt2e genes. Nutrient enrichment had a positive effect on the expression of kfu and uge genes, as well as a negative effect on the expression of the csgD gene. NaCl conferred a positive effect on the expression of the uge gene but negatively affected the expression of the csgD gene.
Debido a que los mariscos y los pescados pueden operar como reservorio de diversas bacterias patógenas, es necesario evaluar el riesgo asociado que los alimentos de origen marino conllevan para la salud humana. Con este objetivo, en el presente estudio se aislaron bacterias potencialmente patógenas de los alimentos de origen marino disponibles en Tailandia. Las 40 cepas obtenidas se distinguieron por huellas reconocidas mediante ERIC-PCR y se identificaron a nivel de especie por medio del sistema VITEK 2. Su gen 16S de ARNr tiene una identidad de secuencia de nucleótidos de 97.74 a 99.79% en comparación con las de las bacterias de 11 géneros, entre ellos Aeromonas, Citrobacter, Enterobacter, Escherichia, Hafnia, Klebsiella, Morganella, Salmonella, Proteus, Providencia y Vibrio. A diferencia de los genes cnf2, LTI, ssaO y vt2e, entre los ocho genes de virulencia detectados el gen tlh se expresó en todas las condiciones. El enriquecimiento de nutrientes provocó un efecto positivo en la expresión de los genes kfu y uge, y un efecto negativo en la expresión del gen csgD. El NaCl confirió un efecto positivo a la expresión del gen uge, pero afectó negativamente la expresión del gen csgD.
PALABRAS CLAVE:
1. Introduction
Seafood has numerous positive effects on human health and regular consumption of seafood is recommended by dietary guidelines (Farmery et al., Citation2018). However, the main problem associated with seafood consumption is the contamination by chemicals, heavy metals, toxins, and pathogenic microorganisms. Seafood has been recognized as an important source of food poisoning epidemics worldwide. Microbial contamination in seafood can occur in every step from exposures to marine water and weeds, harvesting and processing, packaging, storing, transporting, and cooking. Examples of important seafood-associated pathogenic bacteria are Clostridium, Enterococcus, Escherichia, Salmonella, Shigella, and Vibrio (Iwamoto et al., Citation2010). These bacteria have been reported as causative agents of foodborne outbreaks and therefore are indicators of microbial contamination in food and water. Within an individual genus, only some species and strains are pathogenic (Duvallet et al., Citation2017; Iwamoto et al., Citation2010). Each species has a broad variety of different degrees of virulence, ranging from avirulence to lethality. The ability of bacteria to cause diseases is controlled by several virulence genes which may be located on transmissible genetic elements including transposons, plasmids, and specific regions of the bacterial chromosomes, termed pathogenicity islands (PAI) (Hacker et al., Citation1997). Virulence genes play roles in controlling related mechanisms such as synthesis and release of virulence factors, adherence factors, survival factors, putative regulators, proteins, enzymes, toxins, iron-binding compounds, and biofilm (Bahador et al., Citation2019; Liu et al., Citation2019; Mannion et al., Citation2018; Zughaier & Cornelis, Citation2018); formation of lesions in intestinal epithelial cells (Capeda-Molero et al., Citation2017); suppression of host immune systems; infection in urinary tracts and bloodstream (Daga et al., Citation2019); adaptation to acidic and anaerobic conditions in intestines; adaptation for survival in the presence of intestinal microbiota (Jubelin et al., Citation2018); host-bacterial interaction (Niu et al., Citation2013); and determination of host specificity (Pan et al., Citation2014). These mechanisms are essential for inhabitation, survival, adaptation, and multiplication in hosts. Virulence genes are consequently important for bacterial infection and pathogenesis. The expression of virulence genes has been found to be affected by several factors such as nutrient starvation (Chandra et al., Citation2017; Paytubi et al., Citation2017); concentrations of NaCl (Larsen & Jespersen, Citation2015), oxygen (Melson & Kendall, Citation2019), and glucose (Valdes et al., Citation2018); temperature (Guijarro et al., Citation2015); growth phase (Koohsari et al., Citation2017); kind of carbon source (Ferrando et al., Citation2014; Kentache et al., Citation2016); pH (Do et al., Citation2019); sodium glycocholate (Joffre et al., Citation2019); and L-glutamine (Haber et al., Citation2017).
The aims of this work was to investigate the presence of virulence genes in seafood-associated bacteria and to determine the environmental conditions that stimulate the expression of virulence genes.
2. Materials and methods
2.1. Isolation of target bacteria from seafood
Thirty-five samples of fresh seafood, which belonged to 15 species listed in , were collected from three central seafood markets in Thailand, including the Talaythai market in Samutsakhon province, Bankhaotakiab market in Prachuapkhirikhun province, and Banphe market in Rayong province, in August 2014. All samples were placed in sterile zip-lock plastic bags, kept in ice buckets prior to isolation of target bacteria belonging to genera Escherichia, Salmonella, Shigella, and Vibrio. The isolation was performed within 6 h after sample collection (Da Silva et al., Citation2012). Twenty-five grams of flesh samples were aseptically excised, transferred to stomacher bags containing 225 ml of peptone water, and then homogenized at 200 rpm for 2 min by using a Seward stomacher 400 circulator (Seward Limited, West Sussex, UK). Homogenates were serially diluted in peptone water to make dilutions of 10−1, 10−2, and 10−3. These dilutions were pour plated on three selective media including eosin methylene blue (EMB) agar (HiMedia Laboratories, Mumbai, India), Salmonella-Shigella (SS) agar (Pronadisa, Madrid, Spain), and thiosulfate citrate bile salts sucrose (TCBS) agar (HiMedia Laboratories, Mumbai, India). Plates were incubated at 35°C for 2 days. Presumptive Escherichia coli formed metallic sheen colonies with diameters of 2–3 mm on EMB agar. Presumptive Salmonella formed colorless, transparent colonies with or without black centers on SS agar. Presumptive Shigella formed colorless, transparent colonies on SS agar. Presumptive Vibrio isolates were considered those displaying yellow, green, or green-blue colonies on TCBS agar. The numbers of presumptive target bacteria in each seafood sample were quantified as a log colony-forming unit (CFU)/g. Pure cultures of selected isolates were maintained on Luria-Bertani (LB) slants at 4°C and in 20% glycerol at −80°C.
Table 1. Targeted virulence genes and related information.
Tabla 1. Genes de virulencia seleccionados e información relacionada
Table 2. Average numbers of presumptive target bacteria per sample of the same seafood species sold in Thailand.
Tabla 2. Número promedio de bacterias presuntamente seleccionadas por muestra de la misma especie de marisco o pescado vendida en Tailandia
2.2. Enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) fingerprinting
ERIC-PCR was performed to analyze genotypic diversity and relatedness among bacterial isolates as well as to distinguish individual strains. Genomic DNA of each isolate was extracted from an exponentially grown culture using a GF-1 bacterial DNA extraction kit (Vivantis Technologies Sdn. Bhd., Selangor Darul Ehsan, Malaysia) and used as a template DNA for PCR using a pair of primers ERIC2 (5ʹ AAG TAA GTG ACT GGG GTG AGC G 3ʹ) and ERIC1R (5ʹ ATG TAA GCT CCT GGG GAT TAC C 3ʹ) as described previously (Ogutcu et al., Citation2009). DNA was amplified in a 25-μl reaction volume containing 2 μl of template DNA, 1.25 mM dNTP, 2.5 μl of 10X buffer, 1.5 mM MgCl2, 2 mM of each primer, and 2.5 units of Taq DNA polymerase. Negative controls (no DNA added) were included in all sets of reactions. PCR cycles consisted of the first denaturation at 95°C for 7 min, and then 30 cycles of denaturation at 95°C for 1 min, annealing at 52°C for 1 min, and extension at 65°C for 8 min, followed by a final extension at 65°C for 16 min. The presence and sizes of the amplified fragments were determined by agarose [1% in Tris-borate-EDTA (TBE) buffer] gel electrophoresis. Gels containing SafeView FireRed gel casting dye (Applied Biological Materials Inc., Richmond, Canada) were visualized with a Molecular Imager® Gel DocTM XR+ system (Bio-Rad Laboratories, Hercules, CA, USA). The unweighted pair groups using mathematical averages (UPGMA) dendrogram was constructed using the Phoretix ID Pro. software (Total Lab Ltd., Newcastle upon Tyne, UK). The strains generating individual ERIC-PCR fingerprints were selected for further steps of the study.
2.3. Sequence analysis of partial 16S rRNA gene
Genomic DNA of each strain was extracted from an exponentially grown culture using a GF-1 bacterial DNA extraction kit (Vivantis Technologies Sdn. Bhd., Selangor Darul Ehsan, Malaysia) and used as a template DNA for PCR using a pair of universal primers UN16S 926 f (5ʹ AAA CTY AAA KGA ATT GAC GG 3ʹ) and UN16S 1392 r (5ʹ ACG GGC GGT GTG TRC 3ʹ) (Lane, Citation1991) as described previously (Pongsilp et al., Citation2002). Partial 16S rRNA gene was amplified in a 25-μl reaction volume containing 2 μl of template DNA, 0.2 mM dNTP, 2.5 μl of 10X buffer, 1.5 mM MgCl2, 2 mM of each primer, and 1.25 units of Taq DNA polymerase. Negative controls (no DNA added) were included in all sets of reactions. PCR cycles consisted of the first denaturation at 95°C for 5 min, and then 34 cycles of denaturation at 95°C for 30 sec, annealing at 62°C for 30 sec, and extension at 72°C for 1 min, followed by a final extension at 72°C for 5 min. The presence and sizes of the amplified fragments were determined as described above. The PCR products were purified using a QIAquick gel extraction kit (Qiagen, Valencia, CA, USA) and subsequently sequenced by Bio Basic, Inc. (Markham, Ontario, Canada). The nucleotide sequences were aligned with the reference 16S rRNA gene sequences using the nucleotide BLAST (BLASTn) program of the National Center for Biotechnology Information (NCBI) (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify the closest genera.
2.4. Species identification by the VITEK 2 system
Strains were identified to the species level by the GN card of the VITEK 2 system version 07.01 (bioMerieux, Inc., Durham, NC, USA).
2.5. Detection and sequence analysis of virulence genes
Genomic DNA of each strain was extracted as described above and used as a template DNA for PCR to detect the presence of 22 virulence genes listed in . The rationale for selecting these virulence genes was based on their prevalence in the identified bacterial genera and their potential impact on human and animal health. The presence and sizes of the amplified fragments were determined as described above. The PCR products were purified using a QIAquick gel extraction kit (Qiagen, Valencia, CA, USA) and subsequently sequenced by Bio Basic, Inc. (Markham, Ontario, Canada). The nucleotide sequences were aligned with reference sequences using the BLASTn program of NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.6. Factors affecting the expression of virulence genes
Representative strains harboring virulence genes were selected to monitor the expression of virulence genes by a reverse-transcription PCR (RT-PCR) assay. The factors that we examined were as follows: 1) concentrations of NaCl (0 M, 0.17 M, and 0.3 M); 2) nutrient enrichment [LB medium and minimal M63 medium (Skyberg et al., Citation2007)]; 3) aeration conditions (microaerophilic and aerobic conditions); 4) temperatures (25°C and 37°C); and 5) growth phases [early exponential phase (6 h of growth), mid-exponential phase (12 h of growth)], and stationary phase (16 h of growth). Overnight cultures were subcultured by inoculating 1/100 volumes into LB broth without NaCl. Subcultures were grown at 37°C without aeration for 12–16 h and then inoculated into either LB broth or M63 broth. Cultures were grown under the tested conditions. The 10 conditions investigated were as follows: 1) LB broth without NaCl under the microaerophilic condition at 37°C for 12 h; 2) LB broth with 0.17 M NaCl under the microaerophilic condition at 37°C for 12 h; 3) LB broth with 0.3 M NaCl under the microaerophilic condition at 37°C for 12 h; 4) M63 broth without NaCl under the microaerophilic condition at 37°C for 12 h; 5) M63 broth with 0.17 M NaCl under the microaerophilic condition at 37°C for 12 h; 6) M63 broth with 0.3 M NaCl under the microaerophilic condition at 37°C for 12 h; 7) LB broth with 0.17 M NaCl under the aerobic condition at 37°C for 12 h; 8) LB broth with 0.17 M NaCl under the microaerophilic condition at 25°C for 12 h; 9) LB broth with 0.17 M NaCl under the microaerophilic condition at 37°C for 6 h; and 10) LB broth with 0.17 M NaCl under the microaerophilic condition at 37°C for 16 h. After cultivation, total RNA was extracted from cell pellets using a ZR Fungal/Bacterial RNA Miniprep. kit (Zymo Research Corporation, Irvine, CA, USA) and immediately used as a template DNA for RT-PCR. RT-PCR was performed using a SuperScriptTM III One-Step RT-PCR System with PlatinumTM Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) and pairs of primers specific to each virulence gene. The PCR mixtures were prepared according to the manufacturer’s instructions. Negative controls (DNase, RNase free ultrapure distilled water) and positive controls (PCR products of each gene) were included in all sets of reactions. The presence and sizes of the synthesized cDNA were determined by agarose (1% in TBE buffer) gel electrophoresis as described above.
2.7. Statistical analysis
Experiments were conducted in triplicate. Means and standard deviations were calculated by the Excel program version 16.0.
3. Results and discussion
3.1. Prevalence of presumptive target bacteria
Among 35 seafood samples, the numbers of presumptive E. coli, Salmonella-Shigella, and Vibrio were above the detection limit (1.2 log CFU/g) in seven (20.0%), 13 (37.1%), and 10 (28.6%) samples, respectively. The numbers of presumptive E. coli, Salmonella-Shigella, and Vibrio per sample that were above the detection limit ranged from 1.5 ± 1.1 to 1.9 ± 0.9, from 2.3 ± 1.3 to 2.4 ± 1.3, and from 1.2 ± 0.6 to 2.2 ± 1.5 log CFU/g, respectively. The average numbers of presumptive target bacteria per sample of the same seafood species sold in three central seafood markets in Thailand are shown in . As reported in our previous publication, 89% (31/35) of these seafood samples contained presumptive enterobacteria, as estimated by total plate counts using violet red bile glucose (VRBG) agar. The average numbers of presumptive enterobacteria per sample ranged between 1.3 ± 0.2 and 5.4 ± 0.1 log CFU/g (Pongsilp & Nimnoi, Citation2018). Forty-two presumptive isolates from three media were selected from different seafood species and collection sites. The isolates were purified and stored. The isolates from EMB agar, SS agar, and TCBS agar were abbreviated as SFEMB, SFSS, and SFTCBS, respectively.
The numbers of these potentially pathogenic bacteria are considered as an important issue for food safety. Among pathogenic bacterial species, the minimum infective doses (MID), which cause diseases, vary considerably. The MIDs are relatively high for different serovars of Salmonella (>105 cells), E. coli (>105 cells), and Vibrio cholerae (103 –108 cells). By contrast, the MIDs are apparent low for enterohemorrhagic E. coli (EHEC) (about 10 cells) and some Shigella (less than 10 cells) (Kothary & Babu, Citation2001; Schmid-Hempel & Frank, Citation2007).
3.2. ERIC-PCR fingerprinting of presumptive isolates of target bacteria
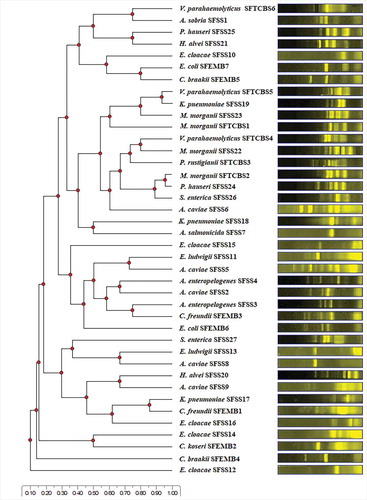
The 42 presumptive isolates of target bacteria generated 40 distinct ERIC-PCR patterns ranging in number from 1–7 and in size from approximately less than 500–4,000 base pairs (bp). This result suggests that 40 individual strains were obtained. The UPGMA dendrogram constructed from ERIC-PCR patterns of 40 individual strains is presented in .
3.3. Identification of the closest genera based on partial 16S rRNA gene sequences
Partial 16S rRNA gene sequences (approximately 500 bp) of 40 presumptive strains were employed for the identification of their closest genera. Alignments of the obtained sequences (GenBank accession numbers MT422214-MT422253) revealed 97.74–99.79% identities to those of potentially pathogenic bacteria in 11 genera including Aeromonas (nine strains), Enterobacter (seven strains), Citrobacter (five strains), Morganella (four strains), Klebsiella (three strains), Vibrio (three strains), Escherichia (two strains), Hafnia (two strains), Proteus (two strains), Salmonella (two strains), and Providencia (one strain). The sequences of the gene encoding small-subunit ribosomal RNA (16S rRNA) provide valuable data and the sequence comparisons have been generally employed as a powerful tool for the identification of bacteria to the genus level at a sequence identity threshold of 95% (Pongsilp, Citation2012; Schloss & Handelsman, Citation2005). The obtainment of potentially pathogenic bacteria in 11 genera suggests that EMB agar, SS agar, and TCBS agar were not sufficiently specific to isolate only E. coli, Salmonella-Shigella, and Vibrio, respectively. Both Citrobacter and Escherichia exhibited similar colonial characteristics on EMB agar. Seven genera, including Aeromonas, Enterobacter, Hafnia, Klebsiella, Morganella, Proteus, and Salmonella, that formed colonies with similar morphologies, were isolated from SS agar. Providencia and Vibrio grew with similar characteristics on TCBS agar.
3.4. Species identification based on the VITEK 2 system
The VITEK 2 system was employed for species identification of 40 strains. The VITEK results are consistent with the 16S rRNA gene sequencing results. The VITEK results revealed that the strains had 91–99% probabilities of being members of 17 species including Aeromonas caviae (five strains), Aeromonas enteropelogenes (two strains), Aeromonas salmonicida (one strain), Aeromonas sobria (one strain), Citrobacter braakii (two strains), Citrobacter freundii (two strains), Citrobacter koseri (one strain), Enterobacter cloacae (five strains), Enterobacter ludwigii (two strains), E. coli (two strains), Hafnia alvei (two strains), Klebsiella pneumoniae (three strains), Morganella morganii (four strains), Proteus hauseri (two strains), Providencia rustigianii (one strain), Salmonella enterica (two strains), and Vibrio parahaemolyticus (three strains).
The 11 bacterial genera identified in this study have been reported as severe pathogens. Their occurrence in seafood indicates poor sanitary quality and therefore represents a risk to public health. Acceptable limits of some of these bacteria have been regulated by international and national health organizations. For example, the Ministry of Public Health, Thailand, has announced microbiological standards for seafood. Salmonella spp., V. cholerae, and V. parahaemolyticus shall not be detected in 25 g while E. coli must not exceed 3 CFU/g. Previous studies have reported the contamination of seafood by some of these bacteria. The highest prevalence of Aeromonas hydrophila was observed in fish (19.5%), followed by shrimp (9.2%), lobster (9.3%), and crab (6.7%) caught off the south coast of Iran, in which 57 strains out of 62 (91.9%) harbored the cytolytic enterotoxin gene (Rahimi et al., Citation2014). The prevalence of Citrobacter (45.56%), Proteus (31.79%), E. coli (7.64%), Shigella (7.64%), Salmonella (3.35%), Klebsiella (2.44%), and Enterobacter (1.52%) in horse mackerel (Trachurus trachurus) sold in Istanbul, Turkey was annually recorded (Tosun et al., Citation2016). All 20 seafood samples from landing centers and retail markets in Mumbai, India were contaminated with fecal coliforms ranging in number from 150 to 1.1 × 103 CFU/100 g. Among 329 isolates of E. coli, 175 isolates (53.19%) harbored virulence genes such as Shiga toxin 1 (stx1), Shiga toxin 2 (stx2), intimin (eaeA), and hemolysin (hlyA) (Prabhakar et al., Citation2017). The prevalence of Shiga toxin-producing E. coli (STEC) in seafood from the Americas region, the European region, and the Western Pacific region was 0.42%, 1.70%, and 0.00%, respectively [WHO & FAO, Citation2018]. The prevalence of Vibrio in 160 retail seafood samples in Berlin, Germany was 55% (Vu et al., Citation2018). Vibrio was characterized as one of the top 10 most abundant genera among 4,953 operational taxonomic units (OTUs) derived from seawater in nine sampling sites in the upper gulf of Thailand (Nimnoi & Pongsilp, Citation2020).
3.5. Presence of virulence genes among strains
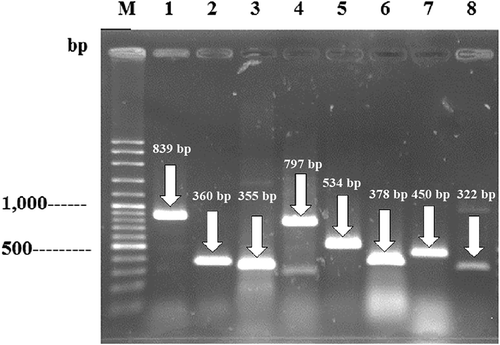
Detection of the 22 virulence genes by PCR revealed the presence of eight genes including cnf2, csgD, kfu, LTI, ssaO, tlh, uge, and vt2e. The numbers of positive strain in each genus or species that possessed at least one virulence gene are presented in . The PCR products of the detected virulence genes are shown in . Sequencing of PCR-amplified fragments from a representative strain of each species (a total of 11 strains) reinforced the presence of these genes based on 82.82–100% identities to the reference sequences. The 17 sequences of virulence genes were deposited in NCBI under GenBank accession numbers MT483947-MT483963. presents the detected virulence genes and their % identities to the reference sequences. Bacteria that have been previously reported to harbor these virulence genes are listed in . In this study, 24 strains out of 40 (60.0%) possessed at least one virulence gene. The cnf2 and vt2e genes were present in both strains of E. coli. The csgD gene was distributed among six species of three genera including Citrobacter (all five strains of C. braakii, C. freundii, and C. koseri), Enterobacter (all seven strains of E. cloacae and E. ludwigii), and K. pneumoniae (two strains out of three). Three species including C. braakii (one strain out of two), E. coli (both strains), and H. alvei (both strains) harbored the LTI gene. E. cloacae (three strains out of five), E. ludwigii (one strain out of two), and K. pneumoniae (one strain out of three) harbored the uge gene. While the kfu, ssaO, and tlh genes were confined to K. pneumoniae (one strain out of three), S. enterica (both strains), and V. parahaemolyticus (all three strains), respectively. The cooccurrence of three virulence genes, including the cnf2, LTI, and vt2e, was found in both strains of E. coli. The existence of virulence genes provides a proper mean of risk assessment of potentially pathogenic bacteria.
Table 3. The number of strain in each genus or species that possessed virulence gene.
Tabla 3. Número de cepas de cada género o especie que poseen el gen de virulencia
Table 4. Detected virulence genes and % identities to the reference sequences.
Tabla 4. Genes de virulencia detectados y porcentaje de identidades respecto a las secuencias de referencia
Figure 2. The PCR products of the detected virulence genes.
Figura 2. Productos de la PCR de los genes de virulencia detectados
Previous studies have reported the impact of these detected virulence genes on disease progression and their presence in potentially pathogenic bacteria associated with seafood. The cnf2 gene was associated with severe dysenteric syndromes caused by necrotoxic E. coli (NTEC). The csgD gene has been reported to have a great impact on the natural lifestyle of Salmonella such as the rdar morphotype that is correlated with invasion of the intestinal epithelial cells (Mackenzie et al., Citation2019). The csgD gene was detected in all 14 isolates of seafood-associated S. enterica serovar Weltevreden (Bhowmick et al., Citation2011a). The kfu and uge genes have been reported as ones of the important virulence genes in invasive K. pneumoniae strains which cause mastitis (Osman et al., Citation2014). The LTI gene has been reported as a signature gene responsible for the virulence of enterotoxigenic E. coli (ETEC) which causes diarrhea (Tomar et al., Citation2015). The vt2e gene was associated with edema disease caused by E. coli (Mallorquí et al., Citation2018). The ssaO gene was involved in systematic virulence of Salmonella in a mouse typhoid fever model. The binding of EIIAGlc protein to type three secretion system 2 (TTSS-2), which also included SSaO protein, switched Salmonella from growth arrest to acute virulence through activation of virulence factor secretion (Maze et al., Citation2014). The ssaO gene was detected in all 57 seafood-associated serovars of Salmonella (Bhowmick et al., Citation2011b). The tlh gene encodes thermolabile hemolysin (TLH) which causes the lysis of red blood cells and therefore contributes to pathogenicity (Praja & Safnurbati, Citation2018). Although the tlh gene is considered species-specific for V. parahaemolyticus, it is also widespread in other vibrios including Vibrio alginolyticus, Vibrio anguillarum, Vibrio diabolicus, Vibrio fischeri, Vibrio harveyi, Vibrio mimicus, Vibrio natriegens, Vibrio proteolyticus, and Vibrio vulnificus (Wang et al., Citation2007; Xie et al., Citation2005; Yanez et al., Citation2015). The tlh gene was employed for molecular confirmation of 22 V. parahaemolyticus isolates from seafood in the south-west coast of India (Meparambu Prabhakaran et al., Citation2020) and 104 V. parahaemolyticus isolates from seafood in Polish market (Lopatek et al., Citation2018). The association of virulence genes with outbreaks caused by V. parahaemolyticus has been proposed. The tdh and trh genes are considered to be connected to the ability to cause illness. The considerable difference in genetic marker distribution between clinical and environmental strains of V. parahaemolyticus in Thailand during 1998–1999 has been reported. Either or both of the tdh and trh genes were found in 92.78% of the clinical strains, whereas they were found in only 0.68% of the environmental strains (FAO & WHO, Citation2020).
3.6. Factors influencing the expression of virulence genes
To determine the influence of factors on the expression of virulence genes, six representative strains of different genera that harbored most of the virulence genes were selected. The representative strains were as follows: 1) C. braakii SFEMB5 that harbored the csgD and LTI genes; 2) E. ludwigii SFSS13 that harbored the csgD and uge genes; 3) E. coli SFEMB6 that harbored the cnf2, LTI, and vt2e genes; 4) K. pneumoniae SFSS17 that harbored the csgD and kfu genes; 5) S. enterica SFSS26 that harbored the ssaO gene; and 6) V. parahaemolyticus SFTCBS4 that harbored the tlh gene. The cDNA PCR products synthesized from total RNA from cells grown under varied conditions would represent gene expression. presents the expression of virulence genes under the desired conditions. Among the 10 conditions tested, the tlh gene was expressed under all conditions, which, contrary to the cnf2, LTI, ssaO, and vt2e genes. The remaining three genes (csgD, kfu, and uge) were expressed under some conditions. The csgD-inducing condition was growth in M63 medium without NaCl, suggesting that nutrient and NaCl deprivation had a positive effect on the expression of the csgD gene. The kfu gene was expressed under six conditions in which the strains were grown in LB medium but was not expressed under all three conditions in which the strains were grown in M63 medium. Additionally, the expression of the kfu gene occurred when cells were in mid-exponential and stationary growth phases. Therefore, the kfu-inducing condition was growth during mid-exponential and stationary phases in the enriched medium. While concentrations of NaCl (0 M, 0.17 M, and 0.3 M), aeration conditions (microaerophilic and aerobic conditions), temperatures (25°C and 37°C) had no effect on the expression of the kfu gene. The uge gene was expressed under two conditions. The factors influencing its expression included NaCl, nutrient enrichment, aeration condition, temperature, and growth phase. The uge-inducing condition was growth during the mid-exponential phase in enriched medium containing 0.17 M or 0.3 M NaCl under the microaerophilic condition at 37°C.
Table 5. Expression of virulence genes under the desired conditions.
Tabla 5. Expresión de los genes de virulencia en las condiciones deseadas
Previous studies have reported the factors stimulating the expression of virulence genes. Low temperature (9°C) had both negative and positive effects on the expression of the tlh gene in different strains of V. parahaemolyticus, indicating the expression was irregular depending on variations in environmental conditions (Zhao et al., Citation2015). The csgD gene was expressed under different environmental conditions such as growth at low temperature (<32°C), low osmolarity, and starvation (Gerstel & Romling, Citation2001). The expression of the csgD gene was low in the exponential growth phase but increased in the stationary growth phase (Ogasawara et al., Citation2010). The osmotic agents (NaCl and sucrose) had a negative effect on the expression of the csgD gene (Jubelin et al., Citation2005; Prigent-Combaret et al., Citation2001; Zhou et al., Citation2013). Our finding supports the previous reports that nutrient and NaCl deprivation stimulate the expression of the csgD gene. In this study, the expression of the csgD gene could occur during growth at 37°C when cells were grown under nutrient and NaCl deprivation conditions. The kfu-inducing and uge-inducing condition of pathogenic bacteria has not been previously described. In addition, it has been reported that environmental conditions trigger virulence gene expression and therefore contribute to the development of the disease. For example, an increase to 37°C and a depletion of extracellular iron are universal invasion signals to the bacteria and enable fine-tuning of virulence factor expression, resulting in the promotion of survival and proliferation of bacteria within their hosts (Lam et al., Citation2014).
4. Conclusion
This study underlines a potential risk of seafood-associated bacteria. Seafood was found to harbor diverse groups of potentially pathogenic bacteria, of which a large proportion belonged to enterobacteria. Among eight detected virulence genes, the csgD gene was mostly prevalent. The expression of four virulence genes occurred under the possible environmental conditions, especially the tlh gene of V. parahaemolyticus, which was expressed under broad conditions. This may raise more concerns about public health, Nevertheless, V. parahaemolyticus strains found in this study still remained a characteristic of environmental strains as they lacked the tdh and trh genes which are virulence marker genes for clinical strains.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Aher, T., Roy, A., & Kumar, P. (2012). Molecular detection of virulence genes associated with pathogenicity of Klebsiella spp. isolated from the respiratory tract of apparently healthy as well as sick goats. Israel Journal of Veterinary Medicine, 67(4), 249–252. http://www.ijvm.org.il/sites/default/files/klebsiella_spp..pdf
- Akbari, M., Bakhshi, B., Peerayeh, S. N., & Behmanesh, M. (2015). Detection of curli biogenesis genes among Enterobacter cloacae isolated from blood cultures. International Journal of Enteric Pathogens, 3(4), e28413. https://doi.org/10.17795/ijep28413
- Bahador, N., Shoja, S., Faridi, F., Dozandeh-Mobarrez, B., Qeshmi, F. I., Javadpour, S., & Mokhtary, S. (2019). Molecular detection of virulence factors and biofilm formation in Pseudomonas aeruginosa obtained from different clinical specimens in Bandar Abbas. Iranian Journal of Microbiology, 11(1), 25–30. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6462266/pdf/IJM-11-25.pdf
- Bhowmick, P. P., Devegowda, D., Ruwandeepika, H. A. D., Fuchs, T. M., Srikumar, S., Karunasagar, I., & Karunasagar, I. (2011a). gcpA (stm1987) is critical for cellulose production and biofilm formation on polystyrene surface by Salmonella enterica serovar Weltevreden in both high and low nutrient medium. Microbial Pathogenesis, 50(2), 114–122. https://doi.org/10.1016/j.micpath.2010.12.002
- Bhowmick, P. P., Devegowda, D., Ruwandeepika, H. A. D., Karunasagar, I., & Karunasagar, I. (2011b). Presence of Salmonella pathogenicity island 2 genes in seafood-associated Salmonella serovars and the role of the sseC gene in survival of Salmonella enterica serovar Weltevreden in epithelial cells. Microbiology, 157(Pt1), 160–168. https://doi.org/10.1099/mic.0.043596-0
- Blanco, M., Lazo, L., Blanco, J. E., Dahbi, G., Mora, A., Lopez, C., Gonzalez, E. A., & Blanco, J. (2006). Serotypes, virulence genes, and PFGE patterns of enteropathogenic Escherichia coli isolated from Cuban pigs with diarrhea. International Microbiology, 9(1), 53–60. http://scielo.isciii.es/pdf/im/v9n1/07blanco.pdf
- Capeda-Molero, M., Berger, C. N., Walsham, A. D. S., Ellis, S. J., Wemyss-Holden, S., Schuller, S., Frankel, G., & Fernandez, L. A. (2017). Attaching and effacing (A/E) lesion formation by enteropathogenic E. coli on human intestinal mucosa is dependent on non-LEE effectors. PLoS Pathogens, 13(10), e1006706. https://doi.org/10.1371/journal.ppat.1006706
- Chandra, K., Garai, P., Chatterjee, J., & Chakravortty, D. (2017). Peptide transporter YjiY influences the expression of the virulence gene mgtC to regulate biofilm formation in Salmonella. FEMS Microbiology Letters, 364(24), fnx236. https://doi.org/10.1093/femsle/fnx236
- Da Silva, N., Taniwaki, M. H., Junqueira, V. C. A., Silveira, N., Da Silva Do Nascimento, M., & Gomes, R. A. R. (2012). Microbiology examination methods of food and water: A laboratory manual (1st ed.). CRC Press.
- Daga, A. P., Koga, V. L., Soncicni, J. G. M., De Matos, C. M., Perrugini, M. R. E., Pelisson, M., Kobayashi, R. K. T., & Vespero, E. C. (2019). Escherichia coli bloodstream infections in patients at a university hospital: Virulence factors and clinical characteristics. Frontiers in Cellular Infection Microbiology, 9, 191. https://doi.org/10.3389/fcimb.2019.00191
- Do, H., Makthal, N., VanderWal, A. R., Saavedra, M. O., Olsen, R. J., Musser, J. M., & Kumaraswami, M. (2019). Environmental pH and peptide signaling control virulence of Streptococcus pyogenes via a quorum-sensing pathway. Nature Communications, 10(1), 2586. https://doi.org/10.1038/s41467-019-10556-8
- Duvallet, C., Gibbons, S. M., Gurry, T., Irizarry, R. A., & Alm, E. J. (2017). Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nature Communications, 8(1), 1784. https://doi.org/10.1038/s41467-017-01973-8
- FAO & WHO. (2020). Risk assessment tools for Vibrio parahaemolyticus and Vibrio vulnificus associated with seafood. Microbiological risk assessment series 20. http://www.fao.org/3/ca7240en/ca7240en.pdf
- Farmery, A. K., Hendrie, G. A., O’Kane, G., McManus, A., & Green, B. S. (2018). Sociodemographic variation in consumption patterns of sustainable and nutritious seafood in Australia. Frontiers in Nutrition, 5, 118. https://doi.org/10.3389/fnut.2018.00118
- Ferrando, M. L., van Baarlen, P., Orru, G., Piga, R., Bongers, R. S., Wels, M., De Greeff, A., Smith, H. E., & Wells, J. M. (2014). Carbohydrate availability regulates virulence gene expression in Streptococcus suis. PLoS One, 9(3), e89334. https://doi.org/10.1371/journal.pone.0089334
- Gerstel, U., & Romling, U. (2001). Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environmental Microbiology, 3(10), 638–648. https://doi.org/10.1046/j.1462-2920.2001.00235.x
- Guijarro, J. A., Cascales, D., Garcia-Torrico, A. I., Garcia-Dominguez, M., & Mendez, J. (2015). Temperature-dependent expression of virulence genes in fish-pathogenic bacteria. Frontiers in Microbiology, 6, 700. https://doi.org/10.3389/fmicb.2015.00700
- Gutierrez West, C. K., Klein, S. L., & Lovell, C. R. (2013). High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Applied and Environmental Microbiology, 79(7), 2247–2252. https://doi.org/10.1128/AEM.03792-12
- Haber, A., Friedman, S., Lobel, L., Burg-Golani, T., Sigal, N., Rose, J., Livnat-Levanon, N., Lewinson, O., & Herskovits, A. A. (2017). L-glutamine induces expression of Listeria monocytogenes virulence genes. PLoS Pathogens, 13(1), e1006161. https://doi.org/10.1371/journal.ppat.1006161
- Hacker, J., Blum-Oehler, G., Muhldorfer, I., & Tschape, H. (1997). Pathogenicity islands of virulent bacteria: Structure, function and impact on microbial evolution. Molecular Microbiology, 23(6), 1089–1097. https://doi.org/10.1046/j.1365-2958.1997.3101672.x
- Ho, W. S., Tan, L. K., Ooi, P. T., Yeo, C. C., & Thong, K. L. (2013). Prevalence and characterization of verotoxigenic-Escherichia coli isolates from pigs in Malaysia. BMC Veterinary Research, 9(1), 109. https://doi.org/10.1186/1746-6148-9-109
- Iwamoto, M., Ayers, T., Mahon, B. E., & Swerdlow, D. L. (2010). Epidemiology of seafood-associated infections in the United States. Clinical Microbiology Reviews, 23(2), 399–411. https://doi.org/10.1128/CMR.00059-09
- Joffre, E., Nicklasson, M., Álvarez-Carretero, S., Xiao, X., Sun, L., Nookaew, I., Zhu, B., & Sjoling, A. (2019). The bile salt glycocholate induces global changes in gene and protein expression and activates virulence in enterotoxigenic Escherichia coli. Scientific Reports, 9(1), 108. https://doi.org/10.1038/s41598-018-36414-z
- Jubelin, G., Desvaux, M., Schüller, S., Etienne-Mesmin, L., Muniesa, M., & Blanquet-Diot, S. (2018). Modulation of enterohaemorrhagic Escherichia coli survival and virulence in the human gastrointestinal tract. Microorganisms, 6(4), 115. https://doi.org/10.3390/microorganisms6040115
- Jubelin, G., Vianney, A., Beloin, C., Ghigo, J. M., Lazzaroni, J. C., Lejeune, P., & Dorel, C. (2005). CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. Journal of Bacteriology, 187(6), 2038–2049. https://doi.org/10.1128/JB.187.6.2038-2049.2005
- Kentache, T., Milochanic, E., Cao, T. N., Mokhtari, A., Ake, F. M., Ma Pham, Q. M., Joyet, P., & Deutsher, J. (2016). Transport and catabolism of pentitols by Listeria monocytogenes. Journal of Molecular Microbiology and Biotechnology, 26(6), 369–380. https://doi.org/10.1159/000447774
- Koohsari, H., Ghaemi, E. A., Mozaffari, N. A., Moradi, A., Sadegh-Sheshpoli, M., & Javid, S. N. (2017). The effect of adding blood on the virulence genes expression of Staphylococcus aureus in exponential and stationary growth phase. Jundishapur Journal of Microbiology, 10(6), e14380. https://doi.org/10.5812/jjm.14380
- Kothary, M. H., & Babu, U. S. (2001). Infective dose of foodborne pathogens in volunteers: A review. Journal of Food Safety, 21(1), 49–73. https://doi.org/10.1111/j.1745-4565.2001.tb00307.x
- Lam, O., Wheeler, J., & Tang, C. M. (2014). Thermal control of virulence factors in bacteria: A hot topic. Virulence, 5(8), 852–862. https://doi.org/10.4161/21505594.2014.970949
- Lane, D. J. (1991). 16S/23S rRNA sequencing. In E. Stackebrandt & M. Goodfellow (Eds.), Nucleic acid techniques in bacterial systematic (pp. 115–175). Wiley.
- Larsen, N., & Jespersen, L. (2015). Expression of virulence-related genes in Listeria monocytogenes grown on Danish hard cheese as affected by NaCl content. Foodborne Pathogens and Disease, 12(6), 536–544. https://doi.org/10.1089/fpd.2014.1930
- Li, J., Ni, X. D., Liu, Y. J., & Lu, C. P. (2011). Detection of three virulence genes alt, ahp and aerA in Aeromonas hydrophila and their relationship with actual virulence to zebrafish. Journal of Applied Microbiology, 110(3), 823–830. https://doi.org/10.1111/j.1365-2672.2011.04944.x
- Liu, Y., Liu, B., Yang, P., Wang, T., Chang, Z., Wang, J., Wang, Q., Li, W., Wu, J., Huang, D., Jiang, L., & Yang, B. (2019). LysR-type transcriptional regulator OvrB encoded in O island 9 drives enterohemorrhagic Escherichia coli O157: H7 virulence. Virulence, 10(1), 783–792. https://doi.org/10.1080/21505594.2019.1661721
- Lopatek, M., Wieczorek, K., & Osek, J. (2018). Antimicrobial resistance, virulence factors, and genetic profiles of Vibrio parahaemolyticus from seafood. Applied and Environmental Microbiology, 84(16), e00537–18. https://doi.org/10.1128/AEM.00537-18
- Mackenzie, K. D., Wang, Y., Musicha, P., Hansen, E. G., Palmer, M. B., Herman, D. J., Feasey, N. A., & White, A. P. (2019). Parallel evolution leading to impaired biofilm formation in invasive Salmonella strains. PLoS Genetics, 15(6), e1008233. https://doi.org/10.1371/journal.pgen.1008233
- Mallorquí, J., Simon-Grifé, M., Ferrer-Soler, L., Roca, M., March, R., & Sitjà, M. (2018). Reduced mortality and morbidity associated with verotoxin 2e-induced edema disease in pigs using a recombinant verotoxin 2e vaccine. Journal of Swine Health and Production, 26(5), 253–261. https://www.aasv.org/shap/issues/v26n5/v26n5p253.pdf
- Mannion, A., Shen, Z., & Fox, J. G. (2018). Comparative genomics analysis to differentiate metabolic and virulence gene potential in gastric versus enterohepatic Helicobacter species. BMC Genomics, 19(1), 830. https://doi.org/10.1186/s12864-018-5171-2
- Maze, A., Glatter, T., & Bumann, D. (2014). The central metabolism regulator EIIAGlc switches Salmonella from growth arrest to acute virulence through activation of virulence factor secretion. Cell Reports, 7(5), 1426–1433. https://doi.org/10.1016/j.celrep.2014.04.022
- Mechri, B., Medhioub, A., Medhioub, M. N., & Aouni, M. (2016). Prevalence of biofilm formation and wide distribution of virulence associated genes among Vibrio spp. strains isolated from the Monastir Lagoon, Tunisia. Polish Journal of Microbiology, 65(3), 307–318. https://doi.org/10.5604/17331331.1215610
- Melson, E. M., & Kendall, M. M. (2019). The sRNA DicF integrates oxygen sensing to enhance enterohemorrhagic Escherichia coli virulence via distinctive RNA control mechanisms. Proceedings of the National Academy of Sciences of the United States of America, 116(28), 14210–14215. https://doi.org/10.1073/pnas.1902725116
- Meparambu Prabhakaran, D., Ramamurthy, T., & Thomas, S. (2020). Genetic and virulence characterisation of Vibrio parahaemolyticus isolated from Indian coast. BMC Microbiology, 20(1), 62. https://doi.org/10.1186/s12866-020-01746-2
- Nhu, N. T. K., Phan, M. D., Peters, K. M., Lo, A. W., Forde, B. M., Chong, T. M., Yin, W. F., Chan, K. G., Chromek, M., Brauner, A., Chapman, M. R., Beatson, S. A., & Schembri, M. A. (2018). Discovery of new genes involved in curli production by a uropathogenic Escherichia coli strain from the highly virulent O45: K1:H7 lineage. mBio, 9(4), e01462–18. https://doi.org/10.1128/mBio.01462-18
- Nimnoi, P., & Pongsilp, N. (2020). Marine bacterial communities in the upper gulf of Thailand assessed by Illumina next-generation sequencing platform. BMC Microbiology, 20(1), 19. https://doi.org/10.1186/s12866-020-1701-6
- Niu, C., Yu, D., Wang, Y., Ren, H., Jin, Y., Zhou, W., Li, B., Cheng, Y., Yue, J., Gao, Z., & Liang, L. (2013). Common and pathogen-specific virulence factors are different in function and structure. Virulence, 4(6), 473–482. https://doi.org/10.4161/viru.25730
- Ogasawara, H., Yamada, K., Kori, A., Yamamoto, K., & Ishihama, A. (2010). Regulation of the Escherichia coli csgD promoter: Interplay between five transcription factors. Microbiology, 156(Pt8), 2470–2483. https://doi.org/10.1099/mic.0.039131-0
- Ogutcu, H., Adiguzel, A., Gulluce, M., Karadayi, M., & Sahin, F. (2009). Molecular characterization of Rhizobium strains isolated from wild chickpeas collected from high altitudes in Erzurum-Turkey. Romanian Biotechnological Letters, 14(2), 4294–4300. http://www.rombio.eu/rbl2vol14/cnt/Lucr-11.pdf
- Osman, K. M., Hassman, H. M., Orabi, A., & Abdelhafez, A. S. T. (2014). Phenotypic, antimicrobial susceptibility profile and virulence factors of Klebsiella pneumoniae isolated from buffalo and cow mastitic milk. Pathogens and Global Health, 108(4), 191–199. https://doi.org/10.1179/2047773214Y.0000000141
- Pan, X., Yang, Y., & Zhang, J. R. (2014). Molecular basis of host specificity in human pathogenic bacteria. Emerging Microbes & Infection, 3(3), e23. https://doi.org/10.1038/emi.2014.23
- Panicker, G., Call, D. R., Krug, M. J., & Bej, A. K. (2004). Detection of pathogenic Vibrio spp. in shellfish by using multiplex PCR and DNA Microarrays. Applied and Environmental Microbiology, 70(12), 7436–7444. https://doi.org/10.1128/AEM.70.12.7436-7444.2004
- Pass, M. A., Odedra, R., & Batt, R. M. (2000). Multiplex PCRs for identification of Escherichia coli virulence genes. Journal of Clinical Microbiology, 38(5), 2001–2004. https://doi.org/10.1128/JCM.38.5.2001-2004.2000
- Paytubi, S., Cansado, C., Madrid, C., & Balsalobre, C. (2017). Nutrient composition promotes switching between pellicle and bottom biofilm in Salmonella. Frontiers in Microbiology, 8, 2160. https://doi.org/10.3389/fmicb.2017.02160
- Pongsilp, N. (2012). Phenotypic and genotypic diversity of rhizobia. Bentham Science Publishers. https://doi.org/10.2174/97816080546191120101
- Pongsilp, N., & Nimnoi, P. (2018). Diversity and antibiotic resistance patterns of enterobacteria isolated from seafood in Thailand. CyTA Journal of Food, 16(1), 793–800. https://doi.org/10.1080/19476337.2018.1479453
- Pongsilp, N., Teaumroong, N., Nuntagij, A., Boonkerd, N., & Sadowsky, M. J. (2002). Genetic structure of indigenous non-nodulating and nodulating populations of Bradyrhizobium in soils from Thailand. Symbiosis, 33(1), 39–58.
- Prabhakar, P., Lekshmi, M., Nayak, B. B., & Kumar, S. (2017). Incidence of potentially pathogenic Escherichia coli in fresh seafood in Mumbai. Pollution Research, 36(3), 541–546.
- Praja, R. K., & Safnurbati, D. P. (2018). The infection of Vibrio parahaemolyticus in shrimp and human. Oceana Biomedicina Journal, 1(1), 44–58. https://doi.org/10.30649/obj.v1i1.6
- Prigent-Combaret, C., Brombacher, E., Vidal, O., Ambert, A., Lejeune, P., Landini, P., & Dorel, C. (2001). Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. Journal of Bacteriology, 183(24), 7213–7223. https://doi.org/10.1128/JB.183.24.7213-7223.2001
- Rahimi, E., Raissy, M., Razzaghimanesh, M., Dastgerdi, A. A., & Shahraki, M. M. (2014). Occurrence of Aeromonas hydrophila in fish, shrimp, lobster and crab in Iran. Kafkas Universitesi Veteriner Fakultesi Dergisi, 20(5), 691–696. https://doi.org/10.9775/kvfd.2014.10892
- Sandner, L., Eguiarte, L. E., Navarro, A., Cravioto, A., & Souza, V. (2001). The elements of the locus of enterocyte effacement in human and wild mammal isolates of Escherichia coli: Evolution by assemblage or disruption? Microbiology, 147(Pt11), 3149–3158. https://doi.org/10.1099/00221287-147-11-3149
- Schloss, P. D., & Handelsman, J. (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Applied and Environmental Microbiology, 71(3), 1501–1506. https://doi.org/10.1128/AEM.71.3.1501-1506.2005
- Schmid-Hempel, P., & Frank, S. A. (2007). Pathogenesis, virulence, and infective dose. PLoS Pathogens, 3(10), e147. https://doi.org/10.1371/journal.ppat.0030147
- Skyberg, J. A., Siek, K. E., Doetkott, C., & Nolan, L. K. (2007). Biofilm formation by avian Escherichia coli in relation to media, source and phylogeny. Journal of Applied Microbiology, 102(2), 548–554. https://doi.org/10.1111/j.1365-2672.2006.03076.x
- Tomar, R. S., Agarwal, M., & Jyoti, A. (2015). Determination of drug resistance and virulent gene signatures in potable water isolates of Escherichia coli in Gwalior city. Journal of Pharmaceutical Sciences and Research, 7(11), 967–971.
- Tosun, S. Y., Alakavuk, D. U., & Mol, S. (2016). Isolation of Salmonella spp. and other members of Enterobacteriaceae from horse mackerel (Trachurus trachurus), sold in public markets of Istanbul, Turkey. Journal of Food and Health Science, 2(2), 82–89. https://doi.org/10.3153/JFHS16009
- Uhlich, G. A., Chen, C. Y., Cottrell, B. J., Hofmann, C. S., Yan, X., & Nguyen, L. (2016). Stx1 prophage excision in Escherichia coli strain PA20 confers strong curli and biofilm formation by restoring native mlrA. FEMS Microbiology Letters, 363(13), fnw123. https://doi.org/10.1093/femsle/fnw123
- Valdes, K. M., Sundar, G. S., Belew, A. T., Islam, E., EI-Sayed, N. M., Breton, Y. L., & McIver, K. S. (2018). Glucose levels alter the Mga virulence regulon in the group A. Streptococcus. Scientific Reports, 8(1), 4971. https://doi.org/10.1038/s41598-018-23366-7
- Vu, T. T. T., Alter, T., & Huehn, S. (2018). Prevalence of Vibrio spp. in retail seafood in Berlin, Germany. Journal of Food Protection, 81(4), 593–597. https://doi.org/10.4315/0362-028X.JFP-17-366
- Wang, S. X., Zhang, X. H., Zhong, Y. B., Sun, B. G., & Chen, J. X. (2007). Genes encoding the Vibrio harveyi haemolysin (VHH) thermolabile haemolysin (TLH) are widespread in Vibrios. Acta Microbiologica Sinica, 47(5), 874–881.
- WHO & FAO. (2018). Shiga toxin-producing Escherichia coli (STEC) and food: Attribution, characterization, and monitoring report. Microbiological risk assessment series 31. Food and Agriculture Organization. https://apps.who.int/iris/bitstream/handle/10665/272871/9789241514279-eng.pdf?ua=1
- Xie, Z. Y., Hu, C. Q., Chen, C., Zhang, L. P., & Ren, C. H. (2005). Investigation of seven Vibrio virulence genes among Vibrio alginolyticus and Vibrio parahaemolyticus strains from the coastal mariculture systems in Guangdong, China. Letters in Applied Microbiology, 41(2), 202–207. https://doi.org/10.1111/j.1472-765X.2005.01688.x
- Yamamoto, S., Terai, A., Yuri, K., Kurazono, H., Takeda, Y., & Yoshida, O. (1995). Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunology and Medical Microbiology, 12(2), 85–90. https://doi.org/10.1111/j.1574-695X.1995.tb00179.x
- Yanez, R., Bastías, R., Higuera, G., Salgado, O., Katharios, P., Romero, J., Espejo, R., & García, K. (2015). Amplification of tlh gene in other Vibrionaceae specie by specie-specific multiplex PCR of Vibrio parahaemolyticus. Electronic Journal of Biotechnology, 18(6), 459–463. https://doi.org/10.1016/j.ejbt.2015.09.007
- Zhao, A., Liu, H., Sun, W., Li, Q., Pan, Y., & Zhao, Y. (2015). Irregular virulence genes expression of Vibrio parahaemolyticus in shrimp or seawater matrix. Research & Reviews: Journal of Microbiology & Biotechnology, 4(2), 26–31.
- Zhou, G., Li, L. J., Shi, Q. S., Ouyang, Y. S., Chen, Y. B., & Hu, W. F. (2013). Effects of nutritional and environmental conditions on planktonic growth and biofilm formation of Citrobacter werkmanii BF-6. Journal of Microbiology and Biotechnology, 23(12), 1673–1682. https://doi.org/10.4014/jmb.1307.07041
- Zhou, G., Peng, H., Wang, Y. S., Huang, X. M., Xie, X. B., & Shi, Q. S. (2017). Complete genome sequence of Citrobacter werkmanii strain BF-6 isolated from industrial putrefaction. BMC Genomics, 18(1), 765. https://doi.org/10.1186/s12864-017-4157-9
- Zogaj, X., Bokranz, W., Nimtz, M., & Romling, U. (2003). Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infection and Immunity, 71(7), 4151–4158. https://doi.org/10.1128/iai.71.7.4151-4158.2003
- Zughaier, S. M., & Cornelis, P. (2018). Editorial: Role of iron in bacterial pathogenesis. Frontiers in Cellular and Infection Microbiology, 8, 344. https://doi.org/10.3389/fcimb.2018.00344