ABSTRACT
This study has employed 16S rDNA analysis to elucidate the characteristics of the microbial community in the surroundings, facilities, and meat samples collected from a pork wholesale market with the aim of revealing bacterial contamination. In addition, the efficacy of sodium hypochlorite disinfectant was assessed by comparing the bacterial communities in the viable samples before and after disinfection. The bacterial abundances in meat, chopping boards, and knives were positively correlated (r > 0.95, p < .05), suggesting potential cross-contamination risks during market transactions. The species richness indices for the external transport vehicles, pork hooks, and floors were significantly different from those of the samples taken after decontamination (p < .05). Linear discriminant analysis Effect Size (LEfSe) analyses showed that Firmicutes, Proteobacteria, and Chloroflexi were significantly more abundant in the transaction period, whereas Bacteroidetes levels decreased compared to the clean-up period. NaClO disinfection altered the composition of bacterial communities.
1. Introduction
Pork is a popular meat in China because of its excellent nutritional content and flavour, accounting for 62.9% of total meat consumption in 2018 (Zhang et al., Citation2021). However, pork is the most frequently contaminated food, contributing to serious meat safety concerns (Wilson et al., Citation2020; Wu et al., Citation2021). It is possible that this microbial contamination originated from the farms, or in the abattoirs during processing for retail sale (Demaître et al., Citation2020; Kang et al., Citation2014; Vojkovska et al., Citation2015). For example, a recent survey of retail markets indicated that Salmonella contamination was present in pork (73.1%), with 30% of these samples also contained Escherichia coli (Zhang et al., Citation2018). Pseudomonas spp., Brochothrix thermosphacta, and lactic acid bacteria have also been detected in retail pork at levels ranging 5.3 to 6.4 log CFU/g (Andritsos et al., Citation2012). 16S rDNA amplicon sequencing has also identified Staphylococcus aureus, Pseudomonas spp., and E. coli as the dominant contaminants in retail pork (Peruzy et al., Citation2020). These previous studies focused on the retail supply chain; however, the degree of microbial contamination at the wholesale market level has not been extensively investigated.
In China, 70–80% of food is distributed through wholesale markets, which is a critical sector which food safety risks can be controlled (Xiao et al., Citation2021; Zhou et al., Citation2022). The meat wholesale market handles pork portioning and distribution and connects the slaughterhouse and grocery store. Hog carcasses are split in half in slaughterhouses and transported to wholesale markets using cold chain vehicles. Once the market opens, carcasses are transported by carts from vehicles to stalls, where they are portioned and sold to retailers. Usually, pork wholesale markets conduct operations in regard to trading from 12 am to 8 am and they disinfect floors with sodium hypochlorite (NaClO) after closing (~9 am). NaClO is recommended by the World Health Organization (WHO) for disinfecting surfaces owing to its broad-spectrum antimicrobial activity and is widely used in food production facilities (Sato et al., Citation2019). However, whether bacterial contamination associated with meat transaction facilities is altered following NaClO decontamination has not yet been evaluated. This information enables the identification of potential microbial risks in pork products.
Although traditional bacterial culture and plate methods are the gold standards for microbiological diagnosis, these approaches generally focus on mapping the distribution of specific environmentally transmitted pathogenic bacteria, such as Salmonella, E. coli, S. aureus (Cobo Díaz et al., Citation2021; O’Brien, Citation2002). Traditional bacterial culture and plate methods are time-consuming, largely underestimate the abundance of species, and have limitations in characterizing complex microbial communities (Cobo Díaz et al., Citation2021; Peruzy et al., Citation2020). High-throughput sequencing is a technique widely used to assess microbial community structures in different environments (S. Xu et al., Citation2016) and enables the identification of organisms belonging to previously unknown taxa. For example, 16S rDNA gene sequencing has been used to characterize the microbial composition of pork during storage (Bassey et al., Citation2021; Li et al., Citation2019; Zhao et al., Citation2022).
The focus of the current study was to assess the effect of disinfection with NaClO on the bacterial species composition using 16S rDNA amplicon sequencing. These results not only assess the effectiveness of decontamination but also expand our knowledge on the routes of microbial transmission.
2. Materials and methods
2.1. Sampling
Samples were collected from a pork wholesale market in Yuhang District, Hangzhou, China, which processes approximately 10,000 pigs daily. In this market, regular disinfection operations were implemented following operational closure. This involved spraying a NaClO solution with a chlorine concentration of 500 mg/L over the surfaces of the surroundings and facilities. In total, 104 samples were collected from different sites during daily market activities (). A total of 62 samples were collected during the market transaction period at 2 a.m. and were obtained from external transport vehicles (BEV, n = 6), local transport vehicles (NLV, n = 6), carts (NC, n = 12), pork hooks (NPK, n = 3), meat (NM, n = 10), knives (NK, n = 10), chopping boards (NCB, n = 10), and floors (NF, n = 5). Disinfection was performed after the market closed at 11 a.m., 42 disinfected samples were collected from external transport vehicles (DEV, n = 6), local transport vehicles (DLV, n = 6), carts (DC, n = 12), pig hooks (DPK, n = 3), chopping boards (DCB, n = 10), and floors (DF, n = 5). We attempted to gather samples before and after the disinfection process, except for meat and knives, which were only present during pork transactions. These samples were examined to determine the attributes of the bacterial communities. Samples were collected using a sterile absorbent gauze pre-moistened with phosphate -buffered saline (1 × PBS, pH 7.2). When a surface with a sufficient size was available (e.g. chopping boards), ~ 1 m2 was sampled by swabbing first horizontally, then vertically, and finally by diagonally turning the swab around each direction. For other surfaces, such as knives, individual units were swabbed. Single-use disposable gloves were used during sampling and the gloves were changed after each sample was collected to avoid cross-contamination. Sample swabs were placed in tubes containing 15 mL PBS, placed in a cooling box containing ice packs, and transported to the laboratory within 2 h for sample processing.
Table 1. Sample information in the pork wholesale market.
2.2. DNA extraction
Swab samples were individually added to tubes containing 15 mL PBS and centrifuged at 10,000 rpm for 5 min. The supernatants were filtered through 0.22 μm polycarbonate membranes (GE Osmonics, Minnetonka, MN, U.S.A.) and stored at −80°C. DNA was extracted using a TIANamp Stool DNA Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. Final DNA concentration and purity were measured using a Nanodrop One spectrophotometer (Thermo Fisher, Pittsburgh, PA, U.S.A.), and DNA quality was confirmed by 1% agarose gel electrophoresis. Each sample was processed in duplicate and stored at −20°C until Polymerase Chain Reaction (PCR) analysis.
2.3. PCR amplification and sequencing
Polymerase chain reaction (PCR) was performed by amplifying the V3-V4 hypervariable region of 16S rDNA using the universal bacterial-specific primers 16S-341F (5’-CCTAYGGGRBGCASCAG-3’) and 16S-806 R (5’-GGACTACNNGGGTWTCTAAT-3’), where the barcode is an eight-base sequence unique to each sample. The cycling reaction parameters were as follows: 5 min at 95°C and 27 cycles of 30 s at 95°C, 30 s at 55°C, 45 s at 72°C and a final extension for 10 min at 72°C. PCR amplification was performed in a total volume of 25 μL of the reaction mixture, containing 25 ng of template DNA, 12.5 μL of PCR premix, 2.5 μL of each primer and PCR-grade water to adjust the volume. Amplification was carried out using an ABI GeneAmp 9700 instrument (Thermo Scientific) with FastPfu polymerase (Transgen Biotech, Beijing, China) and 10 ng of template DNA, according to the manufacturer’s protocol. The pooled PCR products were purified by 2% agarose gel electrophoresis. Bands corresponding to 300 bp were excised and purified using the GeneJET Gel Extraction Kit (Thermo Fisher Scientific). The purified amplified fragments were subjected to equimolar and paired-end sequencing (2 × 250) using an Illumina PE250 platform (Shanghai, China). Raw data were uploaded to the NCBI BioProject database and we obtained the following ID number: PRJNA823853.
2.4. Sequence data analysis
Raw data generated from 16S rDNA MiSeq sequencing were analyzed by Biozeron Biotechnology (Shanghai, China). Amplicon sequence quality control was performed using the Divisive Amplicon Denoising Algorithm2 (DADA2) to identify indel mutations and substitutions, which were processed with QIIME2 (ver. 2020.11) (Callahan et al., Citation2016). Amplicon sequence variants (ASVs) were generated using paired sequence merging and chimera filtering. To obtain the species taxonomic information corresponding to each ASV, high-performance clustering, alignment, and search algorithms (UCLUST, ver. 1.2.22q) was used to analyze the phylogenetic affinities of representative ASV sequences in the Silva (SSU132) database, with a confidence threshold of 0.8. Rarefaction analysis, based on Mothur v.1.35.1 was conducted to reveal the diversity. Alpha and beta diversities were calculated by random normalization of the same sequences. The Chao1 index was selected for the alpha diversity analysis, and the complexity of species diversity was calculated using QIIME (De Filippis et al., Citation2018). Principal coordinate analysis (PCoA) was used in order to explore the effects of variable factors, and ADONIS was used to measure the Bray-Curtis distances. Phyla and ASVs and relative abundance levels > 1% and 0.1%, respectively, were defined as predominant and sorted for further comparison.
Linear discriminant analysis Effect Size (LEfSe) analysis was used to determine significant differences in the relative abundance of bacteria before and after disinfection. LEfSe analysis of microbial abundance was performed by pooling all samples between the N (transaction period) and D (clean-up period) groups to differentiate microbial communities. Additionally, LEfSe analysis was performed at different locations before and after the disinfection was performed. The chi-square and Kruskal-Wallis rank-sum tests were used to investigate the differences resulting from the distribution of multiple populations. Linear discriminant analysis (LDA) effect size (LEfSe) was applied to discover the biomarker panels that possessed organismal features for differentiating the microbial communities specific to a particular treatment (Li et al., Citation2020).
Microbial functions were predicted based on 16S rDNA sequences using PICRUSt2 (Langille et al., Citation2013). The Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2, v2.2.0-b) (https://github.com/picrust/picrust2) program based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to predict the functional pathways used by the microbiota in different samples. The representative ASVs in FASTA format and a Biological Observation Matrix (BIOM) table of the abundance of each ASV across each sample were used as inputs for PICRUSt2. A BIOM table was generated using the Python package, Biom (v.2.1.7). The minimum mapping rate of ASV representative sequences to representative sequences in the KEGG database was set to 0.6, a value below this meant they did not participate in the functional prediction. Functional differences among cultivars were analyzed using the Kruskal-Wallis H test in the R project Vegan package. STAMP software (v2.1.3) was used for statistical analyses and visualization of the identified pathways.
2.5. Statistical analysis
GraphPad Prism 8 software (San Diego, CA, U.S.A.) was used for data analysis and generation of the figures. A significant difference was established at p < .05. Correlation analyses were performed using the Pearson correlation coefficient, calculated with the rcorr function in the R package Hmisc.
3. Results
3.1. Taxonomic profiles for sample surfaces
A total of 6,362,065 high-quality validated gene sequences clustered into 14,780 ASVs were obtained from the environmental and meat samples. The total numbers of ASVs and reads were higher during market transactions, particularly for the external transport vehicles, local transport vehicles, and carts. The number of reads in the floor samples decreased after disinfection was performed (). Sparse curves indicate sufficient sequencing data and species abundances in the sample. As shown in Figure S1, the dilution curves flattened with increasing sequencing depth, suggesting that the amount of sequencing data was reasonable.
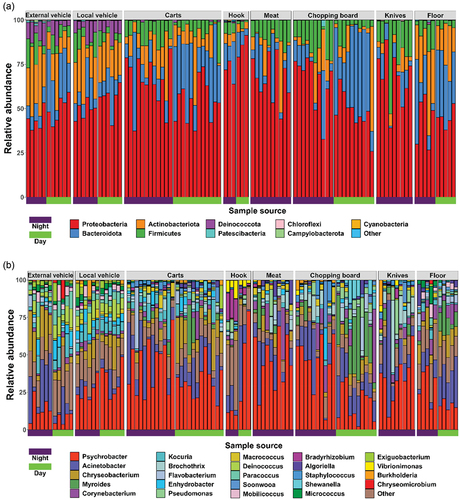
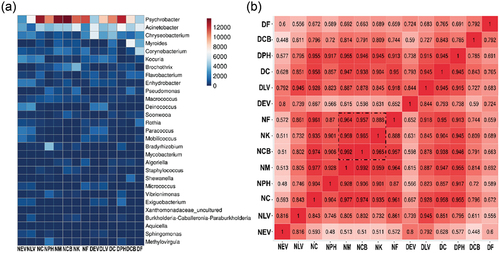
The microbiota on the surfaces of the different samples varied, with 31 phyla, 75 classes, 179 orders, 287 families, 749 genera, and 1798 species. The stacked bar chart shows the dominant phyla and genera in each sample. At the phylum level, the primary phyla identified were Proteobacteria (56.9%), Bacteroidota (17.7%), and Actinobacteria (13.1%), with different distributions on the various sample surfaces. Proteobacteria, Bacteroidota, Actinobacteriota, and Firmicutes were the dominant phyla in all samples; interestingly, Deinococcota was dominant only in vehicle samples (). At the genus level, we also identified 30 genera as the most abundant colonizers; Psychrobacter, Acinetobacter, and Chryseobacterium possessed average abundance levels of 29.7, 13.6 and 6.4%, respectively (). Our findings are consistent with the literature (Li et al., Citation2019; Tassou et al., Citation2021) in that Acinetobacter is the most commonly found bacterium in pork samples (Emamjomeh et al., Citation2023).
3.2. Differences in taxonomic profiles following NaClO decontamination
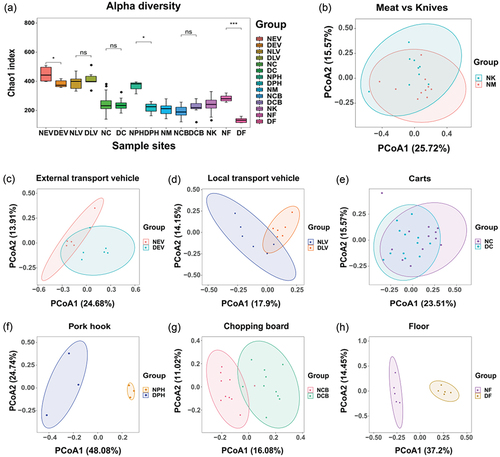
Alpha-diversity analyses showed that the greatest species abundance was found in transport vehicles, whereas samples from the internal environment of the wholesale market, such as meat, chopping boards, and knife surfaces, had relatively small species abundance. As expected, the average values of the species richness indices for the external transport vehicles, pork hooks, and floors were significantly different from those of the samples taken after decontamination (p < .05). Floor samples showed the lowest alpha diversity after the decontamination was performed (). Bray – Curtis distance-based PCoA plots were generated in order to examine the differences in microbial community structure before and after disinfection of these samples. There were no significant differences in the microbiological composition between the meat and knife samples (p > .05), indicating potential cross-contamination (). The disinfection process had a significant effect on the microbiological composition of external transport vehicles, local transport vehicles, carts, pork hooks, chopping boards, and floors (). The vehicle samples were significantly separated from the other samples in coordinate space, indicating significant differences in their microbial community structures (Figures S2 and S3). These results indicated that the NaClO disinfectant was effective in altering the taxonomic profiles of the microorganisms in the pork wholesale market in terms of abundance and composition.
Figure 2. Analysis of the alpha diversity and beta diversity, (a) Chao1 index of bacterial flora in each sampling site. (b) PCoA analysis of meat and knife samples, (c) external transport vehicles, (d) local transport vehicles, (e) carts, (f) pork hook, (g) chopping boards, and(H) floor samples for the two sampling periods.
Differences in bacterial composition between groups N and D were identified using LEfSe analyses, which showed that bacteria were classified into four distinct taxa at the phylum level; Firmicutes, Proteobacteria, and Chloroflexi were found to be significantly more abundant in the N (transaction period) group, whereas Bacteroidetes levels decreased when compared to those in the D (clean-up period) group (). When a logarithmic LDA score of 3.0 was used as the cut-off point, 23 differential genera were identified, with Escherichia-Shigella being significantly higher in group D and Methylobacterium being the most abundant genus in group N (). Listeriaceae were found to be abundant on the chopping board, and Enterobacterales made a large contribution to the floor samples (Figure S4-S9). In addition, we counted pathogen- and spoilage-associated species at the genus level. The presence of potential pathogens and spoilage-associated bacteria identified at the genus level differed between the meat trading period (37.65%) and following decontamination (1.13%). Meat samples contained the highest abundance levels of pathogen contamination (12.9%), and knives contained the highest abundance levels of spoilage-associated bacteria (0.24%). The relative abundance levels of potential pathogens and spoilage-associated bacteria on floors were decreased from 0.01 to 0% and 0.37 to 0.15%, respectively, after market decontamination (). The criteria used to designate specific microbes as pathogens or spoilage-causing bacteria () were based on previous studies (). At the species level, the pathogenic and spoilage bacteria of concern () indicated a high abundance of Brochothrix thermosphacta, Pseudomonas fragi, E. coli, and S. aureus in the samples taken during market transactions. These became less dominant after the market closed, and the subsequent decontamination occurred (). B. thermosphacta and P. fragi were the most prevalent spoilage bacteria on the knives when the market was on trading ().
Figure 3. LEfSe analysis diagram of each dominant taxa between the N group (before disinfection) and the D groups (after disinfection). (a) Cladogram of microbial communities (b) LDA score of size differentiation using a threshold of 3 in groups D and N.
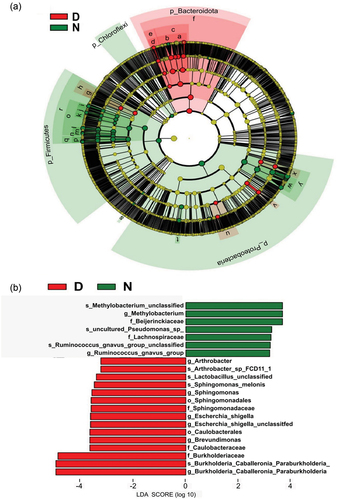
Figure 4. Distribution of pathogenic and spoilage bacteria on different sample surfaces. Percentage of abundance of (a) pathogenic and (c) spoilage bacteria at the genus level. The most abundant species of (b) pathogenic and (d) spoilage bacteria.
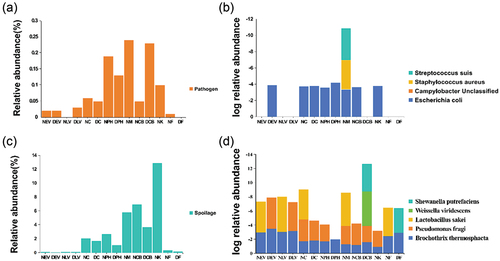
Table 2. The cardinal symptoms and corruption phenomena caused by potential pathogens and spoilage-associated bacteria.
3.3. Bacterial cross-contamination in the pork wholesale market
We examined the cross-contamination profiles of each sample surface, as shown in , Psychrobacter was higher at group N than group D samples, especially at meat and chopping board surfaces, where the relative abundance of Psychrobacter was significantly higher than at other sample surfaces (). Correlations between bacterial contamination during the two sampling periods were examined using Spearman’s correlation analysis. Bacterial abundance in meat was highly positively correlated with chopping boards (r = 0.99, p < .05), suggesting that these tow sample types were potential sources of bacterial cross-contamination. During the clean-up period, the correlation between DLV, DC, and DPH was only found to be high in Group D (r > 0.7) and it decreased significantly in the remaining samples. The abundance of bacteria in the floor samples showed only a low correlation with other samples, indicating that NaClO was an effective decontaminant. However, targeted disinfection measures may not be sufficient ().
3.4. Microbial function prediction
PICRUSt2 was used to obtain the relative KEGG pathway abundance information derived from 16S rDNA amplicon sequencing data, which revealed several pathways were significantly different in terms of NaClO disinfection (Group D) and without disinfection (Group N) (). RNA degradation (tadA) and glycosphingolipid biosynthesis (gloB) were enhanced in Group D. Biosynthetic pathways for streptomycin (strA, strB) and vancomycin (vraS) displayed an increase after disinfection. Correlations were found between the streptomycin gene (strB) and novobiocin biosynthesis genes (SQD, sqdB) (). This suggests that NaClO promotes antibiotic resistance. Group N samples that were not exposed to the disinfectant yielded higher gene expression levels of S. aureus (isdG and isdl), which coincided with the abundance determinations for this bacterium.
Figure 6. PICRUSt2 prediction analysis. (a) Influence of NaClO disinfection on bacterial abundance assessed using KEGG pathways of bacterial communities, only pathways that were significantly affected are presented. (b) Network plots of correlation analysis between genes associated with these pathways, only correlations ≥ 0.7 were presented.

4. Discussion
Over the past ten years, the application of high-through put sequencing (HTS) methods, specifically targeting the 16S rDNA gene, has facilitated the analysis of microbial composition and detection of pathogenic and spoilage bacteria (Cauchie et al., Citation2020; Zhao et al., Citation2015). HTS provides a reliable method for comprehensively describing the dynamics of bacterial communities and analyzing the correlations between microbial metabolic pathways and spoilage bacteria (Charmpi et al., Citation2020; Xu et al., Citation2022; Yang et al., Citation2016; Zhong et al., Citation2021). In this study, NaClO disinfection altered the bacterial community structures, and heterogeneous taxonomic profiles were found between the surface types and sampling times. B. thermosphacta, P. fragi, E. coli, and S. aureus were the most abundant bacterial species found during the meat transaction period. However, this species become less dominant after decontamination, and the most abundant bacteria causing spoilage in pork were B. thermosphacta, Enterobacteriaceae, and Pseudomonas. Their metabolic activities can lead to a sour flavor, discoloration, gas and slime production, and a decrease in pH (Bassey et al., Citation2021; Ercolini et al., Citation2010; Stellato et al., Citation2016; Wickramasinghe et al., Citation2021; Zhao et al., Citation2015, Citation2022). Pseudomonas, Streptococcus, and Staphylococcus were found to be the dominant genera in another study of fresh pork samples from retail markets (Cheung et al., Citation2008; Zhang et al., Citation2021). In this study, Pseudomonas, Streptococcus and Staphylococcus were also detected. Among them, Pseudomonas fragi was detected as a relatively high abundance spoilage-associated bacteria during the meat trade. Staphylococcus spp. and Streptococcus spp. were also detected in 11 and 14 species, respectively. Staphylococcus can generate enterotoxins and biofilms, and certain strains can withstand high temperatures and develop resistance to antibiotics (Kadariya et al., Citation2014; Khatoon et al., Citation2018). Pseudomonas is a taxonomic group of great interest in the meat industry because of its prominent role in meat spoilage and is among the most frequently reported taxa found after sanitation of processed surfaces from all types of food production chains (Zhang et al., Citation2021). Its persistence is most likely due to its ability to grow at low temperatures, to form biofilms and tolerance to biocides (Møretrø & Langsrud, Citation2017). P. fragi is a lipolytic and proteolytic species of the Enterobacteriaceae family that produces volatile organic compounds and other undesirable metabolites, such as biogenic amines. These contribute to meat spoilage. Brochothrix and Pseudomonas are abundant in the aerobic microbial population of fresh meat, and Brochothrix is associated with sourcing rather than with putrefaction (Illikoud et al., Citation2019; Peruzy et al., Citation2020).
The knive and cutting board samples used for meat partitioning exhibited a significant presence of spoilage and pathogenic bacteria, similar to the meat samples, suggesting that meat partitioning could potentially cause cross-contamination ( and S2). Bacteria can thrive on these surfaces, resulting in the transmission of foodborne pathogens to humans (Del Blanco et al., Citation2017). Therefore, it is imperative to implement appropriate measures to prevent the spread of microbes during the process of dividing meat portions and disinfecting surroundings and equipment in wholesale market. The effectiveness of disinfectants depends on factors such as material and protein load (Naïtali et al., Citation2009; Wang et al., Citation2020). The wooden chopping board surface exhibits greater roughness and can significantly increase the thickness and compactness of the biofilm, thereby increasing its resistance to disinfectants (Zhu et al., Citation2021). Under unhygienic conditions, a high load may reduce the efficacy against Gram-positive bacteria (Bessems, Citation1998; Tong, Hu, Chen, Li, Li, et al., Citation2021).
Our gene functional annotation analysis using PICRUSt2 suggests that NaClO disinfection during the clean-up period increased the abundance of genes related to RNA degradation and glycosphingolipid biosynthesis. RNA was used as the indicator of cell viability, and the expression of target genes was induced only in viable cells. Previous studies have revealed that the destruction of the cell membrane leads to the extrusion of macromolecules (e.g. ATP, proteins, and RNA) (Xu et al., Citation2017). Bacterial glycosphingolipids are structural elements tightly linked to environmental stress survival (Johansson et al., Citation2020). This indicates that NaClO provides selective pressure for bacteria (Tong, Hu, Chen, Li, Aifeng, et al., Citation2021). A previous study reported that the EC 3.4.16.4 serine-type d-Ala-d-Ala carboxypeptidase is significantly more abundant in pork samples and is a crucial component in regard to vancomycin resistance (Aráoz et al., Citation2000). The PWY-3781 aerobic respiration I (cytochrome c) genes are abundant in poultry and can contribute to human infections (Nearing et al., Citation2019).
Culture-dependent analyses coupled with typing of recovered isolates using molecular techniques are widely used to identify routes of microbial contamination between surfaces (Cobo Díaz et al., Citation2021). In contrast, 16S rDNA methods are culture-independent, convenient, rapid, and accurate and can be used to study microbial ecology and evolution (Ercolini, Citation2013; Knight et al., Citation2018). However, it is not possible to determine whether the functional genes detected were possessed by living bacteria, and it cannot be accurately determined whether these genes are functioning as described for the human gut microbiota (Lagier et al., Citation2016). Culture-dependent and-independent methods can complement each other in the study of microbial populations in food. On the other hand, the correlation analysis is a statistical measure often used in studies to show an association between variables. The limitations of correlation analysis are its assumption of a linear association and its sensitivity to a range of observations. The value of the correlation coefficient is also influenced by the measurement error. Additional PCR or culture procedures must be conducted to validate the findings of correlation analysis (Camara, Citation2018; Janse et al., Citation2021).
5. Conclusion
This is the first study to characterize the differences in bacterial contamination during the transaction and clean-up periods of a large wholesale pork market. NaClO disinfectants altered the bacterial community compositions. Chopping boards and meat have also been shown to be potential sources of bacterial cross-contamination. Functional predictions revealed that RNA degradation, glycosphingolipid levels, and antibiotic biosynthesis were enhanced during the clean-up period characterized by NaClO disinfection. This study demonstrates that bacterial community similarities and functional variations are highly correlated with NaClO disinfection.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Andritsos, N., Mataragas, M., Mavrou, E., Stamatiou, A., & Drosinos, E. (2012). The microbiological condition of minced pork prepared at retail stores in Athens, Greece. Meat Science, 91(4), 486–11. https://doi.org/10.1016/j.meatsci.2012.02.036
- Aráoz, R., Anhalt, E., René, L., Badet-Denisot, M.-A., Courvalin, P., & Badet, B. (2000). Mechanism-based inactivation of VanX, a d-alanyl-d-alanine dipeptidase necessary for vancomycin resistance. Biochemistry, 39(51), 15971–15979. https://doi.org/10.1021/bi001408b
- Bassey, A., Chen, Y., Zhu, Z., Odeyemi, O., Gao, T., Olusola, O., Ye, K., Li, C., & Zhou, G. H. (2021). Evaluation of spoilage indexes and bacterial community dynamics of modified atmosphere packaged super-chilled pork loins. Food Control, 130, 108383. https://doi.org/10.1016/j.foodcont.2021.108383
- Bessems, E. (1998). The effect of practical conditions on the efficacy of disinfectants. International Biodeterioration & Biodegradation, 41(3), 177–183. https://doi.org/10.1016/S0964-8305(98)00022-5
- Brooks, B. W., Devenish, J., Lutze-Wallace, C. L., Milnes, D., Robertson, R. H., & Berlie-Surujballi, G. (2004). Evaluation of a monoclonal antibody-based enzyme-linked immunosorbent assay for detection of Campylobacter fetus in bovine preputial washing and vaginal mucus samples. Veterinary Microbiology, 103(1), 77–84. https://doi.org/10.1016/j.vetmic.2004.07.008
- Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., & Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods, 13(7), 581–583. https://doi.org/10.1038/nmeth.3869
- Camara, P. G. (2018). Methods and challenges in the analysis of single-cell RNA-sequencing data. Current Opinion in Systems Biology, 7, 47–53. https://doi.org/10.1016/j.coisb.2017.12.007
- Cauchie, E., Delhalle, L., Taminiau, B., Tahiri, A., Korsak, N., Burteau, S., Fall, P. A., Farnir, F., Baré, G., & Daube, G. (2020). Assessment of spoilage bacterial communities in food wrap and modified atmospheres-packed minced pork meat samples by 16S rDNA metagenetic analysis [Original Research]. Frontiers in Microbiology, 10, 10. https://doi.org/10.3389/fmicb.2019.03074
- Charmpi, C., Van der Veken, D., Van Reckem, E., De Vuyst, L., & Leroy, F. (2020). Raw meat quality and salt levels affect the bacterial species diversity and community dynamics during the fermentation of pork mince. Food Microbiology, 89, 103434. https://doi.org/10.1016/j.fm.2020.103434
- Cheung, P.-Y., Lo, K., Cheung, T., Yeung, W., Leung, P., & Kam, K. (2008). Streptococcus suis in retail markets: How prevalent is it in raw pork? International Journal of Food Microbiology, 127(3), 316–320. https://doi.org/10.1016/j.ijfoodmicro.2008.08.006
- Cobo Díaz, J., Alvarez-Molina, A., Oniciuc, E., Walsh, C., Mencía-Ares, O., Puente-Gómez, P., Likotrafiti, E., Fernández-Gómez, P., Prieto, B., Crispie, F., Ruiz, L., Gonzalez-Raurich, M., López, M., Prieto, M., Cotter, P., & Alvarez-Ordóñez, A. (2021). Microbial colonization and resistome dynamics in food processing environments of a newly opened pork cutting industry during 1.5 years of activity. Microbiome, 9(1), 204. https://doi.org/10.1186/s40168-021-01131-9
- Correia Peres Costa, J., Bover-Cid, S., Bolívar, A., Cosano, G., & Pérez-Rodríguez, F. (2019). Modelling the interaction of the sakacin-producing Lactobacillus sakei CTC494 and listeria monocytogenes in filleted gilthead sea bream (Sparus aurata) under modified atmosphere packaging at isothermal and non-isothermal conditions. International Journal of Food Microbiology, 297, 72–84. https://doi.org/10.1016/j.ijfoodmicro.2019.03.002
- De Filippis, F., Storia, A., Villani, F., & Ercolini, D. (2018). Strain-level diversity analysis of Pseudomonas fragi after in situ pangenome reconstruction shows distinctive spoilage-associated metabolic traits clearly selected by different storage conditions. Applied and Environmental Microbiology, 85. https://doi.org/10.1128/AEM.02212-18
- Del Blanco, A., Caro, I., Quinto, E. J., & Mateo, J. (2017). Quality changes in refrigerated stored minced pork wrapped with plastic cling film and the effect of glucose supplementation. Meat Science, 126, 55–62. https://doi.org/10.1016/j.meatsci.2016.12.007
- Demaître, N., Van Damme, I., De Zutter, L., Geeraerd, A. H., Rasschaert, G., & De Reu, K. (2020). Occurrence, distribution and diversity of listeria monocytogenes contamination on beef and pig carcasses after slaughter. Meat Science, 169, 108177. https://doi.org/10.1016/j.meatsci.2020.108177
- Emamjomeh, M., Mohd Hashim, A., Abdul-Mutalib, N., Khairil Mokhtar, N., Mustapha, N. A., Maeda, T., & Amin Nordin, S. (2023). Profiling bacterial communities and foodborne pathogens on food-associated surface following contact with raw beef, chicken and pork using 16S amplicon metagenomics. Food Control, 149, 109698. https://doi.org/10.1016/j.foodcont.2023.109698
- Ercolini, D. (2013). High-throughput sequencing and metagenomics: Moving forward in the culture-independent analysis of food microbial ecology. Applied and Environmental Microbiology, 79(10), 3148–3155. https://doi.org/10.1128/AEM.00256-13
- Ercolini, D., Casaburi, A., Nasi, A., Ferrocino, I., DiMonaco, R., Ferranti, P., Mauriello, G., & Villani, F. (2010). Different molecular types of Pseudomonas fragi have the same overall behaviour as meat spoilers. International Journal of Food Microbiology, 142(1–2), 120–131. https://doi.org/10.1016/j.ijfoodmicro.2010.06.012
- Illikoud, N., Rossero, A., Chauvet, R., Courcoux, P., Pilet, M.-F., Charrier, T., Jaffrès, E., & Zagorec, M. (2019). Genotypic and phenotypic characterization of the food spoilage bacterium brochothrix thermosphacta. Food Microbiology, 81, 22–31. https://doi.org/10.1016/j.fm.2018.01.015
- Janse, R. J., Hoekstra, T., Jager, K. J., Zoccali, C., Tripepi, G., Dekker, F. W., & van Diepen, M. (2021). Conducting correlation analysis: Important limitations and pitfalls. Clinical Kidney Journal, 14(11), 2332–2337. https://doi.org/10.1093/ckj/sfab085
- Johansson, M., Azzouz, M., Häggendal, B., Säljö, K., Malmi, H., Zavialov, A., & Teneberg, S. (2020). Glycosphingolipids recognized by Acinetobacter baumannii. Microorganisms [Internet], 8(4), 612. https://doi.org/10.3390/microorganisms8040612
- Kadariya, J., Smith, T. C., & Thapaliya, D. (2014). Staphylococcus aureus and Staphylococcal Food-Borne disease: An ongoing challenge in public health. Biomed Research International, 2014, 827965. https://doi.org/10.1155/2014/827965
- Kameník, J., Dušková, M., Šedo, O., Saláková, A., Pavlík, Z., Zdráhal, Z., & Karpíšková, R. (2015). Lactic acid bacteria in hot smoked dry sausage (non-fermented salami): Thermal resistance of Weissella viridescens strains isolated from hot smoked dry sausages. LWT - Food Science and Technology, 61(2), 492–495. https://doi.org/10.1016/j.lwt.2014.12.012
- Kang, G., Seong, P.-N., Moon, S., Cho, S., Ham, H.-J., Park, K., Kang, S. M., & Park, B.-Y. (2014). Distribution channel and microbial characteristics of pig by-products in Korea. Korean Journal for Food Science of Animal Resources, 34(6), 792–798. https://doi.org/10.5851/kosfa.2014.34.6.792
- Khatoon, Z., McTiernan, C. D., Suuronen, E. J., Mah, T.-F., & Alarcon, E. I. (2018). Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon, 4(12), e01067. https://doi.org/10.1016/j.heliyon.2018.e01067
- Knight, R., Vrbanac, A., Taylor, B., Aksenov, A., Callewaert, C., Debelius, J., González, A., Kosciolek, T., McCall, L.-I., McDonald, D., Melnik, A., Morton, J., Navas, J., Quinn, R., Sanders, J., Swafford, A., Thompson, L., Tripathi, A. … Caporaso, J. G. (2018). Best practices for analysing microbiomes. Nature Reviews Microbiology, 16(7), 410–422. https://doi.org/10.1038/s41579-018-0029-9
- Lagier, J.-C., Khelaifia, S., Tidjani Alou, M., Ndongo, S., Niokhor, D., Hugon, P., Caputo, A., Cadoret, F., Traore, S., Seck, E., Dubourg, G., Durand, G., Gaël, M., Guilhot, E., Togo, A., Bellali, S., Bachar, D., Cassir, N. … Levasseur, A. (2016). Culture of previously uncultured members of the human gut microbiota by culturomics. Nature Microbiology, 1(12). https://doi.org/10.1038/nmicrobiol.2016.203
- Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., Clemente, J. C., Burkepile, D. E., Vega Thurber, R. L., Knight, R., Beiko, R. G., & Huttenhower, C. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology, 31(9), 814–821. https://doi.org/10.1038/nbt.2676
- Li, N., Zhang, Y., Wu, Q., Gu, Q., Chen, M., Zhang, Y., Sun, X., & Zhang, J. (2019). High-throughput sequencing analysis of bacterial community composition and quality characteristics in refrigerated pork during storage. Food Microbiology, 83, 86–94. https://doi.org/10.1016/j.fm.2019.04.013
- Li, Y., Tan, X., Zhao, X., Xu, Z.-F., Dai, W., Duan, W., Huang, S., Zhang, E., Liu, J., Zhang, S., Yin, R., Shi, X., Lu, Z., & Pan, Y. (2020). Composition and function of oral microbiota between gingival squamous cell carcinoma and periodontitis. Oral Oncology, 107, 104710. https://doi.org/10.1016/j.oraloncology.2020.104710
- Lim, S.-M., Lee, N.-K., & Paik, H.-D. (2019). Antibacterial and anticavity activity of probiotic lactobacillus plantarum 200661 isolated from fermented foods against streptococcus mutans. LWT, 118, 108840. https://doi.org/10.1016/j.lwt.2019.108840
- Mahros, M., Abd-Elghany, S., & Sallam, K. (2021). Multidrug-, methicillin-, and vancomycin-resistant Staphylococcus aureus isolated from ready-to-eat meat sandwiches: An ongoing food and public health concern. International Journal of Food Microbiology, 346, 109165. https://doi.org/10.1016/j.ijfoodmicro.2021.109165
- Møretrø, T., & Langsrud, S. (2017). Residential bacteria on surfaces in the food industry and their implications for food safety and quality: Residential bacteria in food industry …. Comprehensive Reviews in Food Science and Food Safety, 16(5), 1022–1041. https://doi.org/10.1111/1541-4337.12283
- Naïtali, M., Dubois-Brissonnet, F., Cuvelier, G., & Bellon-Fontaine, M.-N. (2009). Effects of pH and oil-in-water emulsions on growth and physicochemical cell surface properties of listeria monocytogenes: Impact on tolerance to the bactericidal activity of disinfectants. International Journal of Food Microbiology, 130(2), 101–107. https://doi.org/10.1016/j.ijfoodmicro.2009.01.008
- Nearing, J. T., Connors, J., Whitehouse, S., Van Limbergen, J., Macdonald, T., Kulkarni, K., & Langille, M. G. I. (2019). Infectious complications are associated with alterations in the gut microbiome in pediatric patients with acute lymphoblastic leukemia [Original Research]. Frontiers in Cellular and Infection Microbiology, 9, 9. https://doi.org/10.3389/fcimb.2019.00028
- O’Brien, T. (2002). Emergence, spread, and environmental effect of antimicrobial resistance: How use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America, 34(Suppl 3), S78–84. https://doi.org/10.1086/340244
- Peruzy, M. F., Houf, K., Joossens, M., Yu, Z., Therese, Y., Proroga, R., & Murru, N. (2020). Evaluation of microbial contamination of different pork carcass areas through culture-dependent and independent methods in small-scale slaughterhouses. International Journal of Food Microbiology, 336. https://doi.org/10.1016/j.ijfoodmicro.2020.108902
- Sato, Y., Ishihara, M., Nakamura, S., Fukuda, K., Kuwabara, M., Takayama, T., Hiruma, S., Murakami, K., Fujita, M., & Yokoe, H. (2019). Comparison of various disinfectants on bactericidal activity under organic matter contaminated environments. Biocontrol Science, 24(2), 103–108. https://doi.org/10.4265/bio.24.103
- Stellato, G., La Storia, A., De Filippis, F., Borriello, G., Villani, F., Ercolini, D., & Elkins, C. A. (2016). Overlap of spoilage-associated microbiota between meat and the meat processing environment in small-scale and large-scale retail distributions. Applied and Environmental Microbiology, 82(13), 4045–4054. https://doi.org/10.1128/AEM.00793-16
- Tassou, C., Argyri, A., Doulgeraki, A., Nychas, G.-J., Chorianopoulos, N., Grounta, A., Dourou, D., & Spyrelli, E. (2021). Microbiota of chicken breast and thigh fillets stored under different refrigeration temperatures by next generation sequencing. Foods, 10(4), 10. https://doi.org/10.3390/foods10040765
- Tong, C., Hu, H., Chen, G., Li, Z., Aifeng, L., & Zhang, J. (2021). Chlorine disinfectants promote microbial resistance in pseudomonas sp. Environmental Research, 199, 111296. https://doi.org/10.1016/j.envres.2021.111296
- Tong, C., Hu, H., Chen, G., Li, Z., Li, A., & Zhang, J. (2021). Disinfectant resistance in bacteria: Mechanisms, spread, and resolution strategies. Environmental Research, 195, 110897. https://doi.org/10.1016/j.envres.2021.110897
- Tong, S., Ma, L., Ronholm, J., Hsiao, W., & Lu, X. (2021). Whole genome sequencing of campylobacter in agri-food surveillance. Current Opinion in Food Science, 39, 130–139. https://doi.org/10.1016/j.cofs.2020.12.020
- Velez, F. J., Bosilevac, J. M., & Singh, P. (2021). Validation of high-resolution melting assays for the detection of virulent strains of Escherichia coli O26 and O111 in beef and pork enrichment broths. Food Control, 128, 108123. https://doi.org/10.1016/j.foodcont.2021.108123
- Vojkovska, H., Karpiskova, R., Kajsiková, M., & Drahovska, H. (2015). Characterization of cronobacter spp. isolated from food of plant origin and environmental samples collected from farms and from supermarkets in the Czech Republic. International Journal of Food Microbiology, 217, 130–136. https://doi.org/10.1016/j.ijfoodmicro.2015.10.017
- Wang, Z., Fang, Y., Zhi, S., Simpson David, J., Gill, A., McMullen Lynn, M., Neumann Norman, F., Gänzle Michael, G., & Björkroth, J. (2020). The locus of heat resistance confers resistance to chlorine and other oxidizing chemicals in Escherichia coli. Applied and Environmental Microbiology, 86(4), e02123–02119. https://doi.org/10.1128/AEM.02123-19
- Wickramasinghe, N. N., Ravensdale, J., Coorey, R., Dykes, G. A., & Chandry, P. S. (2021). Transcriptional profiling of biofilms formed on chilled beef by psychrotrophic meat spoilage bacterium, Pseudomonas fragi 1793. Biofilm, 3, 100045. https://doi.org/10.1016/j.bioflm.2021.100045
- Wilson, C. N., Pulford, C. V., Akoko, J., Perez Sepulveda, B., Predeus, A. V., Bevington, J., Duncan, P., Hall, N., Wigley, P., Feasey, N., Pinchbeck, G., Hinton, J. C. D., Gordon, M. A., Fevre, E. M., & Bourret, T. J. (2020). Salmonella identified in pigs in Kenya and Malawi reveals the potential for zoonotic transmission in emerging pork markets. PloS Neglected Tropical Diseases, 14(11), e0008796. https://doi.org/10.1371/journal.pntd.0008796
- Wu, J., Li, R., Zhang, M., Shan, K., Jia, X., Zhao, D., Nian, Y., & Li, C. (2021). Microbiota changes on the surface of pig carcasses during refrigerated transportation and marketing. Food Materials Research[Cdata[food materials research]], 1(1), 1–9. https://doi.org/10.48130/FMR-2021-0004
- Xiao, X., Wang, W., Zhang, J., Liao, M., Rainwater, C., Yang, H., & Li, Y. (2021). A quantitative risk assessment model of salmonella contamination for the yellow-feathered broiler chicken supply chain in China. Food Control, 121, 107612. https://doi.org/10.1016/j.foodcont.2020.107612
- Xu, L., Zhang, C. M., & Xu, P. (2017). Mechanisms of ultraviolet disinfection and chlorination of Escherichia coli: Culturability, membrane permeability, metabolism, and genetic damage. Journal of Environmental Sciences, 65. https://doi.org/10.1016/j.jes.2017.07.006
- Xu, S., Lu, W., Liu, Y., Ming, Z., Liu, Y., Meng, R., & Wang, H. (2016). Structure and diversity of bacterial communities in two large sanitary landfills in China as revealed by high-throughput sequencing (MiSeq). Waste Management, 63, 41–48. https://doi.org/10.1016/j.wasman.2016.07.047
- Xu, Y., Yang, G., Xu, J., Guan, X., Li, R., & Wang, S. (2022). Influence of the combination of cinnamon essential oil nanoemulsions and epsilon-polylysine on microbial community and quality of pork during refrigerated period and radio frequency cooking. International Journal of Food Microbiology, 381, 109911. https://doi.org/10.1016/j.ijfoodmicro.2022.109911
- Yang, C., Che, Y., Qi, Y., Liang, P., & Song, C. (2016). High-throughput sequencing of viable microbial communities in raw pork subjected to a fast cooling process. Journal of Food Science, 82. https://doi.org/10.1111/1750-3841.13566
- Yi, Z., Yan, J., Ding, Z., & Xie, J. (2022). The HD-GYP domain protein of shewanella putrefaciens YZ08 regulates biofilm formation and spoilage activities. Food Research International, 157, 111466. https://doi.org/10.1016/j.foodres.2022.111466
- Zhang, L., Fu, Y., Xiong, Z., Ma, Y., Wei, Y., Qu, X., Zhang, H., Zhang, J., & Liao, M. (2018). Highly prevalent multidrug-resistant salmonella from chicken and pork meat at retail markets in Guangdong, China. Frontiers in Microbiology, 9, 9. https://doi.org/10.3389/fmicb.2018.02104
- Zhang, Y., Hou, W., Yue, Q., Zhang, Y., Yi, Y., Min, T., & Wang, H. (2021). Effects of environmental humidity on microbial community and quality of chilled fresh pork. SSRN Electronic Journal. https://doi.org/10.2139/ssrn.3964470
- Zhao, F., Wei, Z., Zhou, G., Kristiansen, K., & Wang, C. (2022). Effects of different storage temperatures on bacterial communities and functional potential in pork meat. Foods, 11(15), 2307. https://doi.org/10.3390/foods11152307
- Zhao, F., Zhou, G., Ye, K., Wang, S., Xu, X., & Li, C. (2015). Microbial changes in vacuum-packed chilled pork during storage. Meat Science, 100, 145–149. https://doi.org/10.1016/j.meatsci.2014.10.004
- Zhong, A., Chen, W., Duan, Y., Li, K., Tang, X., Tian, X., Wu, Z., Li, Z., Wang, Y., & Wang, C. (2021). The potential correlation between microbial communities and flavors in traditional fermented sour meat. LWT, 149, 111873. https://doi.org/10.1016/j.lwt.2021.111873
- Zhou, J., Jin, Y., & Liang, Q. (2022). Effects of regulatory policy mixes on traceability adoption in wholesale markets: Food safety inspection and information disclosure. Food Policy, 107, 102218. https://doi.org/10.1016/j.foodpol.2022.102218
- Zhu, Z., Shan, L., Zhang, X., Hu, F., Zhong, D., Yuan, Y., & Zhang, J. (2021). Effects of bacterial community composition and structure in drinking water distribution systems on biofilm formation and chlorine resistance. Chemosphere, 264, 128410. https://doi.org/10.1016/j.chemosphere.2020.128410