ABSTRACT
Group 1 Innate Lymphoid Cells (which include Natural Killer cells and ILC1s) aid in gut anti-bacterial defense through the production of IFNγ, which is critical for mobilizing protective responses against enteric pathogens. When intestinal epithelial barrier integrity is compromised, commensal bacteria are likely to translocate from the gut lumen into the lamina propria. Few studies have addressed the mechanisms by which commensal bacteria impact the function of gut Group 1 ILCs, especially ILC1s. Utilizing an in vitro human colonic lamina propria mononuclear cell (LPMC) model, we evaluated Group 1 ILC cytokine and cytolytic protein production in response to a panel of enteric Gram-positive and Gram-negative commensal and pathogenic bacteria. IFNγ-production by NK cells and ILC1s was significantly increased after LPMC exposure to Gram-negative commensal or pathogenic bacteria, but not after exposure to the Gram-positive bacteria commensals tested. Stimulation of IFNγ production from Group 1 ILCs was not through direct recognition of bacteria by NK cells or ILC1s, but rather required accessory cells within the LPMC population. Myeloid dendritic cells generated IL-12p70, IL-18, and IL-1β upon exposure to enteric bacteria and these cytokines contributed to Group 1 ILC production of IFNγ. Furthermore, Gram-negative commensal or pathogenic bacteria induced significant expression of Granzyme B in NK cells and ILC1s. Overall, these data demonstrate that some enteric commensal bacteria indirectly induce inflammatory cytokine production and cytolytic protein expression from human colonic Group 1 ILCs, a process which could contribute to inflammation in the setting of microbial translocation.
Introduction
Group 1 Innate Lymphoid Cells (ILCs), which include the well-studied Natural Killer (NK) cells and the more recently discovered ILC1 subset, have an established role in anti-viral immunity,Citation1,Citation2 but are also important in anti-bacterial immunityCitation3 particularly in the gastrointestinal tract.Citation4,Citation5 Within the gut mucosa, one mechanism by which Group 1 ILCs control bacterial infection is through the production of IFNγ. For example, depletion of NK cells increased bacterial disseminationCitation6 and increased intestinal pathology (attributed to loss of early IFNγ production) during murine infection with the enteric pathogen Citrobacter rodentium.Citation7 Depletion of NK cells increased susceptibility of mice to infection with Salmonella typhimuriumCitation8,Citation9 and was attributed to loss of protective IFNγ production by murine NK cells.Citation10,Citation11 Using human peripheral blood-derived macrophages infected with S. typhimurium, production of IFNγ and degranulation by NK cells was shown to control infection.Citation8,Citation12 This control was dependent on macrophage secretion of IL-12 and IL-18Citation8 or macrophage secretion of IL-23, IL-18, and IL-1β.Citation12 Although less well studied, ILC1s are also important in limiting intestinal infections. IFNγ production by gut ILC1s was critical for defense during infection with the parasite Toxoplasma gondii,Citation13 and mice lacking IFNγ-expressing ILC1s had increased susceptibility to infection with Clostridium difficile.Citation14 Furthermore, other subsets within the ILC family (Group 3 ILCs) that gained the IFNγ- producing capacity of ILC1s, protected mice during S. typhimurium infection.Citation15 These studies highlight that IFNγ production by Group 1 ILCs in response to enteric pathogens is a protective feature of the gut immune response.
Since Group 1 ILCs produce IFNγ in response to pathogenic bacteria as described above, it is possible that these cells may contribute to localized inflammation during disease states, such as Inflammatory Bowel Disease (IBD) or Human Immunodeficiency Virus (HIV) infection, in which gut bacteria translocate across a damaged epithelium. Supporting the role of Group 1 ILCs in driving disease-associated gut pathology, frequencies of NK cells or ILC1s were increased in Crohn’s disease or Ulcerative colitis (types of IBD) and in some studies, IFNγ-producing Group 1 ILCs were increased.Citation16–Citation19 In the context of HIV, we have reported an increase in IFNγ-producing NKp44+ CD56+ ILCs (a population of cells likely to include both NK cells and ILC1s) in the colonic mucosa of people living with HIV (PLWH) who are naïve to treatment.Citation20 Increases in IFNγ-producing and cytotoxic colonic ILCs (defined as CD3-NKp44+) were also reported in Rhesus macaques infected with SIV (the non-human primate model of HIV).Citation21,Citation22 Conversely, in PLWH on anti-retroviral treatment with viral suppression the percentage of IFNγ-producing ILC1s in the gut mucosa were not different compared to uninfected controls.Citation23 These studies suggest that accumulation of Group 1 ILCs in IBD or untreated HIV may contribute to gut pathology.
Given the large numbers of commensal bacteria resident within the gut lumen, it is likely that commensal bacteria translocate into the lamina propria when the epithelial barrier is damaged. Changes in the gut microbiome, termed dysbiosis, during IBD and HIV have been shown to generally reflect an increase in certain Gram-negative commensal bacteria (e.g., Proteobacteria) including those with pathogenic potential, termed “pathobionts,” and a corresponding decrease in some Gram-positive bacteria with immunomodulatory properties (e.g., Firmicutes).Citation24,Citation25 Studies investigating the effect of the gut microbiome on Group 1 ILC function have demonstrated that murine splenic NK cells from germ-free or antibiotic-treated mice had reduced IFNγ production and impaired cytotoxic function,Citation26 and was shown to be dependent on the interaction between the microbiota and mononuclear phagocytes. In addition, murine oral exposure to commensal bacteria Lactobacillus pentosus induced IFNγ production and cytotoxicity from splenic NK cells.Citation27 Using human peripheral blood, we have previously demonstrated that exposure to commensal Escherichia coli induced IFNγ production from NK cells and was dependent on contact with blood monocytes.Citation28 Others have shown that exposure of human peripheral blood to strains of lactic acid bacteria such as Lactobacillus acidophilus induced IFNγ production by NK cells, whereas exposure to other strains including Bifidobacterium bifidum and Lactobacillus reuteri did not.Citation29 This human peripheral blood NK IFNγ response was linked to the production of IL-12p70 by myeloid dendritic cells (mDCs),Citation29 indicating that differing responses of NK cells to commensal bacteria are possible and may be dependent on the cytokine milieu driven by mDCs. This group additionally demonstrated that exposure of human peripheral blood to lactic acid bacteria stains induced cytolytic NK cell responses.Citation29 There are limited data on the characterization of cytokine production and cytolytic molecule expression by NK cells of the human gut, particularly in response to commensal bacteria. However, it has been demonstrated that in vitro exposure of gut NK cells from patients with Crohn’s disease to Enterococcus faecalis or E. coli enhanced IFNγ production.Citation17 To the best of our knowledge, no studies have investigated the effector response of the other Group 1 subset, ILC1s, to commensal bacteria.
In this study, we sought to understand how gut Group 1 ILCs respond to enteric commensal bacteria, which is of relevance to disease states where commensal bacteria translocate through a compromised gut epithelial barrier. We hypothesized that both pathogenic and commensal enteric bacteria would induce cytokine production (IFNγ) and cytolytic potential (Granzyme B) from Group 1 ILCs. To address this, we utilized an in vitro human colonic lamina propria mononuclear cell (LPMC) model to investigate NK cell and ILC1 cytokine profiles and cytolytic potential after exposure to pathogenic S. typhimurium, as well as, a panel of human enteric Gram-negative and Gram-positive commensal bacteria. Overall, our observations expand on the understanding of how human colonic Group 1 ILCs respond to enteric commensal bacteria and contribute to the gut mucosal immune response.
Results
Identification of Group 1 ILCs in human colonic tissue
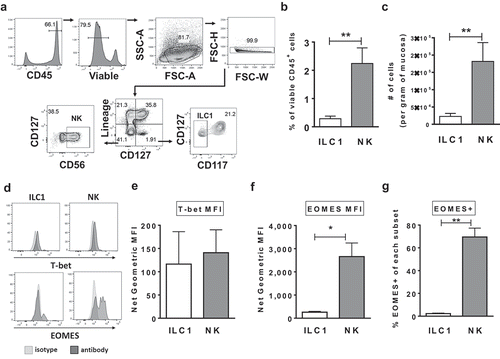
Human Group 1 ILCs were identified in colonic lamina propria mononuclear cells (LPMCs) using multi-color flow cytometry. NK cells were defined within viable, leukocytes (CD45+) as Lineage- (CD3, CD20, CD13, CD123, CD303, CD34, FCεR1α, CD11c, CRTH2) CD127-CD56+ and ILC1s as viable CD45+ Lin-CD127+CD117- ()).Citation30,Citation31 On average NK cells constituted 2.25% ± 0.62 (mean ± S.E.M.) of viable CD45+ colonic LPMCs ex vivo and were more frequent than ILC1s (0.28% ± 0.09) both as a percent ()) and as number of cells per gram of colonic mucosa ()). Examination of transcription factors relevant to IFNγ cytokine production and cytolytic functionCitation31 demonstrated that human colonic LP Group 1 ILCs expressed T-bet ex vivo ()) and expression levels (mean fluorescent intensity) were not significantly different between NK cells and ILC1 ()). By comparison, a large distinct population of NK cells expressed the transcription factor EOMES ex vivo ()) compared to lower expression in a small percentage of ILC1s ().
Figure 1. Characterization of Group 1 ILCs within the human colonic lamina propria. (a) Representative flow cytometry gating strategy to identify NK cells and ILC1s in human colonic tissue. (b) Frequencies of NK cells and ILC1s in the colon lamina propria layer as a percent of viable CD45+ cells and (c) as number of cells per gram of colonic mucosa ex vivo. N = 7. (d) Representative flow cytometry depicting the expression of T-bet and EOMES by NK cells and ILC1s. (e) Net geometric mean fluorescent intensity of NK cells and ILC1s expressing the transcription factors T-bet (N = 7) or (f) EOMES (N = 4) ex vivo. (g) Percentages of NK cells and ILC1s expressing the EOMES (N = 4) ex vivo. Bars are mean + S.E.M. Statistical analysis performed was paired t-test. *p < .05, **p < .01.
S. typhimurium induced IFNγ and TNFα production by human colonic Group 1 ILCs
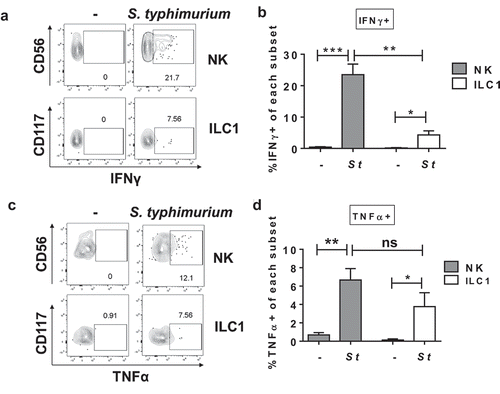
Since human peripheral blood NK cells, splenic murine NK cells, and murine intestinal ILC1s induced IFNγ when exposed to S. typhimurium,Citation8,Citation9,Citation15,Citation28 we determined whether in vitro exposure to S. typhimurium induced IFNγ by human colonic Group 1 ILCs using flow cytometry. Exposure of LPMCs to S. typhimurium for 20 h in the presence of broad-spectrum antibiotics significantly increased the percentage of IFNγ-expressing NK cells and ILC1s above the no stimulation control, and significantly more NK cells than ILC1s expressed IFNγ (). TNFα, another cytokine with proinflammatory potential and reported to be expressed by gut Group 1 ILCsCitation32 was next measured. There was a significant increase in the percentage of TNFα- expressing NK cells and ILC1s upon S. typhimurium exposure () and there was no significant difference between the percentage of NK and ILC1 cells expressing-TNFα ()). On average 14.9% ± 2.0 and 17.4% ± 14.1 of the IFNγ-expressing NK or ILC1s expressed TNFα, respectively (data not shown). These data indicate that exposure to the enteric pathogen S. typhimurium induced pro-inflammatory cytokine production by human colonic Group 1 ILCs, dominated by higher frequencies of IFNγ-expressing than TNFα-expressing NK cells and ILC1s.
Figure 2. S. typhimurium induces IFNγ and TNFα production by human colonic Group 1 ILCs. (a) Representative flow cytometry demonstrating cytokine staining for IFNγ gated on NK cells or ILC1s after LPMC exposure to S. typhimurium (St) or no bacterial control (-) in vitro. (b) Percentages of IFNγ+ NK cells or ILC1s after LPMC exposure to S. typhimurium or no bacterial control. N = 10. (c) Representative flow cytometry demonstrating cytokine staining for TNFα gated on NK cells or ILC1s after LPMC exposure to S. typhimurium or no bacterial control in vitro. (d) Percentages of TNFα+ NK cells or ILC1s after LPMC exposure to S. typhimurium or no bacterial control. N = 7. Bars are mean + S.E.M. Statistical analysis performed was one-way ANOVA comparing the mean of each column. *p < .05, **p < .01, ***p < .001, n.s. = not significant.
IL-12p70, IL-18, and IL-1β contributed to induction of IFNγ by Group 1 ILCs in response to S. typhimurium
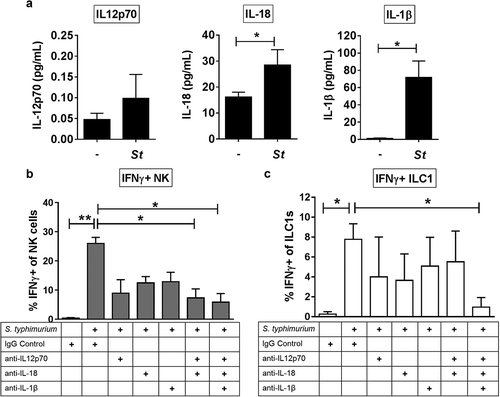
IFNγ production by NK cells in human peripheral blood has been demonstrated to depend on IL-12p70 and variably with co-stimulation from either IL-18 or IL-1β.Citation8,Citation12,Citation33 Therefore, we first asked whether LPMC exposure to recombinant IL-12p70, IL-18 or IL-1β induced IFNγ-producing gut Group 1 ILCs. The addition of individual recombinant cytokines to LPMCs increased the average percentage of NK cells (IL-12p70; 25.9 fold, IL-18; 23.9 fold, and IL-1β; 25.4 fold) and ILC1s (IL-12p70; 21.0 fold, IL-18; 10.0 fold, and IL-1β; 9.12 fold) expressing IFNγ (Figure S1). To determine if S. typhimurium induced production of IL-12p70, IL-18, and IL-1β from human colonic LPMCs, secretion of these cytokines was measured in culture supernatants after bacterial exposure. S. typhimurium exposure induced a modest increase (2.1 fold) in IL-12p70 from LPMCs, and significantly increased IL-18 (1.7 fold) and IL-1β (64.0 fold) production by LPMCs ()). Antibody-mediated blocking of either IL-12p70, IL-18, or IL-1β soluble cytokines reduced the frequencies of IFNγ-producing NK cells and ILC1s generated in response to S. typhimurium (,)). Blocking the combination of IL-12p70 and IL-18 significantly reduced the percentage of IFNγ-producing NK cells in response to S. typhimurium by 72% ± 20.4 and blocking all three cytokines, IL-12p70, IL-18, and IL-1β significantly reduced the percentage of IFNγ-producing NK cells by 82.7% ± 14.3 ()). Frequencies of IFNγ-producing ILC1s were significantly reduced by 86.6% ± 13.3 when blocking the combination of IL-12p70, IL-18 and IL-1β ()). Together these data indicate that IL-12p70, IL-18, and IL-1β generated in response to the enteric pathogen S. typhimurium in vitro contributed to the induction of IFNγ by human colonic Group 1 ILCs.
Figure 3. The cytokines IL-12p70, IL-18 and IL-1β contribute to the induction of IFNγ by Group 1 ILCs in response to S. typhimurium. (a) Amount (pg/mL) of IL-12p70, IL-18, or IL-1β secreted into the supernatant after LPMC exposure to S. typhimurium (St) or no bacterial control (-) in vitro. N = 6. (b) Percentages of IFNγ+ NK cells or (c) ILC1s after LPMC exposure to no bacteria control or S. typhimurium in the presence of 10ug/mL blocking antibodies targeting IL-12p70, IL-18, IL-1β or the antibody isotype control IgG. N = 3. Bars are mean + S.E.M. Statistical analysis performed was (a) paired t-test or (b) one-way ANOVA comparing the mean of each column with the mean of the control column S. typhimurium + IgG isotype control. *p < .05, **p < .01.
Differential IFNγ induction by Group 1 ILCs in response to a panel of enteric commensal bacteria
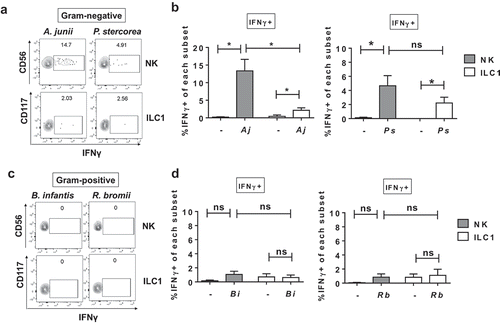
To begin to probe human colonic Group 1 ILC responses to enteric commensal bacteria, we focused on a panel of bacteria that included Gram-negative (Acinetobacter junii, Prevotella stercorea) and Gram-positive (Ruminococcus bromii) bacteria previously shown to be altered in abundance in the colonic mucosa of PLWH,Citation34,Citation35 as well as Gram-positive Bifidobacterium longum sp infantis, a common component of probiotics with reported anti-inflammatory properties.Citation36 Exposure of LPMCs to matched ratios of either A. junii or P. stercorea significantly increased the percentage of IFNγ-expressing Group 1 ILCs (). The frequencies of IFNγ-expressing NK cells generated in response to A. junii were significantly higher than IFNγ-expressing ILC1s ()). In contrast, exposure to matched ratios of R. bromii or B. infantis did not induce a significant increase in the percentage of IFNγ-producing Group 1 ILCs after LPMC exposure (). To determine whether a higher dose of Gram-positive bacteria could induce an IFNγ response from Group 1 ILCs, LPMCs were exposed to increasing amounts of R. bromii. Frequencies of IFNγ-expressing Group 1 ILCs were not significantly increased upon LPMC exposure to 2×, 5×, and 10× the amount of R. bromii used above (Figure S2). These data suggest that Group 1 ILCs differentially responded to enteric commensal bacterial exposure, and Gram-negative bacteria-induced IFNγ production by Group 1 ILCs, whereas the Gram-positive bacteria tested here did not. In keeping with our observations for S. typhimurium (), blocking the cytokines IL-12p70, IL-18 or IL-1β individually reduced the frequency of Group 1 ILCs expressing IFNγ by more than 74.3% ± 25.6 on average for NK cells and 68.5% ± 2.2 for ILC1s during LPMC exposure to a representative commensal A. junii (Figure S3). Additionally, blocking the combination of IL-12p70, IL-18 and IL-1β reduced the percentage of IFNγ-producing NK cells and ILC1s in response to A. junii by 87.6% ± 6.2 and 87.0% ± 12.9, respectively (Figure S3). Thus indicating that IL-12p70, IL-18, and IL-1β are contributing to the induction of IFNγ by human colonic Group 1 ILCs also in response to representative enteric commensal A. junii.
Figure 4. Differential IFNγ induction by Group 1 ILCs in response to a panel of enteric commensal bacteria. (a) Representative flow cytometry demonstrating cytokine staining for IFNγ gated on NK cells or ILC1s after LPMC exposure to Gram-negative A. junii or P. stercorea in vitro. (b) Percentages of IFNγ+ NK cells or ILC1s after LPMC exposure to no bacterial control (-), A. junii (Aj) or P. stercorea (Ps) N = 7. (c) Representative flow cytometry demonstrating cytokine staining for IFNγ gated on NK cells or ILC1s after LPMC exposure to Gram-positive B. infantis or R. bromii in vitro. (d) Percentages of IFNγ+ NK cells or ILC1s after LPMC exposure to no bacterial control or B. infantis (Bi) or R. bromii (Rb). N = 6. Bars are mean + S.E.M. Statistical analysis performed was one-way ANOVA comparing the mean of each column. *p < .05, n.s. = not significant.
IFNγ production by Group 1 ILCs was not mediated by direct recognition of gram-negative bacteria
Gene expression of pattern recognition receptors known to bind extracellular bacterial antigens (TLR2, TLR4, and TLR5) has been reported to be expressed by human peripheral blood NK cellsCitation3 and that TLR2 is expressed on the surface of human duodenum ILCs.Citation32 Therefore, we next examined human colonic LPMCs ex vivo by multicolor flow cytometry for protein expression of TLR2, TLR4, and TLR5. Minimal expression of TLR2, TLR4, and TLR5 was observed ex vivo on Group 1 ILCs in comparison to Lineage-positive cells ()).
Figure 5. IFNγ production was not mediated by Group 1 ILC direct recognition of Gram-negative bacteria. (a) Percentages of LPMCs stained ex vivo for TLR2, TLR4, or TLR5 expression and gated on NK cells or ILC1s, or Lineage-positive cells that are larger on Forward Scatter-area vs Side Scatter-area flow plots.N = 6. (b) Quantification of IFNγ (pg/mL) in the supernatant of purified NK cells or (c) ILC1s exposed to recombinant IL-12p70 and IL-18 (50ng/mL), A. junii (Aj) or R. bromii (Rb) in vitro. N = 3. (d) Normalization of secreted IFNγ (pg/mL) per the amount of plated NK or ILC1s (cells/mL) to determine IFNγ (pg/cell). N = 3. Bars are mean + S.E.M. Statistical analysis performed was paired t-test or one-way ANOVA comparing the mean of each column. **p < .01, ***p < .001, n.s. = not significant.
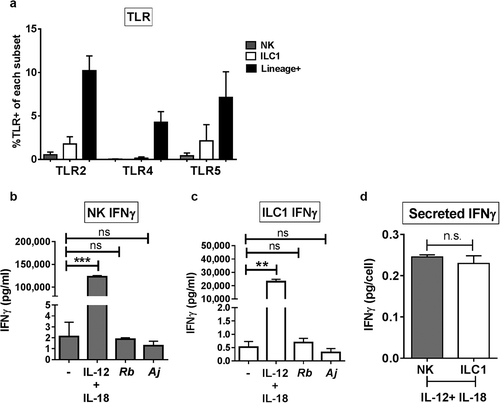
To determine if Group 1 ILCs produced IFNγ through direct recognition of commensal bacteria, ILC1s and NK cells were purified from LPMCs by flow sorting and exposed to either representative Gram-negative (A. junii) or Gram-positive (R. bromii) commensal bacteria or recombinant cytokines IL-12p70 and IL-18. Measurement of secreted IFNγ by purified NK cells demonstrated that NK cells produced IFNγ directly in response to IL-12 and IL-18 but not following exposure to commensal bacteria ()). Similarly, purified ILC1s did not secret IFNγ in response to A. junii or R. bromii, but did in response to IL-12 and IL-18 ()). After normalizing for the amount of cells plated after purification, no significant differences in the amount of IFNγ secreted per cell in response to recombinant IL-12 and IL18 were detected when comparing NK cells and ILC1 ()). These data indicate that Gram-negative bacteria do not directly stimulate IFNγ production by Group 1 ILCs, suggesting a requirement for accessory cells.
Myeloid dendritic cells producing cytokines in response to Gram-negative commensal bacteria promote IFNγ production by Group 1 ILCs
Based on the observation that IL-12p70, IL-18, and IL-1β contributed to Group 1 ILC production of IFNγ in response to S. typhimurium and A. junii (,c) and S3), the cellular source producing these cytokines was next determined after LPMC exposure to commensal bacteria. Gram-negative A. junii exposure significantly increased the percentage of IL-12p70-, IL-18-, and IL-1β-expressing mDCs (defined as CD3-CD19-HLA-DRhighCD11chigh)Citation37–Citation39 ()). Exposure to Gram-positive R. bromii did not significantly increase the frequency of IL-12p70 or IL-18 expressing mDCs ()), and a trend for an increase in IL-1β production by mDCs was observed. No significant induction of these cytokines by macrophages (defined as CD3-CD19-HLA-DR+CD11c−/low) was detected upon R. bromii or A. junii exposure ()).
Figure 6. Myeloid dendritic cells producing cytokines in response to Gram-negative commensal bacteria promote IFNγ production by Group 1 ILCs. (a) Percentages of IL-12p70+, IL-18+, or IL-1β+ mDCs or macrophages after LPMC exposure to no bacteria control (-), A. junii (Aj) or R. bromii (Rb). N = 4. (b) Percentages of IFNγ+ NK cells or ILC1s after LPMC exposure to A. junii with no depletion control (total LPMCs) or mDC depletion. N = 3. Bars are mean + S.E.M. Statistical analysis performed was one-way ANOVA comparing the mean of each column with the mean of the control column (no bacteria). *p < .05, #p = .0637.
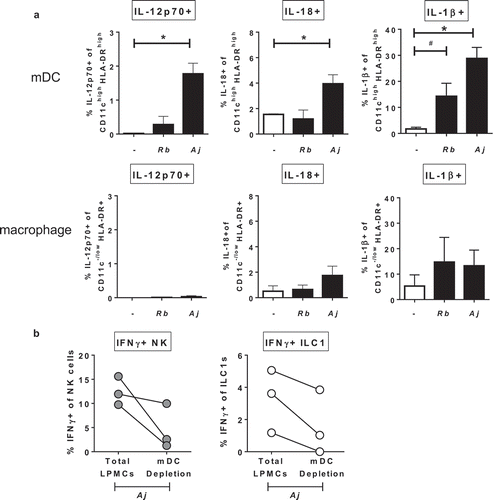
To verify the contribution of mDCs to bacteria-induced IFNγ production by Group 1 ILCs, CD11c+ mDCs were depleted from LPMCs and cytokine production by Group 1 ILCs was determined after exposure to A. junii. Compared to Group 1 ILC cytokine production within total LPMCs, depletion of mDCs reduced the frequencies of IFNγ-producing NK generated in response to A. junii by 62.3% ± 22.9 ()). Similarly, depletion of mDCs reduced the frequency of IFNγ-producing ILC1s by 65.2% ± 22.1 as compared to total LPMCs ()). These data suggest that CD11c+ mDCs within LPMCs are a major producer of cytokines that can drive IFNγ production by human colonic LP Group 1 ILCs in response to commensal bacteria.
Gram-negative bacteria induce Granzyme Bexpression in Group 1 ILCs
Since studies in mice have implicated the gut microbiome in influencing NK cell cytolytic function,Citation26,Citation27 and to the best of our knowledge, no studies have investigated how commensal bacteria impact ILC1 cytolytic function, production of Granzyme B by Group 1 ILCs was next examined. In the absence of bacterial exposure, low percentages of NK cells (5.0%± 1.9) and ILC1s (0.2% ± 0.2) expressing Granzyme B were detected. Intracellular expression of Granzyme B in NK cells and ILC1s increased significantly with exposure to Gram-negative bacteria A. junii and S. typhimurium ()), but did not increase with LPMC exposure to Gram-positive R. bromii. Since effective cytolytic function requires co-expression of Granzyme B and Perforin, the percentage of Group 1 ILCs expressing Perforin were next evaluated. In the absence of bacterial exposure, a distinct population of Perforin-expressing colonic NK cells and ILC1 were detected indicating that a population of Group 1 ILCs could be primed for cytolytic activity ()). However, after LPMC exposure to either R. bromii, A. junii or S. typhimurium there were no significant increases in frequencies of Perforin-expressing NK cells or ILC1s ()). Examination of co-expression of Granzyme B and Perforin ()) demonstrated that there was a significant increase in NK cells and ILC1s co-expressing Granzyme B and IFNγ after LPMC exposure to Gram-negative bacteria ()). Furthermore, the majority of Granzyme B-expressing NK cells (Aj: 92.2% ± 3.4, St: 95.9%± 0.6) and ILC1s (Aj: 72.2% ± 14.7, St: 51.0% ± 12.5) also expressed Perforin in response to Gram-negative bacteria. Thus, commensal and pathogenic Gram-negative bacteria-induced cytolytic potential in both NK and ILC1 cells.
Figure 7. Gram-negative bacteria induce Granzyme B-expressing Group 1 ILCs. (a) Percentages of Granzyme B+ or (b) Perforin+ NK cells or ILC1s after LPMC exposure to no bacteria control (-), R. bromii (Rb), A. junii (Aj) or S. typhimurium (St) in vitro. N = 4. (c) Representative flow cytometry demonstrating co-staining for Granzyme B and Perforin gated on NK cells or ILC1s after LPMC exposure to no bacteria control, R. bromii, A. junii, or S. typhimurium in vitro and (d) Percentages of NK cells or ILC1s expressing both Perforin and Granzyme B. N = 4. (e) Representative flow cytometry demonstrating co-staining for Granzyme B and IFNγ gated on NK cells or ILC1s after LPMC exposure to no bacteria control, R. bromii, A. junii, or S. typhimurium in vitro and (f) Percentages of NK cells or ILC1s expressing both IFNγ and Granzyme B. N = 4. Bars are mean + S.E.M. Statistical analysis performed was one-way ANOVA comparing the mean of each column with the mean of the control column (no bacteria). *p < .05, **p < .01, #p = .0803.
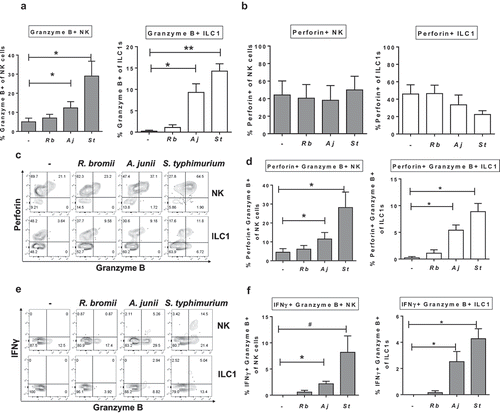
To determine if Group 1 ILCs expressing Granzyme B represented a unique subset, different from the IFNγ-producing cells, co-expression of Granzyme B and IFNγ was next evaluated. Examination of co-expression of Granzyme B and IFNγ ()) demonstrated that there was a significant increase in NK cells and ILC1s co-expressing Granzyme B and Perforin after LPMC exposure to Gram-negative bacteria ()). Additionally, the majority of IFNγ-producing NK cells (Aj: 80.4% ± 7.0, St: 69.2% ± 2.4) or ILC1s (Aj: 55.0% ± 26.3, St: 78.3% ± 15.7) generated in response to A. junii and S. typhimurium exposure were also positive for Granzyme B expression. In keeping with a lack of IFNγ ()) or Granzyme B ()) expression in response to R. bromii, LPMC exposure to R. bromii did not significantly increase the percentages of NK cells or ILC1s co-expressing IFNγ and Granzyme B (). These data suggest that exposure to Gram-negative bacteria can induce polyfunctional Group 1 ILCs.
Discussion
This study is the first to undertake an extensive evaluation of the mechanisms by which in vitro exposure to a representative panel of enteric bacteria including Gram-positive commensals, Gram-negative commensals, and a Gram-negative pathogen drive expression of cytokines from human colonic Group 1 ILCs. Following LPMC exposure to bacteria, IFNγ production by Group 1 ILCs was induced only in response to the Gram-negative bacteria tested here and was driven indirectly, mediated in part by mDC production of the cytokines IL-12p70, IL-18, and IL-1β. Interestingly the Gram-positive bacteria tested did not induce IFNγ which is likely related to differences in mDC cytokine profiles triggered by distinct bacterial species. IFNγ has important anti-viral and anti-bacterial functions in gut immunity, and thus the production of IFNγ by Group 1 ILCs would be a critical component of the innate response to enteric pathogenic challenge. However, in the context of epithelial barrier damage, dysregulation of epithelial tight junctions by IFNγ production from Group 1 ILCs could potentially serve as an additional factor contributing to unregulated gut inflammation in response to translocating commensals.
Here, we add to the body of evidence that ILC1s exist within the human colonCitation16,Citation19 and demonstrate that ILC1s have the ability to produce IFNγ and Granzyme B in response to LPMC exposure to Gram-negative bacteria in vitro. ILC1s were first identified in human tonsils and within the ileum as an IFNγ-producing lymphocyte. The existence of this subset of ILC1s was recently called into question in one study,Citation40 which was unable to detect these cells in any normal and pathological human tissue using CyTOF. However, re-examination of that data set with an alternative gating strategy did identify a unique ILC1 cluster.Citation41 Similar to previous studies reporting varied transcriptional profiles of Group 1 ILCs in both humans and mice,Citation42,Citation43 we noted that the human colon had NK cells and ILC1s with similar levels of T-bet expression, whereas a distinct and larger EOMES-expressing NK cell population was detected relative to that of the ILC1 population.
In addition to expanding what is known about the less-studied ILC1 cell type, we provide a direct functional comparison between NK and ILC1 responses to bacterial stimulation. Our observations of higher frequencies of IFNγ-expressing NK cells versus ILC1s in response to our panel of Gram-negative bacteria suggests that colonic NK cells may be more readily primed to respond than ILC1s. Dissimilarities in IFNγ-producing frequencies may be related to cytokine receptor expression, particularly for IL-12RB2, as higher levels of this receptor have been reported on tonsillar NK cells than ILC1s.Citation42 This difference could translate to a greater response to IL-12 by NK cells than ILC1s. Recent studies examining the developmental progression of these cells in mice have demonstrated that NK cells diverge early from the common innate lymphoid progenitor which later begets the other ILC subsets.Citation13,Citation44 These developmental differences along with differences in that magnitude of cytokine-producing or cytolytic responsesCitation45 have compelled scientists to recently separate these cells into two different groups within the larger ILC familyCitation30 rather than within the same group as was initially established.Citation31 Our work here suggests that in the context of bacterial stimulation, the ability of NK cells and ILC1s to express cytokines and cytolytic molecules is similar, although the degree of response may differ.
An intriguing observation in our data is that of the bacteria tested in this study enteric Gram-negative, but not Gram-positive bacteria-induced IFNγ production from NK cells and ILC1s. Although more work is needed to determine the mechanisms underlying this Gram-negative versus Gram-positive dichotomy, it is tempting to speculate that certain enteric commensal bacteria are inherently more pro-inflammatory. This is in keeping with the characterization of some commensal bacteria as “pathobionts,” which are commensals with the propensity to induce pathological conditions in certain situations, but are otherwise symbionts.Citation46 Here, two of the Gram-negative commensals tested (P. stercorea and A. junii) reside within genera of bacteria that are reported to contain pathobionts,Citation47,Citation48 and thus members of these genera may induce inflammation in the setting of epithelial barrier damage. Interestingly, we have previously shown that these exact species can also induce IL-22 production by human colonic ILC3s,Citation49 which is a cytokine with a role in epithelial barrier maintenance and regulation of intestinal health. However, it is important to note that while we did investigate the effect of four commensal bacteria representing four different phyla on the induction of cytokine production by Group 1 ILCs, we acknowledge that there are a multitude of different bacterial species in the human gut and therefore we cannot conclude that all bacteria would elicit similar responses in Group 1 ILCs.
Previous work has demonstrated that the Gram-positive commensal bacteria L. pentosus is capable of inducing IFNγ production by splenic murine NK cells and was attributed to strong induction of IL-12p70 from CD11c+ DCs by that particular species of bacteria.Citation27 Furthermore, Gram-positive L. acidophilus, but not Gram-positive B. bifidum and L. reuteri stimulated IFNγ by peripheral blood NK cells and this difference in bacterial responses was also linked to levels of IL-12p70 from mDCs.Citation29 In keeping with the concept that IFNγ production by Group 1 ILCs is driven by cytokine production by mDC, here we detected significant increases in mDCs expressing IL-12p70 or IL-18 after exposure to Gram-negative A. junii, but not Gram-positive R. bromii. Although we did not detect increases in macrophages expressing IL-12p70 or IL-18 in our system, we cannot rule out a contributory role of these cells in promoting IFNγ production by Group 1 ILCs during in vitro exposure to bacteria, as has been reported for macrophages in inducing IFNγ from human blood NK cells during in vitro infection with S. typhimurium.Citation8 Our data do demonstrate that bacteria-specific induction of IFNγ via stimulatory cytokines such as IL-12p70 or IL-18 from responding mDCs is likely a critical component of IFNγ signaling in Group 1 ILCs and despite the overarching classification by Gram stain (i.e., cell surface structure), enteric commensal bacteria are not equivalent in their capacity to stimulate immune responses within the GI tract.
The importance of crosstalk between accessory cells and NK cells in response to bacteria has previously been established using human peripheral blood. Monocyte-derived DCs exposed to Helicobacter pylori were critical in driving IFNγ production by NK cellsCitation50 and we have also shown that cell-to-cell contact was required between peripheral blood monocytes and NK cells for induction of IFNγ in response to in vitro stimulation with commensal E. coli.Citation28 In addition, macrophages infected with S. typhimurium were involved in the production of IFNγ and degranulation by NK cells, likely through a contact-dependent mechanism.Citation8 Studies examining the need for crosstalk between APCs and ILC1s are more limited, particularly in relation to bacterial stimuli, although human peripheral blood CD14+ DCs exposed to LPS-induced IFNγ-producing ILC1s.Citation51 In our current study, we demonstrate the need for crosstalk between colonic mucosal mDC and Group 1 ILCs for production of IFNγ in response to enteric commensal bacteria. Although purified NK cells and ILC1s responded directly to recombinant IL-12p70 and IL-18, implying that contact dependence between mDCs and Group 1 ILCs is not a requirement for induction of IFNγ, further studies are required to confirm this in the context of bacterial stimulation.
There are differing reports on the capacity of ILC1s to be cytotoxic,Citation16,Citation19 which may depend on how cytolytic function was defined experimentally (i.e., Granzyme B expression or degranulation). Here we observed that Group 1 ILCs have increased expression of Granzyme B and demonstrated co-expression with Perforin following exposure to Gram-negative bacteria suggesting cytolytic capacity. Intriguingly, the Granzyme B expressing subset also co-expressed IFNγ indicating that NK cells and ILC1s from the human colon may be polyfunctional in the presence of bacterial stimulation. Polyfunctionality has been reported in the context of chronic SIV infection with increased frequencies of colonic IFNγ and CD107a (a marker of cytolytic degranulation)-expressing ILCs compared to frequencies in uninfected primates.Citation21 In addition to the traditional role of Granzyme B in killing, it has been demonstrated that Granzyme B can directly degrade extracellular matrix components,Citation52 cleave pro-IL-18 to generate active IL-18,Citation53 and enhance LPS-induced monocyte release of TNFα.Citation54 Thus, it is possible that Group 1 ILCs may contribute to the inflammatory milieu during microbial translocation through differing mechanisms – including both IFNγ production and Granzyme B production.
Overall, this current work expands on our understanding of the basic biology of human gut Group 1 ILCs and provides insight about the mechanisms by which enteric bacteria trigger a response from Group 1 ILCs. We demonstrate that mDCs and the cytokines IL-12p70, IL-18, and IL-1β are important in eliciting a Group 1 ILC response to bacteria. These in vitro observations highlight that production of IFNγ and induction of Granzyme B by Group 1 ILCs in response to commensal bacteria could be a component of inflammation in different GI diseases in which microbial translocation is a key feature.
Materials and methods
Lamina propria mononuclear cells (LPMC)
Research detailed here was reviewed by the Colorado Multiple Institutional Review Board (COMIRB) at the University of Colorado Anschutz Medical Campus and was granted exempt research status. Deidentified colonic tissue samples that would otherwise be discarded were acquired from human patients undergoing elective abdominal surgery. Colons were categorized as macroscopically normal tissue specimens. The criteria for exclusion of certain tissue specimens include samples from patients that underwent chemotherapy or radiation within 6 weeks before surgery, patients diagnosed with Inflammatory Bowel Disease, patients with HIV infection or patients treated with immunosuppressive drugs within 1 month of surgery. A release was signed by all patients undergoing surgery to allow unrestricted use of discarded tissue. LPMCs were isolated from tissue samples as previously describedCitation49,Citation55,Citation56 and stored in liquid nitrogen until use.
Preparation of bacterial stocks
Salmonella enterica subsp enterica serovar Typhimurium (ATCC 35986) was grown under aerobic conditions at 37°C for 1–2 d on LB agar plates (Sigma-Aldrich). Growth of anaerobic bacteria was performed using a BD GasPak EZ Anaerobe Pouch System (BD Diagnostics). Prevotella stercorea (DSM No. 18206) was grown for 5–7 d under anaerobic conditions in liquid-chopped meat broth (Hardy Diagnostics) supplemented with 1% Trace Minerals (ATCC), 1% Vitamin Supplements (ATCC), 0.05% Tween 80, 29.7mM acetic acid, 8.1 mM propionic acid and 4.4 mM butyric acid (Sigma-Aldrich). Ruminococcus bromii (ATCC# 27255) was grown under anaerobic conditions at 37°C for 1–2 d in liquid-chopped meat broth (Hardy Diagnostics). The long-term stock of Bifidobacterium longum subsp infantis (ATCC 15697) was grown under anaerobic conditions at 37°C for 2–3 d in liquid-chopped meat broth (Hardy Diagnostics) and the working stock was grown on Brucella plates (Teknova) under anaerobic conditions at 37°C for 2–3 d. Acinetobacter junii (ATCC 17908) was grown under aerobic conditions at 26°C for 1–2 d using Nutrient Agar plates (Edge Biologicals). Single-use working stocks of all bacteria were prepared using DPBS and long-term stocks were prepared using 10% glycerol. All bacterial stocks were enumerated using the BD Cell Viability Kit (BD Bioscience) and stored at −80°C until use.
In vitro stimulation of LPMCs with whole bacteria
For the in vitro stimulations, human colon LPMCs were thawed as previously describedCitation55,Citation56 and cultured in RPMI with 1% penicillin/streptomycin/glutamine (Life Technologies), 10% human AB serum (Gemini Bioproducts) and 500 µg/ml Zosyn-Piperacillin and Tazobactam (Wyeth). LPMCs were exposed to a panel of Gram-positive and Gram-negative bacteria. Broad-spectrum antibiotics including Penicillin, Streptomycin, Piperacillin, and Tazobactam were present throughout the time in culture to prevent bacterial overgrowth.
For assays examining Group 1 ILC (ILC1, NK) responses, whole bacteria were added to cultures at a ratio of 2.5 bacteria to 1 LPMC or 50 ng/mL recombinant cytokines IL-12p70 (Biolegend), IL-18 (R& D Systems), or IL-1β (Biolegend) were added to LPMC cultures. LPMCs were then incubated for 16 h at 37°C + 5% CO2, followed by the addition of Golgi Plug Transport Inhibitor (BD Bioscience) for 4 h. Cells were then collected for flow cytometry as described below.
For assays examining antigen-presenting cell responses (mDCs and Macrophages), whole bacteria were added to cell cultures at ratio of 2.5 bacteria to 1 LPMC and incubated for 4 h at 37°C + 5% CO2, followed by the addition of Golgi Plug Transport Inhibitor (BD Bioscience) for 16 h. Cells were then collected for flow cytometry as described below.
For blocking experiments, LPMCs were first exposed to blocking antibodies targeting IL-12p70, IL-18, and/or IL-1β (R & D Systems) at 10 μg/mL for 30 min. S. typhimurium or A. junii was then added to cultures (2.5 bacteria:1 LPMC) and incubated for 16 h at 37°C + 5% CO2, followed by the addition of Golgi Plug Transport Inhibitor (BD Bioscience) for 4 h. Cells were then collected for flow cytometry as described below. The concentration of blocking antibodies was optimized for use by measurement of IFNγ+ NK cells in the presence of recombinant IL-12p70 + (Biolegend) IL-18 (R & D Systems) both at 50 ng/mL (Figure S4). Antibodies and controls used to block are listed in Table S1.
For mDCs depletion experiments, CD11c+ cells were depleted from LPMCs using the EasySep PE Positive Selection Kit according to manufacturer’s instructions (StemCell Technologies) and the antibody PE-CD11c (Biolegend) followed by incubation with bacteria as described above. Greater than 90.25% of CD11c+ mDCs were depleted from total LPMCs.
For the measurement of secreted cytokines, LPMCs were plated at a concentration of 2.0 × 10Citation6 million cells per mL in a 96 well plate and exposed to whole bacteria added to cell cultures at a ratio of 2.5 bacteria to 1 LPMC and incubated for 24 h at 37°C + 5% CO2. Supernatant was collected and saved at −20°C until use. IL-12p70, IL-18, and IL-1β were measured in the supernatant using the U-PLEX Assay according to manufacturer’s instructions and quantified on the QuickPlex SQ 120 Instrument (Mesoscale Discovery).
Flow cytometry protocol for surface and intracellular staining
To identify Group 1 ILCs by flow cytometry viable CD45+ single-cell lymphocytes were initially gated followed by gating for ILC1s as Lineage-CD127+ CD117- and NK cells as Lineage-CD127-CD56+. The cocktail for lineage negative antibodies contained anti-CD3, CD20, CD13, CD123, CD303, CD34, FCεR1α, CD11c, and CRTH2. All flow cytometry data were acquired on an LSRII flow cytometer (BD Biosciences). Routine quality control using the Cytometer Setup and Tracking feature within the BD FACSDiva software version 6.1.2 (BD Biosciences) was performed daily. All antibodies and dyes used for flow cytometry staining are listed in Table S2.
For ex vivo phenotyping of Group 1 ILCs, cells were thawed and surface stained for frequencies of ILC subsets and surface expression of TLR2, TLR4, and TLR5 or intranuclear stained for the transcription factors T-bet and EOMES using the Foxp3/Transcription Factor buffer set according to manufacturer’s instructions (Thermo Fisher Scientific).
For in vitro culture examination of Group 1 ILCs, stimulated LPMCs were collected and surface stained followed by intracellular staining for IFNγ, TNFα, Granzyme B, and Perforin using Medium A and Medium B buffers according to manufacturer’s instructions (Thermo Fisher Scientific).
For in vitro culture examination of antigen-presenting cells, stimulated cells were collected and surface stained to identify mDCs (CD45+ Viable CD3- CD19- HLA-DRhigh CD11chigh) or Macrophages (CD45 + Viable CD3- CD19- HLA-DR+ CD11c−/low)Citation37–Citation39 followed by intracellular staining for cytokines IL-12p70, IL-18, and IL-1β using Medium A and Medium B buffers.
In vitro stimulation of purified ILC1 and NK cells
ILC1s and NK cells were isolated from colonic LPMCs to greater than 95.8% and 91.5% purity, respectively, by sorting using the MoFlo Astrios EQ (Beckman Colter, Indianapolis, IN). ILC1s were sorted from Viable CD45+ single-cell lymphocytes that were Lineage-CD127+CD117- and NK cells were sorted as Viable CD45+ single-cell lymphocytes that were Lineage-CD127-CD56+. Purified NK cells were plated at 500,000 cells per mL and ILC1s were plated at 100,000 cells per mL in a 96 well plate. Group 1 ILCs were then exposed to either 50ng/mL IL-12p70 (Biolegend) and IL-18 (R & D Systems) or whole bacteria (1 cell:1 bacteria) and incubated for 24 h at 37°C + 5% CO2. Supernatant was collected and saved at −20°C until use. IFNγ was measured in the supernatant using the IFNγ U-PLEX Assay according to manufacturer’s instructions and quantified on the QuickPlex SQ 120 Instrument (Mesoscale Discovery).
Data analysis
Each tissue specimen provided by a donor for research was considered a single sample for data analysis. Figure legends indicate how many samples were examined for each assay. Analysis for flow cytometry data was completed using FlowJo v10.0 and only data sets with a minimum number of events (25 events for ILC1s or 25 events for NK cells) were included in analysis. GraphPad Prism v6.00 for Windows (GraphPad Software) was used for statistical analysis and graphing. Statistical differences between conditions were determined by paired t-test or one-way ANOVA as indicated in figure legend.
Author contributions
MC, SD, AC, EB, CW designed the study and interpreted the work. MC, SD, and CW wrote the manuscript. MC and CP performed experiments. MM provided tissue specimens. All authors contributed to manuscript revision, and read and approved the submitted version.
Disclosure of potential conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplemental Material
Download PDF (360.8 KB)Acknowledgments
We are grateful to the Flow Cytometry Shared Resource at the University of Colorado Cancer Center (P30CA046934) for technical assistance with sorting ILC1s and NK cells. We also acknowledge the help of Jay Liu, Jon Kibbie, Sabrina Nesladek, Allison Christians, and other members of the Wilson lab for technical assistance with the processing of the tissue specimens.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Waggoner SN, Reighard SD, Gyurova IE, Cranert SA, Mahl SE, Karmele EP, McNally JP, Moran MT, Brooks TR, Yaqoob F, et al. Roles of natural killer cells in antiviral immunity. Curr Opin Virol. 2016;16:15–17. doi:10.1016/j.coviro.2015.10.008.
- Vashist N, Trittel S, Ebensen T, Chambers BJ, Guzman CA, Riese P. Influenza-activated ilc1s contribute to antiviral immunity partially influenced by differential gitr expression. Front Immunol. 2018;9:505. doi:10.3389/fimmu.2018.00505.
- Souza-Fonseca-Guimaraes F, Adib-Conquy M, Cavaillon JM. Natural killer (NK) cells in antibacterial innate immunity: angels or devils? Mol Med. 2012;18:270–285. doi:10.2119/molmed.2011.00201.
- Fuchs A. Ilc1s in tissue inflammation and infection. Front Immunol. 2016;7:104. doi:10.3389/fimmu.2016.00104.
- Ivanova D, Krempels R, Ryfe J, Weitzman K, Stephenson D, Gigley JP. Nk cells in mucosal defense against infection. Biomed Res Int. 2014;2014:413982. doi:10.1155/2014/413982.
- Hall LJ, Murphy CT, Hurley G, Quinlan A, Shanahan F, Nally K, Melgar S. Natural killer cells protect against mucosal and systemic infection with the enteric pathogen citrobacter rodentium. Infect Immun. 2013;81(2):460–469. doi:10.1128/IAI.00953-12.
- Reid-Yu SA, Small CL, Coombes BK. Cd3(-)nk1.1(+) cells aid in the early induction of a th1 response to an attaching and effacing enteric pathogen. Eur J Immunol. 2013;43(10):2638–2649. doi:10.1002/eji.201343435.
- Lapaque N, Walzer T, Meresse S, Vivier E, Trowsdale J. Interactions between human nk cells and macrophages in response to salmonella infection. J Immunol. 2009;182(7):4339–4348. doi:10.4049/jimmunol.0803329.
- Ashkar AA, Reid S, Verdu EF, Zhang K, Coombes BK. Interleukin-15 and nk1.1+ cells provide innate protection against acute salmonella enterica serovar typhimurium infection in the gut and in systemic tissues. Infect Immun. 2009;77(1):214–222. doi:10.1128/IAI.01066-08.
- Kupz A, Scott TA, Belz GT, Andrews DM, Greyer M, Lew AM, Brooks AG, Smyth MJ, Curtiss R 3rd, Bedoui S, et al. Contribution of thy1+ nk cells to protective ifn-gamma production during salmonella typhimurium infections. Proc Natl Acad Sci U S A. 2013;110(6):2252–2257. doi:10.1073/pnas.1222047110.
- Bao S, Beagley KW, France MP, Shen J, Husband AJ. Interferon-gamma plays a critical role in intestinal immunity against salmonella typhimurium infection. Immunology. 2000;99(3):464–472. doi:10.1046/j.1365-2567.2000.00955.x.
- van de Wetering D, de Paus RA, van Dissel JT, van de Vosse E. Salmonella induced il-23 and il-1beta allow for il-12 production by monocytes and mphi1 through induction of ifn-gamma in cd56 nk/nk-like t cells. PLoS One. 2009;4(12):e8396. doi:10.1371/journal.pone.0008396.
- Klose CSN, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, et al. Differentiation of type 1 ilcs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157(2):340–356. doi:10.1016/j.cell.2014.03.030.
- Abt MC, Lewis BB, Caballero S, Xiong H, Carter RA, Susac B, Ling L, Leiner I, Pamer EG. Innate immune defenses mediated by two ilc subsets are critical for protection against acute clostridium difficile infection. Cell Host Microbe. 2015;18(1):27–37. doi:10.1016/j.chom.2015.06.011.
- Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d’Hargues Y, Goppert N, Croxford AL, Waisman A, Tanriver Y, et al. A t-bet gradient controls the fate and function of ccr6-rorgammat+ innate lymphoid cells. Nature. 2013;494(7436):261–265. doi:10.1038/nature11813.
- Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M. Intraepithelial type 1 innate lymphoid cells are a unique subset of il-12- and il-15-responsive ifn-gamma-producing cells. Immunity. 2013;38(4):769–781. doi:10.1016/j.immuni.2013.02.010.
- Takayama T, Kamada N, Chinen H, Okamoto S, Kitazume MT, Chang J, Matuzaki Y, Suzuki S, Sugita A, Koganei K, et al. Imbalance of nkp44(+)nkp46(-) and nkp44(-)nkp46(+) natural killer cells in the intestinal mucosa of patients with crohn’s disease. Gastroenterology. 2010;139(3):882–892, 892 e881–e883. doi:10.1053/j.gastro.2010.05.040.
- Forkel M, van Tol S, Hoog C, Michaelsson J, Almer S, Mjosberg J. Distinct alterations in the composition of mucosal innate lymphoid cells in newly diagnosed and established crohn’s disease and ulcerative colitis. J Crohns Colitis. 2019;13(1):67–78. doi:10.1093/ecco-jcc/jjy119.
- Bernink JH, Peters CP, Munneke M, Te Velde AA, Meijer SL, Weijer K, Hreggvidsdottir HS, Heinsbroek SE, Legrand N, Buskens CJ, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14(3):221–229. doi:10.1038/ni.2534.
- Dillon SM, Castleman MJ, Frank DN, Austin GL, Gianella S, Cogswell AC, Landay AL, Barker E, Wilson CC. Inflammatory colonic innate lymphoid cells are increased during untreated hiv-1 infection and associated with markers of gut dysbiosis and mucosal immune activation. J Acquir Immune Defic Syndr. 2017;76(4):431–437. doi:10.1097/QAI.0000000000001523.
- Li H, Richert-Spuhler LE, Evans TI, Gillis J, Connole M, Estes JD, Keele BF, Klatt NR, Reeves RK. Hypercytotoxicity and rapid loss of nkp44+ innate lymphoid cells during acute siv infection. PLoS Pathog. 2014;10(12):e1004551. doi:10.1371/journal.ppat.1004551.
- Reeves RK, Rajakumar PA, Evans TI, Connole M, Gillis J, Wong FE, Kuzmichev YV, Carville A, Johnson RP. Gut inflammation and indoleamine deoxygenase inhibit il-17 production and promote cytotoxic potential in nkp44+ mucosal nk cells during siv infection. Blood. 2011;118(12):3321–3330. doi:10.1182/blood-2011-04-347260.
- Kramer B, Goeser F, Lutz P, Glassner A, Boesecke C, Schwarze-Zander C, Kaczmarek D, Nischalke HD, Branchi V, Manekeller S, et al. Compartment-specific distribution of human intestinal innate lymphoid cells is altered in hiv patients under effective therapy. PLoS Pathog. 2017;13(5):e1006373. doi:10.1371/journal.ppat.1006373.
- Loh G, Blaut M. Role of commensal gut bacteria in inflammatory bowel diseases. Gut Microbes. 2012;3(6):544–555. doi:10.4161/gmic.22156.
- Dillon SM, Frank DN, Wilson CC. The gut microbiome and hiv-1 pathogenesis: a two-way street. AIDS. 2016;30(18):2737–2751. doi:10.1097/QAD.0000000000001289.
- Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37(1):171–186. doi:10.1016/j.immuni.2012.05.020.
- Koizumi S, Wakita D, Sato T, Mitamura R, Izumo T, Shibata H, Kiso Y, Chamoto K, Togashi Y, Kitamura H, et al. Essential role of toll-like receptors for dendritic cell and nk1.1(+) cell-dependent activation of type 1 immunity by lactobacillus pentosus strain s-pt84. Immunol Lett. 2008;120(1–2):14–19. doi:10.1016/j.imlet.2008.06.003.
- Dillon SM, Lee EJ, Bramante JM, Barker E, Wilson CC. The natural killer cell interferon-gamma response to bacteria is diminished in untreated hiv-1 infection and defects persist despite viral suppression. J Acquir Immune Defic Syndr. 2014;65(3):259–267. doi:10.1097/01.qai.0000435603.50598.2b.
- Fink LN, Zeuthen LH, Christensen HR, Morandi B, Frokiaer H, Ferlazzo G. Distinct gut-derived lactic acid bacteria elicit divergent dendritic cell-mediated nk cell responses. Int Immunol. 2007;19(12):1319–1327. doi:10.1093/intimm/dxm103.
- Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, et al. Innate lymphoid cells: 10 years on. Cell. 2018;174(5):1054–1066. doi:10.1016/j.cell.2018.07.017.
- Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, et al. Innate lymphoid cells–a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13(2):145–149. doi:10.1038/nri3365.
- Marafini I, Monteleone I, Di Fusco D, Cupi ML, Paoluzi OA, Colantoni A, Ortenzi A, Izzo R, Vita S, De Luca E, et al. Tnf-alpha producing innate lymphoid cells (ilcs) are increased in active celiac disease and contribute to promote intestinal atrophy in mice. PLoS One. 2015;10(5):e0126291. doi:10.1371/journal.pone.0126291.
- Cooper MA, Fehniger TA, Ponnappan A, Mehta V, Wewers MD, Caligiuri MA. Interleukin-1beta costimulates interferon-gamma production by human natural killer cells. Eur J Immunol. 2001;31:792–801.
- Dillon SM, Lee EJ, Donovan AM, Guo K, Harper MS, Frank DN, McCarter MD, Santiago ML, Wilson CC. Enhancement of hiv-1 infection and intestinal cd4+ t cell depletion ex vivo by gut microbes altered during chronic hiv-1 infection. Retrovirology. 2016;13:5. doi:10.1186/s12977-016-0237-1.
- Dillon SM, Lee EJ, Kotter CV, Austin GL, Gianella S, Siewe B, Smith DM, Landay AL, McManus MC, Robertson CE, et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic t-cell activation in untreated hiv-1 infection. Mucosal Immunol. 2016;9(1):24–37. doi:10.1038/mi.2015.33.
- Ruiz L, Delgado S, Ruas-Madiedo P, Sanchez B, Margolles A. Bifidobacteria and their molecular communication with the immune system. Front Microbiol. 2017;8:2345. doi:10.3389/fmicb.2017.02345.
- Bain CC, Mowat AM. The monocyte-macrophage axis in the intestine. Cell Immunol. 2014;291(1–2):41–48. doi:10.1016/j.cellimm.2014.03.012.
- Bain CC, Mowat AM. Macrophages in intestinal homeostasis and inflammation. Immunol Rev. 2014;260(1):102–117. doi:10.1111/imr.12192.
- Smith PD, Smythies LE, Shen R, Greenwell-Wild T, Gliozzi M, Wahl SM. Intestinal macrophages and response to microbial encroachment. Mucosal Immunol. 2011;4(1):31–42. doi:10.1038/mi.2010.66.
- Simoni Y, Fehlings M, Kloverpris HN, McGovern N, Koo SL, Loh CY, Lim S, Kurioka A, Fergusson JR, Tang CL, et al. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity. 2017;46(1):148–161.
- Bernink JH, Mjosberg J, Spits H. Human ilc1: to be or not to be. Immunity. 2017;46(5):756–757. doi:10.1016/j.immuni.2017.05.001.
- Bjorklund AK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, Sandberg R, Mjosberg J. The heterogeneity of human cd127(+) innate lymphoid cells revealed by single-cell rna sequencing. Nat Immunol. 2016;17(4):451–460. doi:10.1038/ni.3368.
- Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, Colonna M, Immunological Genome C. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16(3):306–317. doi:10.1038/ni.3094.
- Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508(7496):397–401. doi:10.1038/nature13047.
- Colonna M. Innate lymphoid cells: diversity, plasticity, and unique functions in immunity. Immunity. 2018;48(6):1104–1117. doi:10.1016/j.immuni.2018.05.013.
- Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23(4):473–480. doi:10.1016/j.coi.2011.07.010.
- Larsen JM. The immune response to prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151(4):363–374. doi:10.1111/imm.12760.
- Shulzhenko N, Dong X, Vyshenska D, Greer RL, Gurung M, Vasquez-Perez S, Peremyslova E, Sosnovtsev S, Quezado M, Yao M, et al. Cvid enteropathy is characterized by exceeding low mucosal iga levels and interferon-driven inflammation possibly related to the presence of a pathobiont. Clin Immunol. 2018;197:139–153. doi:10.1016/j.clim.2018.09.008.
- Castleman MJ, Dillon SM, Purba CM, Cogswell AC, Kibbie JJ, McCarter MD, Santiago ML, Barker E, Wilson CC. Commensal and pathogenic bacteria indirectly induce il-22 but not ifnγ production from human colonic ilc3s via multiple mechanisms. Front Immunol. 2019;10:649. doi:10.3389/fimmu.2019.00649.
- Hafsi N, Voland P, Schwendy S, Rad R, Reindl W, Gerhard M, Prinz C. Human dendritic cells respond to helicobacter pylori, promoting nk cell and th1-effector responses in vitro. J Immunol. 2004;173(2):1249–1257. doi:10.4049/jimmunol.173.2.1249.
- Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, Kofoed-Nielsen M, Munneke JM, Hazenberg MD, Villaudy J, Buskens CJ, et al. Interleukin-12 and -23 control plasticity of cd127(+) group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity. 2015;43(1):146–160. doi:10.1016/j.immuni.2015.06.019.
- Buzza MS, Zamurs L, Sun J, Bird CH, Smith AI, Trapani JA, Froelich CJ, Nice EC, Bird PI. Extracellular matrix remodeling by human granzyme b via cleavage of vitronectin, fibronectin, and laminin. J Biol Chem. 2005;280(25):23549–23558. doi:10.1074/jbc.M412001200.
- Omoto Y, Yamanaka K, Tokime K, Kitano S, Kakeda M, Akeda T, Kurokawa I, Gabazza EC, Tsutsui H, Katayama N, et al. Granzyme b is a novel interleukin-18 converting enzyme. J Dermatol Sci. 2010;59(2):129–135. doi:10.1016/j.jdermsci.2010.05.004.
- Wensink AC, Hack CE, Bovenschen N. Granzymes regulate proinflammatory cytokine responses. J Immunol. 2015;194(2):491–497. doi:10.4049/jimmunol.1401214.
- Dillon SM, Rogers LM, Howe R, Hostetler LA, Buhrman J, McCarter MD, Wilson CC. Human intestinal lamina propria cd1c+ dendritic cells display an activated phenotype at steady state and produce il-23 in response to tlr7/8 stimulation. J Immunol. 2010;184(12):6612–6621. doi:10.4049/jimmunol.1000041.
- Howe R, Dillon S, Rogers L, McCarter M, Kelly C, Gonzalez R, Madinger N, Wilson CC. Evidence for dendritic cell-dependent cd4(+) t helper-1 type responses to commensal bacteria in normal human intestinal lamina propria. Clin Immunol. 2009;131(2):317–332. doi:10.1016/j.clim.2008.12.003.
