ABSTRACT
The commensal bacterium Faecalibacterium prausnitzii plays a key role in inflammatory bowel disease (IBD) pathogenesis and serves as a general health biomarker in humans. However, the host molecular mechanisms that underlie its anti-inflammatory effects remain unknown. In this study we performed a transcriptomic approach on human intestinal epithelial cells (HT-29) stimulated with TNF-α and exposed to F. prausnitzii culture supernatant (SN) in order to determine the impact of this commensal bacterium on intestinal epithelial cells. Moreover, modulation of the most upregulated gene after F. prausnitzii SN contact was validated both in vitro and in vivo. Our results showed that F. prausnitzii SN upregulates the expression of Dact3, a gene linked to the Wnt/JNK pathway. Interestingly, when we silenced Dact3 expression, the effect of F. prausnitzii SN was lost. Butyrate was identified as the F. prausnitzii effector responsible for Dact3 modulation. Dact3 upregulation was also validated in vivo in both healthy and inflamed mice treated with either F. prausnitzii SN or the live bacteria, respectively. Finally, we demonstrated by colon transcriptomics that gut microbiota directly influences Dact3 expression. This study provides new clues about the host molecular mechanisms involved in the anti-inflammatory effects of the beneficial commensal bacterium F. prausnitzii.
Introduction
Inflammatory bowel disease (IBD) is a group of disorders characterized by chronic inflammation in the gastrointestinal tract.Citation1,Citation2 One potential cause (and/or consequence) of IBD is the disruption of the intestinal ecosystem equilibrium. For example, gut microbiota analysis of Crohn’s disease (a type of IBD) patients revealed markedly lower diversity of Firmicutes (in particular of the Clostridium leptum group) compared to healthy individuals.Citation3 These assemblages are also relatively poor in Faecalibacterium prausnitzii, a major member of the C. leptum group and one of the most abundant intestinal bacteria in healthy adults.Citation4,Citation5 Because a potential approach to prevent and treat IBD is the oral administration of probiotic and commensal bacteriaCitation6, F. prausnitzii may represent a relevant target for the development of diagnostic, prognostic, or therapeutic tools.
In different pre-clinical models of IBD, F. prausnitzii efficiently improves intestinal inflammationCitation7,Citation8 and gut barrier function.Citation9 Indeed, through secreted metabolites this bacterium is able to block NF-κB activation and IL-8 production, which both contribute to inflammation.Citation10 In addition, this species produces high quantities of butyrate,Citation4 a short-chain fatty acid (SCFA) which is important in gut physiology.Citation11,Citation12 F. prausnitzii also produces several bioactive molecules that affect inflammation and gut barrier function such as shikimic and salicylic acidsCitation12 and a microbial anti-inflammatory molecule (MAMCitation13). Despite many advances in the study of F. prausnitzii’s anti-inflammatory effects within the hostCitation4, we still do not understand the exact molecular mechanism of these effects, which could represent targets for new therapies. To identify the host receptor and signaling pathways involved in the beneficial effects of this anti-inflammatory bacterium, we performed DNA chip-based transcriptomic analyses in human intestinal epithelial cells (IECs; i.e., HT-29 cells) that were stimulated with the proinflammatory cytokine TNF-α and exposed to F. prausnitzii culture supernatant (SN). These analyses led us to focus on Dact3, a member of the Disheveled Binding Antagonist of beta Catenin (DACT) gene family which negatively regulates the Wnt/JNK signaling pathway.Citation14 We present here evidence from both in vitro and in vivo experiments which provide the first clues about the important role of Dact3 in the host molecular mechanisms involved in the anti-inflammatory effects of the beneficial commensal bacterium F. prausnitzii.
Results
Transcriptomic analysis reveals Dact3 as a target of F. prausnitzii
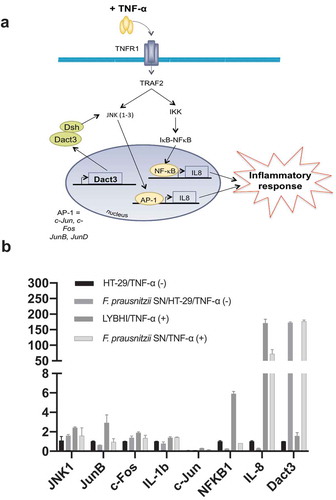
We used TNF-α to stimulate inflammation in HT-29 cells. A specific trait of this stimulation is upregulation of IL-8Citation15–19, which we then measured as a readout for cell inflammatory status. To extend previous research on F. prausnitzii SN in TNF-α-stimulated HT-29 cellsCitation12, we performed a SN dose-effect experiment and determined the stability (i.e., putative degradation) of both IL-8 and TNF-α in the presence of the SN. F. prausnitzii SN had a significant dose–response effect (5% to 30%) while the bacterial culture medium LYBHI did not (Figure S1(a)). Of note, F. prausnitzii SN did not directly degrade either IL-8 (Figure S1(b)) or TNF-α (Figure S1(c)). To elucidate the host molecular mechanisms underlying the anti-inflammatory effects of F. prausnitzii (e.g., specifically the modulatory effects on IL-8 production), we performed a transcriptomic analysis of TNF-α-stimulated HT-29 cells exposed to F. prausnitzii SN (microarray hybridization schema is presented in Figure S2). Before evaluating the effects of the SN, we first determined the regulatory changes due to TNF-α stimulation alone. Compared to control HT-29 cells, 227 genes were upregulated and 60 genes were downregulated in TNF-α-stimulated cells (adj. p < .05, |fold change (FC)| > 1.5). The genes with the largest changes in expression are listed in Table S1. As expected, the pro-inflammatory cytokine IL-8 was among the most upregulated genes. In addition, upstream regulators such as TNF-α and IFN-γ and signaling pathways such as NF-κB were also activated, confirming the inflamed status of the HT-29 cells. Next, we evaluated the effect of the LYBHI medium used to culture F. prausnitzii. Here, only 63 genes were regulated ()), and LYBHI did not seem to reduce cellular inflammation. Finally, we introduced F. prausnitzii SN. There were extensive changes: stimulation and SN treatment specifically modulated 1651 genes ()). Interestingly, these genes are involved in several inflammatory pathways such as NF-κβ, p38 and other ERK/MAPK pathways. Among these, 25% of the genes that had been activated by TNF-α exposure (in our first comparison) were inactivated by treatment with F. prausnitzii SN (70 out of 287). To explore this complex gene regulation network, we submitted the dataset of genes regulated by SN compared to LYBHI to an ingenuity pathway analysis (IPA), which highlighted the SN-affected pathways. This analysis confirmed the ability of F. prausnitzii SN to regulate the ERK/MAPK signaling pathway as the most important pathway related to inflammation modulated specifically by F. prausnitzii SN ()). Notably, 7% of all genes regulated by F. prausnitzii SN were linked to MAPK pathways, including JNK. The 20 genes with the largest fold-change in regulation are shown in ) and Table S2. IL-8 was found to be downregulated, while the most upregulated gene was Dact3 (FC = 17.2). Dact3 belongs to the Dact gene family, whose members interact with the Dsh protein to inhibit Dsh-induced activation of the JNK pathway. The Dact3/JNK pathway is presented in ). Finally, in order to validate our transcriptomic data, the differential expression of some of the most up- and downregulated genes (see Table S3 for the list of genes analyzed) was validated by RT-qPCR using B2M gene as reference for data normalization as previously described.Citation20 As shown in ) all the tested genes displayed the same regulation (e.g., up- or down-) as that observed in the transcriptomic analysis. Strikingly, compared to LYBHI, F. prausnitzii SN led to significant Dact3 upregulation in TNF-α-stimulated HT-29 cells, confirming transcriptomic observations.
Figure 1. Gene and cellular pathways modulation by F. prausnitzii SN. (a) Comparison of the genes modulated by LYBHI+TNF-α vs TNF-α and SN+TNF-α vs TNF- α. The genes specifically modulated by SN and linked to inflammatory pathways have been represented using IPA canonical pathway display: y-axis displays the -log of the p-value which is calculated by Fisher’s exact test right-tailed. The orange points interconnected by a thin line represent the ratio. This ratio is calculated as follows: # of genes in a given pathway that meet the cutoff criteria, divided by the total # of genes that make up that pathway and that are in the reference gene set. (b) IPA canonical pathway display of the genes modulated in the comparison LYBHI+TNF-α vs SN+TNF-α: y-axis displays the -log of p-value which is calculated by Fisher’s exact test right-tailed. The orange and blue colored bars indicate predicted pathway activation, or predicted inhibition, respectively (z-score). Only genes with z-score are represented. The orange points interconnected by a thin line represent the ratio. (c) Histogram of the ten most up- and downregulated genes in the comparison LYBHI+TNF-αvs SN+TNF-α
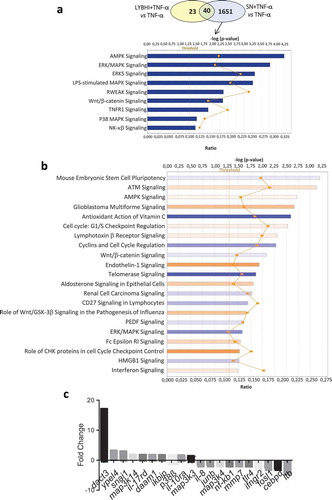
Figure 2. F. prausnitzii SN modulates Dact3 expression in vitro. (a) Schematic representation of Dact3 pathway. (b) RT-qPCR validation of most up- and downregulated genes by F. prausnitzii SN in TNF-α-stimulated HT-29 cells. Results are expressed as the fold change (FC) of gene expression relative to the B2M housekeeping gene
Role of Dact3 in TNF-α-stimulated HT-29 cells
As Dact3 was the most upregulated gene in the transcriptomics analysis, we thus decided to focus our investigations on this gene. To investigate the effect of Dact3 on IL-8 production in TNF-α-stimulated HT-29 cells, we overexpressed or knocked down Dact3 in IECs. First, we tried to overexpress Dact3 using transient transfection in HT-29 cells; unfortunately, we were unable to efficiently transfect these cells (data not shown). This could be due to the strong upregulation of Dact3 that led to massive apoptosis of the cells, as was previously reported for colorectal cancer cells.Citation21 We then used the siRNA technology to knockdown Dact3 mRNA in TNF-α-inflamed HT-29 cells. As shown in ), RT-qPCR analysis confirmed that: i) both the non-targeting (NT) siRNA and LYBHI had no effect on Dact3 expression (FC = 1); ii) F. prausnitzii SN strongly upregulated Dact3 (FC = 30) in presence of NT siRNA; and iii) Dact3 siRNA significantly reduced the abundance of Dact3 mRNA induced by F. prausnitzii SN. In addition, we confirmed knockdown of Dact3 at level protein by Western blot analyses as shown in Figure S3. Besides, treatment with Dact3 siRNA in TNF-α-stimulated HT-29 cells tends to increase IL-8 production ()) suggesting an important role of Dact3 activation in intestinal homeostasis. Moreover, the inhibitory effect of F. prausnitzii SN on IL-8 production was partially lost in Dact3-silencing condition siRNA.
Figure 3. Modulation of Dact3 expression in TNF-α-stimulated HT-29 cells under different conditions. (a) Dact3 expression and (b) IL-8 production by HT-29 cells transfected with either Dact3 or non-targeting (NT) siRNA and treated with both F. prausnitzii SN and TNF-α. Dact3 expression was analyzed by RT-qPCR and IL-8 production by ELISA. Results are represented as the FC of Dact3 expression relative to LYBHI with NT siRNA and IL-8 have been normalized to DMEM medium (used as a control) under NT siRNA conditions (values for DMEM+TNF-a conditions were set to 100%). (c) Dact3 expression and (d) IL-8 production by TNF-α-stimulated HT-29 cells co-incubated with 0.5 µM of TSA or 12.5 µM of DZNep, or both. Results are represented as the FC of Dact3 expression relative to LYBHI and IL-8 values have been normalized to control DMEM. All experiments were performed in triplicate. Non-parametric Kruskal-Wallis and Dunn’s post hoc test *p < .05; **p < .01; ***p < .001
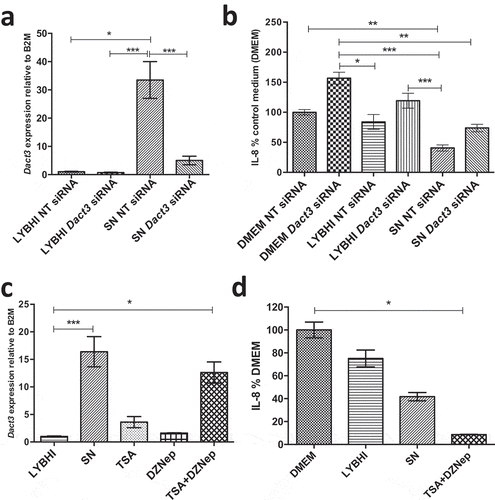
Next, we evaluated the effect of factors modulating Dact3 expression on IL-8 production. As histone modification has been reported to modulate Dact3 expressionCitation21, we used a mix of TSA (a histone deacetylase inhibitor) and DZNep (a histone methylation inhibitor) drugs as previously described.Citation21 These drugs led to a strong upregulation of Dact3 similar to that obtained with F. prausnitzii SN ()). Furthermore, after drug treatment and TNF-α stimulation, Dact3 upregulation strongly inhibited IL-8 production in HT-29 cells, to an even stronger extent than was observed with F. prausnitzii SN ()).
Identification of the F. prausnitzii-effectors responsible for Dact3 modulation
To identify the bacterial effector(s) responsible for Dact3 modulation, as F. prausnitzii abundantly produces butyrate, which reduces IL-8 production in TNF-α-stimulated HT-29 cellsCitation12, we decided to test the effect of this SCFA on Dact3 modulation. As shown in ), treatment with 1 mM of butyrate (the concentration present in the F. prausnitzii SNCitation22) led to strong upregulation of Dact3 and a decrease in IL-8 production ()). Altogether, these results reveal that butyrate is one of the F. prausnitzii produced-metabolites responsible for Dact3 modulation.
Figure 4. Effect of butyrate on Dact3 expression. (a) Dact3 expression and (b) IL-8 production by HT-29 cells co-incubated with 1 mM of butyrate. Results are represented as the FC of Dact3 expression relative to LYBHI and IL-8 have been normalized to DMEM. All experiments were performed in triplicate. Non-parametric Kruskal-Wallis and Dunn’s post hoc test *p < .05; **p < .01; ***p < .001. (c) Dact3 expression and (d) IL-8 production in TNF-α-stimulated HT-29 cells and treated with the SN of different bacterial strains producing or not butyrate. LYBHI and MRS were used as negative control of the SN from the different bacterial strains. Results are expressed as the FC of Dact3 expression relative to LYBHI and IL-8 have been normalized to DMEM. All experiments were performed in triplicate. Non-parametric Kruskal-Wallis and Dunn’s post hoc test *p < .05
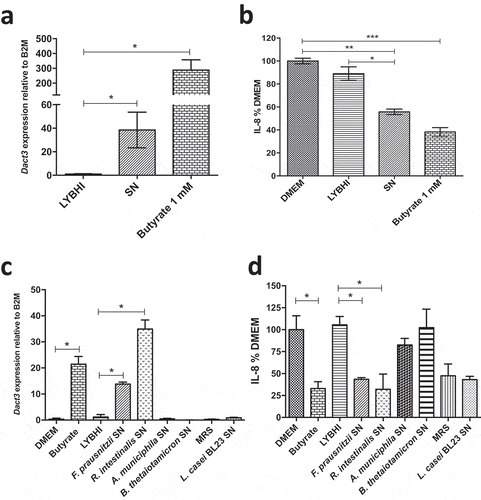
In order to validate this hypothesis, we tested other gut bacterial strains (e.g. Roseburia intestinalis, Akkermansia muciniphila and Bacteroides thetaiotaomicron), known to produce or not butyrate (Table S4), for their capacities to upregulate Dact3 ()) and downregulate IL-8 ()). We also tested a probiotic lactic acid bacterium (LAB) with anti-inflammatory capacities, L. casei BL23, that does not produce butyrate. Strikingly, the SN from the only other butyrate-producing bacterium (R. intestinalis) highly upregulated Dact3 expression and thus downregulated IL-8 production. In contrast, the three other bacterial strains that do not produce butyrate (A. muciniphila, B. thetaiotaomicron and L. casei BL23) did not have any effect on neither Dact3 nor IL-8. Despite L. casei BL23 seems to reduce IL-8 levels, this effect is more due to its culture medium (e.g. MRS) than the bacterium itself since the same inhibition in IL-8 levels was observed with the medium alone. This phenomenon could be explained by the presence of serpin in MRS, a protease inhibitor well known to display immunomodulatory effects.Citation23
In vivo validation of Dact3 modulation
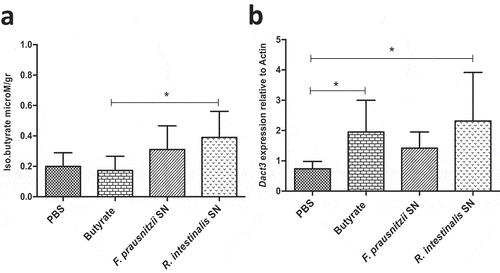
Dact3 modulation was first investigated in healthy mice orally administered with F. prausnitzii SN and sacrificed at different time points (e.g. 0, 3, 6, and 9 hours after administration) to evaluate its effects in normal physiological conditions (Figure S4(a)). F. prausnitzii SN led to a significant increase in Dact3 mRNA in colonic samples 9 h after its administration (Figure S4(b)). Other groups of mice were orally administered with either F. prausnitzii SN, R. intestinalis SN or butyrate (1 mM) and euthanized 9 h later. As shown in ), butyrate was detected in colonic samples of mice treated with either F. prausnitzii SN or R. intestinalis SN but not with butyrate itself, suggesting that soluble butyrate is rapidly absorbed in the colon. Strikingly, all treatments, including butyrate, results in Dact3 upregulation in samples from proximal colon ()).
Figure 5. Modulation of Dact3 expression in vivo by F. prausnitzii SN. Animals were intragastrically administered with either butyrate (1 mM) F. prausnitzii SN or R. intestinalis SN and sacrificed 9 after. (a) Quantification of butyrate in samples from proximal colon of treated mice. (b) FC of Dact3 expression in colonic samples relative to actin housekeeping gene. Non-parametric Kruskal-Wallis and Dunn’s post hoc test *p < .05
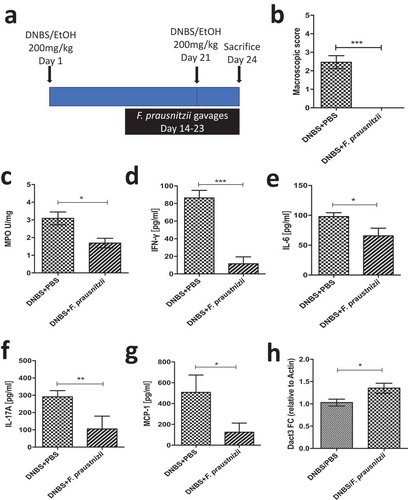
We then investigated in vivo Dact3 modulation in an inflammatory context, using a murine model in which chronic moderate inflammation was induced by intrarectal injection of dinitrobenzene sulfonic acid (DNBS). Animals were given daily oral gavages of live F. prausnitzii ()), as previously described.Citation8 As expected, F. prausnitzii administration led to significant reductions in markers of colitis with improvements in weight loss and a decrease in macroscopic scores ()), MPO activity ()) and a reduction of the pro-inflammatory cytokines IFN-γ ()), IL-6 ()), IL-17A ()) and the chemokine MCP-1 ()). Moreover, Dact3 expression was upregulated in the colon of F. prausnitzii-treated mice compared to the PBS-treated mice ()), confirming that Dact3 is induced by F. prausnitzii in an inflammatory context too.
Figure 6. Modulation of Dact3 expression in vivo by F. prausnitzii. (a) Experimental protocol used for the analysis of the in vivo effects of F. prausnitzii in a mouse model of chronic inflammation (as previously describedCitation8): (b) Macroscopic scores; (c) MPO activity; (d–g) colonic pro-inflammatory cytokine and chemokine concentrations and (h) FC of Dact3 expression in colonic samples relative to actin housekeeping gene. Non-parametric Mann-Whitney U test *p < .05
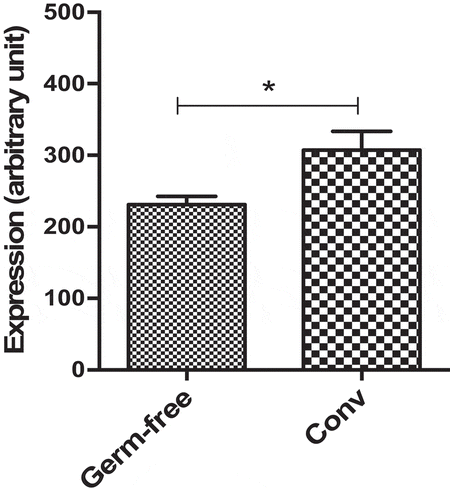
Finally, to assess the effect of the gut microbiota on Dact3 modulation in in vivo physiological conditions, we investigated a dataset that we previously published comparing the colonic transcriptome of germ-free and conventionalized (Conv) mice.Citation24 We found that Dact3 expression was significantly reduced in germ-free compared to Conv mice (). These observations confirm that gut microbiota has a major impact on Dact3 expression.
Discussion
F. prausnitzii is a commensal bacterium well known for its immunomodulatory properties and more specifically for its anti-inflammatory effects both in vitroCitation7,Citation12,Citation13 and in vivo.Citation4,Citation7,Citation9,Citation12,Citation13,Citation25 Here, we demonstrated that F. prausnitzii SN is able to block IL-8 production in TNF-α-activated HT-29, but this effect is not due to the proteolytic degradation of either TNF-α or IL-8. Thus, and because F. prausnitzii is extremely oxygen-sensitive (EOS) and cannot be easily cultured with human cellsCitation26, we decided to further study the immunomodulatory effects of its SN. F. prausnitzii SN modulated the expression of a massive number of genes, in fact, more than the TNF-α treatment itself (Table S1). Among the inflammation-related pathways regulated by F. prausnitzii SN, we found enrichment in genes related to MAPKs. In particular, Dact3, a gene implicated in Wnt/JNK regulation, was among the top-upregulated genes by F. prausnitzii SN in non-stimulated as well as in TNF-α-stimulated cells. The Dact gene family was initially reported in studies of embryonic development.Citation14,Citation27 Although Dact3 is involved in postnatal development (adult Dact3−/- mice show a mild reduction in body weight), the fact that Dact3−/- mice are viable means that this gene is not essential for mouse embryogenesis, postnatal survival, and reproduction.Citation28 Dact3 has a postnatal role as a regulator in the Wnt/β-catenin signaling pathwayCitation21 and has been associated with several types of cancer: colorectalCitation21, breastCitation29, ovarianCitation30, lungCitation31, papillary thyroidCitation32, and renal fibrosis.Citation28 In colorectal cancer cell lines, Dact3 transcription is epigenetically downregulated by a bivalent histone modification.Citation21 In particular, drugs that target both histone methylation (DZNep) and deacetylation (TSA) strongly induce Dact3 expressionCitation21, a result that we confirmed here. Moreover, we also observed that these drugs reduced IL-8 production by TNF-α-stimulated HT-29 cells in a way that was similar to the effect of F. prausnitzii SN. These results show that Dact3 has an important function in the IL-8 pathway and that IL-8 inhibition by F. prausnitzii SN is, at least partly, mediated via Dact3 upregulation. Dact3 could thus be an epigenetic regulator of inflammation and play a key role in intestinal homeostasis. To further decipher this, we evaluated the effect of Dact3 silencing in HT-29 using siRNA. Silencing of Dact3 in IECs prevented F. prausnitzii SN from blocking IL-8 production.
Dact3 histone modulation could provide clues about the bacterial effectors responsible for this regulation. In this study, only butyrate was able to both upregulate Dact3 expression and to block IL-8 production by TNF-α-stimulated HT-29 cells. Indeed, butyrate-induced reduction in IL-8 production was similar to that observed with the F. prausnitzii SN, but Dact3 upregulation was higher in the presence of butyrate than of SN. This is probably because Dact3 is also modulated by histone deacetylasesCitation21 which are known to be inhibited by butyrate.Citation11 In this context, our results confirm those obtained by Fung et al.Citation33, who found that Dact3 appeared to be differentially modulated in HT-29 cells treated with butyrate. However, these authors did not further explore Dact3 in their study.
Altogether, our results provide evidence that F. prausnitzii SN regulates Dact3/IL-8 production and suggest that butyrate produced by F. prausnitzii is the main actor in this regulation. Butyrate is well known to display immunoregulatory effects on IECs; however, these effects are dose and time-dependent in addition to be dependent of the IEC used in vitro (e.g., Caco-2 or HT-29 cells). Indeed, in an interesting study the authors found that butyrate decreases IL-8 secretion in IL-1β-stimulated Caco-2 cells, but increases IL-8 production in HT-29 cells, all of intestinal origin.Citation34 In another study, it was shown that long-term incubations (~24 h) result in a proinflammatory profile of butyrate in HT-29 cells in an NF-κβ-SEAP reporter system stimulated by TNF-αCitation35 while butyrate has been shown to inhibit TNF-α-induced nuclear translocation of NF-κβ (a pro-inflammatory transcription factor) after 30 minutes of TNF- α stimulation.Citation36 Thus, as butyrate displays important pleiotropic effects in the intestinal cell life cycle and numerous beneficial effects for human healthCitation11, it is plausible that butyrate production is a means by which F. prausnitzii affects its host physiological functions and homeostasis to maintain health. However, further studies are necessary to confirm this hypothesis. In particular, genetic manipulation of F. prausnitzii to inactivate the gene encoding the butyril-CoA synthase involved in butyrate metabolism would be helpful to answer to this question. Indeed, O’Cuiv et al.Citation37 have isolated F. prausnitzii transconjugants using metaparental mating and this strategy opens promising perspectives to manipulate F. prausnitzii.
Finally, in vitro observations were validated in vivo in two different models: first, in healthy mice that were orally administered F. prausnitzii SN one time (Figure S4) and second, in inflamed mice that were given live F. prausnitzii orally for 10 days (). In both in vivo models, either F. prausnitzii or F. prausnitzii SN positively regulated Dact3, confirming the key effect of F. prausnitzii on this gene.
In conclusion, we propose in this study a new role for Dact3 as a master regulator of intestinal homeostasis, particularly in inflamed cells. Although it is expressed in IECs at a low level, Dact3 seems to be essential for intestinal homeostasis, as its downregulation or loss leads to a global increase in inflammation. We hypothesize that Dact3 upregulation inhibits the AP-1 transcription factor, which in turn leads to a global downregulation of genes encoding for pro-inflammatory cytokines such as IL-2, IL-6, and IL-8.Citation38 Additionally, the ability of F. prausnitzii SN to modulate other genes involved in cancer pathways (as revealed in the transcriptomic analysis) represents a novel potential beneficial effect of this commensal anti-inflammatory bacterium, which is currently being investigated by our laboratory.
Our study provides the first clues on one of the host molecular targets involved in the anti-inflammatory effects of F. prausnitzii in IECs. Moreover, these results point out Dact3 as a potential master regulator of inflammation in IECs. As there is increasing interest in exploring new alternatives for IBD treatment, this research suggests at least three potential opportunities: i) use of F. prausnitzii itself, ii) use of drugs to modulate Dact3 expression (such as histone deacetylases) and iii) heterologous delivery of Dact3 (either as a cDNA or a protein) using food-grade live vectors.Citation39,Citation40 For these, further studies on Dact3-knockout mice will be necessary to understand the physiological functions of Dact3.
Materiel and methods
Bacterial strains
Faecalibacterium prausnitzii strain A2-165 (DSM N°17677, DSMZ collection, Braunschweig, Germany) was grown in LYBHI medium (BHI, Difco, Detroit, USA) supplemented with 0.5% yeast extract, 1 mg/ml cellobiose (Sigma-Aldrich Chemie GmbH, Buchs, Switzerland), 1 mg/ml maltose and 0.5 mg/ml cysteine (Sigma-Aldrich), at 37°C in an anaerobic chamber. F. prausnitzii supernatant (SN) was recovered by centrifugation and filtered through 0.45-μm-pore-size filters (VWR, Haasrode, Belgium) and stored at −80°C.
Cell lines and co-incubations
The human colon carcinoma cell line HT-29 (ATCC HTB-38) was grown in Dulbecco’s Modified Eagle’s minimal essential medium with 4.5 g/L glucose (DMEM) (Sigma-Aldrich), supplemented with 10% (w/v) heat-inactivated fetal calf serum (FCS) (GibcoBRL, Eragny, France), 4 mM L-glutamine, and penicillin G/streptomycin (5000 IU/mL, 5000 µg/mL) (Sigma-Aldrich). Cultures were incubated in 25-cm2 tissue culture flasks (Nunc, Roskilde, Denmark) at 37ºC in a 10% (v/v) CO2 atmosphere until confluence.
For co-culture experiments, HT-29 cells were seeded in 24-well culture plates (Nunc) in DMEM supplemented with 10% heat-inactivated FCS-1% glutamine at 37°C in a 10% CO2-air atmosphere. Culture medium was changed every day. Experiments began on day 7 after seeding, when cells were at confluence (approx. 1.83 × 106 cells/well). On day 6, 24 h before co-culture with F. prausnitzii SN, the culture medium was changed to one with 5% heat-inactivated FCS and 1% glutamine. The day of the co-culture, either SN or LYBHI medium was added at a concentration of 10% (v/v) in a total volume of 500 µl. Cells were stimulated simultaneously with recombinant human TNF-α (5 ng/ml; Peprotech, NJ, USA) for 6 h at 37°C in 10% CO2. All samples were analyzed in triplicate.
ELISA
Supernatants of mock non-stimulated and TNF-α-stimulated HT-29 cells used for transcriptomic analysis were first validated for IL-8 modulation by ELISA. Supernatants for ELISA and cells for RNA extractions and transcriptomic analysis were collected at the same time and from the same culture plates. IL-8 concentrations were determined by ELISA (Biolegend, San Diego, CA), according to the manufacturer’s instructions. Results were reported as the mean values of duplicate ELISA wells.
RNA isolation
After co-incubation with either LYBHI or F. prausnitzii SN, HT-29 cells were collected and RNA-purified for DNA transcriptomic hybridization. Total RNA was extracted from cells using the RNeasy Mini Kit (Qiagen, USA) and purified by on-column digestion of DNA with DNase I as recommended by the manufacturer to eliminate residual genomic DNA. RNA concentration was determined by Nanodrop quantification (Thermo Fisher Scientific Inc., France). RNA quality was checked on an Agilent 2100 Expert Bioanalyzer (Agilent Technologies, France). Only RNAs with a RIN > 8 were used for transcriptomic and qRT-PCR experiments.
Microarray hybridization
A reference design with complete dye-swap including two biological replicates was used to compare HT-29 cells among different treatments, for a total of 6 Agilent 4x44K Whole Human Genome Microarrays (Agilent Technologies, France). Scheme design is available in Figure S2. For labeling, 100 ng of total RNA was reverse-transcribed and stained with Cy3 or Cy5 using the two-color Low Input Quick Amp Gene Expression Labeling Kit (Agilent Technologies, France) according to the manufacturer’s instructions. The CyDye-labeled cRNAs were then purified using the RNeasy Mini Kit (Qiagen, France). cRNA quantity was determined by Nanodrop and quality was checked on the Agilent 2100 Expert Bioanalyzer. Yield and specific activity were determined per manufacturer’s instructions. We mixed 825 ng of Cy3-labeled cRNA from one treatment with the same amount of Cy5-labeled cRNA from another treatment. cRNAs were hybridized to the Agilent 4x44K Whole Human Genome Microarray (Agilent Technologies, France) at 65°C for 17 h in a rotating incubator. After hybridization, slides were washed and then scanned using an Agilent G2565CA scanner (Agilent Technologies, France). Raw data were extracted using Feature Extraction software version 10.5.1.1 (Agilent Technologies, France).
Microarray analysis
R 3.0.2 software and the LIMMA packageCitation17 were used to analyze microarray data (within-array normalization by loess method followed by between-array normalization by quantile method) and to generate lists of differentially expressed genes. Microarray data were deposited in the NCBI-GEO database (accession number GSE72048). To create gene lists, we filtered by expression levels (|(FC)| > 1.5 as a cutoff) as well as by adjusted p-values, using a 0.05 threshold with the Benjamini and Hochberg false discovery rateCitation18 as multiple testing correction. Selected gene lists (log ratio and p-value data) were loaded into Ingenuity Pathway Analysis (IPA) and Multiarray Experiment ViewerCitation41 to analyze pathways and generate data displays.
Quantitative real-time RT-PCR (qRT-PCR)
Three μg of DNase I-treated total RNAs were reverse-transcribed using Oligo(dT) primers and 1 µl of SuperScript II reverse transcriptase (Invitrogen, France). The resulting cDNAs were quantified by Nanodrop (Thermo Fisher Scientific Inc., France) and diluted to a working concentration of 100 ng/μl. Reactions were performed in a final volume of 25 μl with 500 ng cDNA, 10 pM primers, and SYBR Green PCR Master Mix (Applied Biosystems, USA), using a Mastercycler Realplex (Eppendorf, France). Primers are listed in Table S4. The genes B2M (for human analysis), HMBS or actin (for mice analysis) were used as internal referencesCitation42 and the 2−ΔΔCT methodCitation43 was used to calculate the FC in gene expression.
Dact3 siRNA in HT-29 cells
HT-29 cells were cultured as described above. siGENOME® Human Dact3 siRNA-SMARTpool®, Dact3 siRNA D-015690-01, D-015690-02, D-015690-03, D-015690-17, and control siRNA (Non-Targeting siRNA D001136-01-05 and Cyclophilin B D-001210-02-05) siRNAs were transfected into HT-29 cells using Dharmafect 1 Transfection Reagent (Dharmacon, USA) following manufacturer’s instructions with some modifications. A total of 1 × 105 cells were plated in 12-well plates and transfected using 30 nmol siRNA and 2 µL of Dharmafect 1 Transfection Reagent per well in DMEM containing 5% FCS and 1% L-Glutamine. After 24 h, the medium was changed. After another 24 h, control medium, F. prausnitzii SN, or LYBHI medium was added at a concentration of 10% (v/v) in a total volume of 1 ml. Cells were simultaneously stimulated with recombinant human TNF-α (5 ng/ml; Peprotech, NJ, USA) at 37°C in 10% CO2. After 6 h of co-culture, supernatants were tested for IL-8 production by ELISA and RNA was isolated from cells for Dact3 expression analysis, as described above. All samples were generated and analyzed in triplicate.
Dact3 overexpression in HT-29 cells
HT-29 cells were cultured as above. The S-adenosylhomocystein hydrolase inhibitor 3-Deazaneplanocin A (DZNep) and Trichostatin A (Sigma-Aldrich Chemie GmbH, Buchs, Switzerland) were used as described inCitation21 with some modifications. Cells were plated at 0.5 × 105 cells/well; 24 h later, 12.5 M DZNep was added, and 48 hours after that, 0.5 µM TSA was added for the final 24 h of culture. After treatment, cells were stimulated with TNF-α (5 ng/ml; Peprotech, NJ, USA) and F. prausnitzii SN or LYBHI (added at 10% v/v). After 6 h of co-incubation, supernatants were tested by ELISA for IL-8 production and RNA was isolated from cells for Dact3 expression analysis. All samples were performed in triplicate.
Dact3-microbiota modulation in vivo
Microarray data comparing germ-free and conventionalized mice were described and published previously.Citation24 GEO accession number: GSE63299 Conventionalization of germ-free C3H/HeN mice was performed with fresh stools from C3H/HeN donor mice.
Dact3-F. prausnitzii SN modulation in vivo
C57BL/6 mice (males, 6–8 weeks of age; Janvier, Le Genest Saint Isle, France) were maintained at the animal care facilities of the National Institute of Agricultural Research (IERP, INRA, Jouy-en-Josas, France) under specific pathogen-free conditions. Mice were housed under standard conditions for a minimum of 1 week before experimentation. All experiments were performed in accordance with European Community rules and approved by the animal care committee COMETHEA (Comité d’Ethique en Expérimentation Animale du Center INRA de Jouy-en-Josas et AgroParisTech, Jouy-en-Josas, France). All assays were carried out under agreement N°3445-2016010615159974.
The protocol for Dact3 modulation and quantification in vivo is illustrated in Figure S4(a). Three groups of mice (n = 8) were intragastrically administered 200 µl of F. prausnitzii SN and sacrificed 3 h (T3), 6 h (T6) or 9 h (T9) after gavage. Control (T0) mice were not treated. Colitis was induced as described inCitation4, by intrarectal injection of 200 mg/kg of DNBS solution (Sigma-Aldrich) in 30% ethanol. Fourteen days following the first injection, a 200-µl solution of either 1 × 109 CFU of F. prausnitzii or PBS was administered intragastrically for 10 days. Colitis was reactivated 21 days after the first DNBS injection with a second injection of 200mg/kg of DNBS solution. Mice were sacrificed 3 days after the second DNBS injection (Figure S4(a)) by cervical dislocation and different parameters of inflammation were recorded as previously described.Citation8
Colon samples (1 cm from distal colon) were frozen in 500 µl of RNAlater solution (Ambion, France) in liquid nitrogen and stored at −80°C until use. Total RNA was extracted from individual samples with the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions, using homogenization with Tissue Lyser (Qiagen) and purification by on-column digestion of DNA with DNase I. Total RNA was determined by Nanodrop quantification. Dact3 expression was analyzed by qRT-PCR.
Statistical analysis
Statistical analysis, with the exception of transcriptomic analysis, was completed using GraphPad (GraphPad Software, La Jolla, CA, USA). A p-value of less than 0.05 was considered significant. Significant differences in the relative expression values of the target genes were tested with REST software using pairwise-fixed reallocation randomization.Citation44
Authors’ contributions
Author contributions: L.G.B.H. and F.C. designed research; M.L., R.M., E.T.M., S.C. and F.C. performed research; H.S. and P.G.D contributed analytic tools; M.L., R.M., H.S., F.C., and L.G.B.H., analyzed data; and M.L., R.M., H.S., P.L., F.C., and L.G.B.H wrote the paper. All read and approved the final manuscript.
Disclosure of potential conflicts of interest
The authors declare that they have no competing interests.
Supplemental Material
Download Zip (729.8 KB)Acknowledgments
The authors would like to thank Marco Moroldo, Jérôme Lecardonnel, and Déborah Jardet from the GABI/CRB GABI platform, and Valentin Loux and all IERP personnel for their help. We also thank Dr. Hélène Bierne for her precious advices.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317.
- Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi:10.1056/NEJMra0804647.
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Doré J, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15(8):1183–1189. doi:10.1002/ibd.20903.
- Miquel S, Martin R, Bridonneau C, Robert V, Sokol H, Bermudez-Humaran LG, Thomas M, Langella P. Ecology and metabolism of the beneficial intestinal commensal bacterium Faecalibacterium prausnitzii. Gut Microbes. 2014;5(2):146–151. doi:10.4161/gmic.27651.
- Miquel S, Martin R, Rossi O, Bermudez-Humaran LG, Chatel JM, Sokol H, Thomas M, Wells JM, Langella P. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16(3):255–261. doi:10.1016/j.mib.2013.06.003.
- Martin R, Miquel S, Ulmer J, Kechaou N, Langella P, Bermudez-Humaran LG. Role of commensal and probiotic bacteria in human health: a focus on inflammatory bowel disease. Microb Cell Fact. 2013;12:71. doi:10.1186/1475-2859-12-71.
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux -J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731–16736. doi:10.1073/pnas.0804812105.
- Martin R, Chain F, Miquel S, Lu J, Gratadoux -J-J, Sokol H, Verdu EF, Bercik P, Bermúdez-Humarán LG, Langella P, et al. The commensal bacterium Faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflamm Bowel Dis. 2014;20(3):417–430. doi:10.1097/01.MIB.0000440815.76627.64.
- Martin R, Miquel S, Chain F, Natividad JM, Jury J, Lu J, Sokol H, Theodorou V, Bercik P, Verdu EF, et al. Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol. 2015;15(1):67. doi:10.1186/s12866-015-0400-1.
- Martin R, Bermudez-Humaran LG, Langella P. Searching for the bacterial effector: the example of the multi-skilled commensal bacterium faecal bacterium prausnitzii. Front Microbiol. 2018;9:346. doi:10.3389/fmicb.2018.00346.
- Leonel AJ, Alvarez-Leite JI. Butyrate: implications for intestinal function. Curr Opin Clin Nutr Metab Care. 2012;15(5):474–479. doi:10.1097/MCO.0b013e32835665fa.
- Miquel S, Leclerc M, Martin R, Chain F, Lenoir M, Raguideau S, Hudault S, Bridonneau C, Northen T, Bowen B, et al. Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. MBio. 2015;6(2). doi:10.1128/mBio.00300-15.
- Quevrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, Miquel S, Carlier L, Bermúdez-Humarán LG, Pigneur B, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut. 2016 Mar;65(3):415-425. doi:10.1136/gutjnl-2014-307649.
- Fisher DA, Kivimae S, Hoshino J, Suriben R, Martin P-M, Baxter N, Cheyette BNR. Three Dact gene family members are expressed during embryonic development and in the adult brains of mice. Developmental Dynamics. 2006;235(9):2620–2630. doi:10.1002/dvdy.20917.
- Kim H, Jung BJ, Jung JH, Kim JY, Chung SK, Chung DK. Lactobacillus plantarum lipoteichoic acid alleviates TNF-alpha-induced inflammation in the HT-29 intestinal epithelial cell line. Mol Cells. 2012;33:479–486. doi:10.1007/s10059-012-2266-5.
- Kechaou N, Chain F, Gratadoux -J-J, Blugeon S, Bertho N, Chevalier C, Le Goffic R, Courau S, Molimard P, Chatel JM, et al. Identification of one novel candidate probiotic Lactobacillus plantarum strain active against influenza virus infection in mice by a large-scale screening. Appl Environ Microbiol. 2013;79:1491–1499. doi:10.1128/AEM.03075-12.
- Eckmann L, Jung HC, Schurer-Maly C, Panja A, Morzycka-Wroblewska E, Kagnoff MF. Differential cytokine expression by human intestinal epithelial cell lines: regulated expression of interleukin 8. Gastroenterology. 1993;105:1689–1697. doi:10.1016/0016-5085(93)91064-O.
- Gross V, Andus T, Daig R, Aschenbrenner E, Scholmerich J, Falk W. Regulation of interleukin-8 production in a human colon epithelial cell line (HT-29). Gastroenterology. 1995;108:653–661. doi:10.1016/0016-5085(95)90436-0.
- Andoh A, Fujiyama Y, Sumiyoshi K, Sakumoto H, Okabe H, Bamba T. Tumour necrosis factor-alpha up-regulates decay-accelerating factor gene expression in human intestinal epithelial cells. Immunology. 1997;90:358–363. doi:10.1111/j.1365-2567.1997.00358.x.
- van Rijn SJ, Riemers FM, van den Heuvel D, Wolfswinkel J, Hofland L, Meij BP, Penning LC. Expression stability of reference genes for quantitative RT-PCR of healthy and diseased pituitary tissue samples varies between humans, mice, and dogs. Mol Neurobiol. 2014;49:893–899.
- Jiang X, Tan J, Li JS, Kivimaee S, Yang XJ, Zhuang L, Lee PL, Chan MTW, Stanton LW, Liu ET, et al. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell. 2008;13:529–541. doi:10.1016/j.ccr.2008.04.019.
- Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. Acetate utilization and butyryl coenzyme A (CoA): acetate-CoA-transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol. 2002;68:5186–5190. doi:10.1128/AEM.68.10.5186-5190.2002.
- Urdaci MC, Sanchez B. Some immunomodulatory effects of probiotic bacteria might be due to porcine neutrophil elastase inhibitor, a serpin present in MRS broth. Immunol Lett. 2009;122:99–100. doi:10.1016/j.imlet.2008.11.004.
- Hoffmann TW, Pham H-P, Bridonneau C, Aubry C, Lamas B, Martin-Gallausiaux C, Moroldo M, Rainteau D, Lapaque N, Six A, et al. Microorganisms linked to inflammatory bowel disease-associated dysbiosis differentially impact host physiology in gnotobiotic mice. Isme J. 2016;10(2):460–477. doi:10.1038/ismej.2015.127.
- Laval L, Martin R, Natividad JN, Chain F, Miquel S, Desclee de Maredsous C, Capronnier S, Sokol H, Verdu EF, van Hylckama Vlieg J, et al. Lactobacillus rhamnosus CNCM I-3690 and the commensal bacterium Faecalibacterium prausnitzii A2-165 exhibit similar protective effects to induced barrier hyper-permeability in mice. Gut Microbes. 2015;6:1–9. doi:10.4161/19490976.2014.990784.
- Duncan SH, Hold GL, Harmsen HJM, Stewart CS, Flint HJ. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol. 2002;52:2141–2146.
- Mandal A, Waxman J. Retinoic acid negatively regulates dact3b expression in the hindbrain of zebrafish embryos. Gene Expression Patterns. 2014;16:122–129. doi:10.1016/j.gep.2014.09.003.
- Xue H, Xiao ZC, Zhang J, Wen J, Wang Y, Chang Z, Zhao J, Gao X, Du J, Chen Y-G, et al. Disruption of the Dapper3 gene aggravates ureteral obstruction-mediated renal fibrosis by amplifying Wnt/beta-catenin signaling. J Biol Chem. 2013;288:15006–15014. doi:10.1074/jbc.M113.458448.
- Beltran AS, Russo A, Lara H, Fan C, Lizardi PM, Blancafort P. Suppression of breast tumor growth and metastasis by an engineered transcription factor. PLoS One. 2011;6(9):e24595. doi:10.1371/journal.pone.0024595.
- Li H, Bitler BG, Vathipadiekal V, Maradeo ME, Slifker M, Creasy CL, Tummino PJ, Cairns P, Birrer MJ, Zhang R, et al. ALDH1A1 is a novel EZH2 target gene in epithelial ovarian cancer identified by genome-wide approaches. Cancer Prev Res. 2012;5(3):484–491. doi:10.1158/1940-6207.CAPR-11-0414.
- Xi SC, Yang MC, Tao YG, Xu H, Shan JG, Inchauste S, Zhang M, Mercedes L, Hong JA, Rao M, et al. Cigarette smoke induces C/EBP-beta-mediated activation of miR-31 in normal human respiratory epithelia and lung cancer cells. PLoS One. 2010;5(10):e13764. doi:10.1371/journal.pone.0013764.
- Neta G, Brenner AV, Sturgis EM, Pfeiffer RM, Hutchinson AA, Aschebrook-Kilfoy B, Yeager M, Xu L, Wheeler W, Abend M, et al. Common genetic variants related to genomic integrity and risk of papillary thyroid cancer. Carcinogenesis. 2011;32(8):1231–1237. doi:10.1093/carcin/bgr100.
- Fung KYC, Kerr C, Henderson S, Ilka P, Shaw J, Buckley MJ, Lockett T, Head R, Cosgrove L. Mechanisms associated with acquisition of resistance to butyrate-induced apoptosis in colorectal cancer cells using gene expression analysis. J Proteomics Genomics Res. 2014;1:16–30. doi:10.14302/.2326-0793.jpgr-14-598.
- Bocker U, Nebe T, Herweck F, Holt L, Panja A, Jobin C, Rossol S, Sartor R, Singer MV. Butyrate modulates intestinal epithelial cell-mediated neutrophil migration. Clin Exp Immunol. 2003;131(1):53–60. doi:10.1046/j.1365-2249.2003.02056.x.
- Lakhdari O, Tap J, Beguet-Crespel F, Le Roux K, de Wouters T, Cultrone A, Nepelska M, Lefèvre F, Doré J, Blottière HM, et al. Identification of NF-kappaB modulation capabilities within human intestinal commensal bacteria. J Biomed Biotechnol. 2011;2011:282356. doi:10.1155/2011/282356.
- Yin L, Laevsky G, Giardina C. Butyrate suppression of colonocyte NF-kappa B activation and cellular proteasome activity. J Biol Chem. 2001;276:44641–44646.
- Cuiv PO, Smith WJ, Pottenger S, Burman S, Shanahan ER, Morrison M. Isolation of genetically tractable most-wanted bacteria by metaparental mating. Sci Rep. 2015;5:13282. doi:10.1038/srep13282.
- Cheyette BNR, Waxman JS, Miller JR, Takemaru KI, Sheldahl LC, Khlebtsova N, Fox EP, Earnest T, Moon RT. Dapper, a dishevelled-associated antagonist of beta-catenin and JNK signaling, is required for notochord formation. Developmental Cell. 2002;2:449–461. doi:10.1016/S1534-5807(02)00140-5.
- Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermudez-Humaran LG, Smirnova N, Bergé M, Sulpice T, Lahtinen S, et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med. 2011;3:559–572. doi:10.1002/emmm.201100159.
- Benbouziane B, Ribelles P, Aubry C, Martin R, Kharrat P, Riazi A, Langella P, Bermúdez-Humarán LG. Development of a stress-inducible controlled expression (SICE) system in Lactococcus lactis for the production and delivery of therapeutic molecules at mucosal surfaces. J Biotechnol. 2013;168(2):120–129. doi:10.1016/j.jbiotec.2013.04.019.
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374+.
- Matouskova P, Bartikova H, Bousova I, Hanusova V, Szotakova B, Skalova L, Hribal ML. Reference genes for real-time PCR quantification of messenger RNAs and MicroRNAs in mouse model of obesity. PLoS One. 2014;9(1):e86033. doi:10.1371/journal.pone.0086033.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi:10.1006/meth.2001.1262.
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST (c)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:36e–36. doi:10.1093/nar/30.9.e36.