ABSTRACT
Increasing evidence suggests a significant role for microbiota dependent metabolites and co-metabolites, acting as aryl hydrocarbon receptor (AHR) ligands, to facilitate bidirectional communication between the host and the microbiota and thus modulate physiology. Such communication is particularly evident within the gastrointestinal tract. Through binding to or activating the AHR, these metabolites play fundamental roles in various physiological processes and likely contribute to the maintenance of intestinal homeostasis. In recent years, tryptophan metabolites were screened to identify physiologically relevant AHR ligands or activators. The discovery of specific microbiota-derived indole-based metabolites as AHR ligands may provide insight concerning how these metabolites affect interactions between gut microbiota and host intestinal homeostasis and how this relates to chronic GI disease and overall health. A greater understanding of the mechanisms that modulate the production of such metabolites and associated AHR activity may be utilized to effectively treat inflammatory diseases and promote human health. Here, we review microbiota-derived AHR ligands generated from tryptophan that modulate host-gut microbiota interactions and discuss possible intervention strategies for potential therapies in the future.
Introduction
The human gastrointestinal (GI) tract is a bioreactor with an incredibly diverse and dynamic microbial community referred to as the gut microbiota, which can be considered a functional organ. Over the past decade, tremendous strides have been taken toward understanding the composition and functional capacity of the gut microbiota that resides in the human GI tractCitation1. The mechanisms by which these microorganisms contribute to host health have been extensively investigated, and the gut microbiota has been recognized to play a critical role in human health by generating metabolites, which act both locally and systemically to influence human physiology and disease.Citation2 Modern analytical methods have revealed that thousands of low molecular weight metabolites are produced that may mediate host-gut microbiota interactions.Citation3 Many of these microbiota-dependent metabolites and co-metabolites have been demonstrated to correlate, either positively or negatively with human health or disease. However, a causal mechanism of action has only been identified for a small fraction of the microbiota-dependent metabolites identified thus far. Currently, the most studied categories of metabolites involved in host-gut microbiota interactions, include short-chain fatty acids (SCFA), such as acetate, propionate, and butyrate, produced from the fermentation of dietary fiber, bile acids secreted into the intestinal tract, and tryptophan-derived microbial metabolites.Citation4,Citation5 The biological function of these classes of metabolites continues to be refined. Recent evidence has highlighted that many gut derived microbial metabolites modulate aryl hydrocarbon receptor (AHR) activity, which facilitates metabolic communication between the host and gut microbiota.Citation6 The mechanisms underpinning such AHR-microbiota communication are multi-factorial, involving modulation of immune tolerance and response,Citation7 intestinal homeostasis,Citation8 carcinogenesis,Citation9 and intestinal barrier integrity.Citation10
AHR, a ligand-activated transcription factor, localized in the cytoplasm in a transcriptionally inactive state as a multimeric complex with two molecules of heat shock protein 90 (HSP90), one molecule of X-associated protein 2 (XAP2, also known as AIP), and one molecule of the HSP90 co-chaperone p23 (P23).Citation11 After binding ligand, the activated AHR translocates into the nucleus where it dimerizes with AHR nuclear translocator (ARNT) protein and forms a functional DNA-binding transcription factor. AHR and ARNT, belonging to the family of basic helix-loop-helix-Per-ARNT-Sim (bHLH-PAS) proteins, consisting of the bHLH domain, the PAS domain and the transactivation domain (TAD) where coactivators and corepressors interact ().Citation12 Ligand binding to AHR occurs within the PAS B domain. However, the PAS B region of ARNT is not able to bind ligands. It is now well established that activated AHR/ARNT complex (AHRC) following ligand binding is capable of recruiting multiple coactivator complexes, such as steroid receptor coactivator 1 (SRC-1),Citation13 CREB binding protein (CBP/p300),Citation14 nuclear coactivator 2 (NCoA2) and p/CIP,Citation15 receptor-interacting protein 140 (RIP140),Citation16 coiled-coil coactivator (CoCoA),Citation17 GAC63,Citation18 NcoA4, and TRIP23,Citation19,Citation20 which play significant roles in promoting AHR-responsive gene expression. These coactivator proteins have been reported to be involved in regulation of the transcriptional machinery and remodeling of chromatin structure.Citation11 In addition to these classic transcriptional regulators, AHR is recruited by other transcription factors during transcription, such as estrogen receptor-α (ERα),Citation21 and nuclear factor-κB (NF-κB).Citation22,Citation23 NF-κB containing members of RelB, RelA, c-Rel, p50, and p52,Citation24 transcription factors that modulate inflammatory responses. The canonical NF-κB pathway is triggered through an induction of toll-like receptor (TLR) signaling by bacterial products such as lipopolysaccharide (LPS) or cytokine production such as tumor necrosis factor α (TNFα) and IL1, leading to activation of RelA/p50 heterodimers mediating expression of proinflammatory and cell survival genes.Citation25 The interaction between AHR and NF-κB is bidirectional. AHR agonists has been shown to suppress NF-κB mediated-gene expression.Citation26 Likewise, RelA/p50 also repressed AHR transcriptional activity.Citation27 Interestingly, a direct physical interaction between AHR and RelB was also observed.Citation28 Alternative NF-κB pathway is activated by ligands of a subset of the tumor necrosis factor receptor (TNFR) superfamily members, such as lymphotoxin β receptor (LTβR), B-cell activating factor receptor (BAFFR), cluster of differentiation 40 (CD40), or receptor activator of NF-κB (RANK). The release of NF-κB-inducing kinase (NIK) activates and cooperates with IκB Kinase α (IKKα) to induce p100 phosphorylation ().Citation29 RelB/p52 dimer is produced and translocates into nucleus through the processing of p100.Citation30 RelB/AHR binding element (RelBAHRE) is formed by the interaction between AHR and RelB, regulating the expression of cytokines and chemokines.Citation31 Several reports have shown that multiple inflammatory genes, including chemokines such as monocyte attracting protein (MCP), interleukin 8 (IL8), and CC-chemokine ligand 1 (CCL1), are regulated in an AHR-dependent manner.Citation32–34 Upon exposure to AHR ligands, the AHR/ARNT heterodimer binding to dioxin response elements (DRE) alters numerous AHR target genes, such as cytochrome P450 (CYP) Cyp1a1 and Cyp1b1, and Ahrr ().Citation35 CYP1A1 and CYP1B1 encoded by Cyp1a1 and Cyp1b1 respectively, belong to the cytochrome P450 superfamily of enzymes and have been shown to have a profound impact on the metabolism of xenobiotics.Citation36 Expression of the target gene Ahrr is enhanced by activation of AHR. Interestingly, the generation of AHR repressor (AHRR) serves as an important negative feedback mechanism to limit the availability of ARNT to form AHR/ARNT dimers.Citation37 Additionally, targeted protein degradation of the AHR downregulates AHR signaling. After being exported out of the nucleus, AHR is rapidly degraded in the cytoplasmic compartment by the proteasome.Citation38 Transactivation activity of AHR is also repressed by post-translational modifications such as its SUMOylation.Citation39 AHR was initially discovered as a primary mediator of the toxic effects arising from exposure to man-made environmental contaminants, such as polycyclic or halogenated aromatic hydrocarbons (PAHs or HAHs). For example, AHR was initially identified as the high-affinity receptor for the environmental toxicant 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), a type of HAH.Citation40 The role of the AHR as a xenobiotic sensor responsible for mediating such toxicity and tumor promotion has dominated research on this receptor for decades. Consequently, the underlying physiological ‘raison d’êtra’ of AHR is only now being appreciated.
Figure 1. Structures of AHR and ARNT. AHR and ARNT belong to the family of basic helix-loop-helix-Per-ARNT-Sim (bHLH-PAS) proteins. AHR contains a bHLH, a PAS, and transactivation domain. The transactivation domain is divided into three modular transcriptional domains, namely, an acidic region enriched with glutamic and aspartic acid residues, a glutamine-rich region (Q-rich), and a P/S/T region rich in proline/serine/threonine residues. ARNT has the similar structure with AHR. The function of each domain is illustrated above
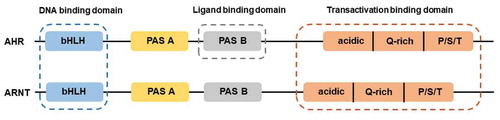
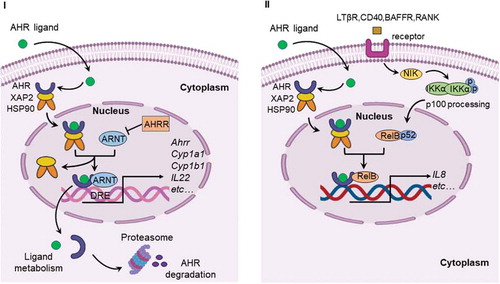
Figure 2. Canonical AHR signaling pathway and alternative AHR/RelB pathway. I. Canonical signaling pathway of AHR. Inactive AHR is retained in the cytoplasm in a multi-protein complex containing chaperone proteins, such as HSP90 and XAP2. AHR translocates into the nucleus after ligand binding and interacts with ARNT to form an AHR/ARNT complex. The AHR/ARNT dimer binds promoter regions containing DRE that regulate expression of numerous target genes, such as Ahrr, Cyp1a1, Cyp1b1 and IL22. II. AHR can also interact with RelB to induce the expression of cytokines and chemokines. The alternative NF-κB pathway is induced by ligands of TNFR superfamily members and is an IKKα-dependent kinase cascade. Activation of this cascade mediates phosphorylation of NIK leading to phosphorylation of IKKα, and subsequent phosphorylation of the p100 NF-κB subunit. This subunit is then cleaved to p52 leading to the formation of the p52/RelB complex. Following translocation of p52/RelB complex to the nucleus, AHR can interact with RelB to form RelB/AhR response element (RelBAHRE) regulating the expression of cytokines and chemokines. AHRR, AHR repressor; ARNT, AHR nuclear translocator; Cyp1a1, cytochrome P450 1A1; Cyp1b1, cytochrome P450 1B1; DRE, dioxin response element; HSP90, heat shock protein 90; IL22, interleukin 22; XAP2, X-associated protein 2; NF-κB, nuclear factor-κB; LTβR, lymphotoxin β receptor; RANK, receptor activator of NF-κB; BAFFR, B-cell activating factor receptor; CD40, cluster of differentiation 40; NIK, NF-κB-inducing kinase; IKK, IKB kinase; IL8, interleukin 8; RelBAHRE, RelB/AHR response element
As stated previously, activation of AHR has immunomodulatory effects through interaction with NF-κB. Compelling evidence has accumulated supporting a role for the AHR in regulating adaptive immune responses relevant to the pathogenesis of diseases, such as inflammatory bowel disease (IBD), multiple sclerosis (MS), rheumatoid arthritis (RA), cancer, and obesity.Citation7,Citation41–44 These observations have led to the hypothesis that the AHR participates in innate immune responses to microbial invasion of barrier tissues. The combination of a TLR ligand (e.g. LPS) and a microbially generated metabolite that activates the AHR could lead to an enhanced inflammatory response.
Thus, the study of AHR regulation and function is likely to reveal unknown biological processes highly related to the development of inflammatory disorders and may realize the potential of an AHR as a novel therapeutic intervention for clinical use. There are a number of reviews published that focus on the identification of exogenous and endogenous AHR ligands.Citation45,Citation46 While here, we review the most recent insights related to microbial AHR ligands produced from host-gut microbiota metabolism with a focus on tryptophan and indole metabolites. Also, we discuss the role of these endogenous or microbially generated metabolites via ligand-dependent activation of AHR to regulate inflammation in multiple chronic diseases.
AHR ligands produced by host-gut microbiota interaction
Ligand-dependent activation of the AHR is mediated by diverse sources, including environmental pollutants, dietary components, and endogenous metabolites. Endogenous mechanisms of AHR activation, independent of xenobiotics, were recognized by the development of Ahr−/- mice, which revealed developmental defects in the immune system.Citation47,Citation48 Increasingly, evidence indicates that the AHR has a wider role than previously recognized, mediating diverse biological processes. Thus, it is critical to further characterize endogenous molecules that influence receptor activity and how that informs us about the AHR in physiological processes. Much interest has focused on a search for putative endogenous AHR ligands. In recent years, many studies have shown that gut microbiota residing in the GI tract provides a rich source of pseudo-endogenous AHR ligands. Interestingly, endogenous AHR ligands have largely been identified as metabolites of tryptophan and indole metabolism (). Furthermore, production of pseudo-endogenous AHR ligands in the GI tract appears to be dependent on the types of bacteria present.Citation49,Citation50 However, although tryptophan itself neither exhibits AHR-binding capacity nor the ability to induce AHR activity, a number of endogenous metabolites of tryptophan are recognized as AHR ligands, including tryptamine (TrA), indole, 2-oxindole, 3-methylindole (skatole), indoxyl sulfate (IS), indole-3-acetic acid (I3A), indole-3-propionic acid (IPA), indole-3-pyruvate (IPyA), indole acrylic acid (IAA), Indole-3-aldehyde (IAld), indole-3-lactic acid (ILA), 5-hydroxyindole-3-acetic acid (5-HIAA), kynurenine, kynurenic acid (KA), and xanthurenic acid (XA).Citation46 The majority of previous studies have focused on the ability of endogenous metabolites to activate AHR, largely without regard to whether the concentration present in the intestinal tract is at a concentration that is physiologically relevant in terms of activating the AHR. Thus, an understanding of key endogenous AHR ligands within the GI tract has not been properly defined, especially in non-pathological conditions. Such knowledge could be beneficial in investigating the physiological roles of the AHR in host-gut microbiota interactions. In a recent study, a panel of tryptophan metabolites capable of activating AHR were quantified in cecal contents from mice and human stool under physiologically “normal” conditions.Citation51 Importantly, indole, 2-oxindole, I3A, and KA derived from tryptophan metabolism pathways by gut microbiota were recognized as the primary sources of AHR activation within the GI tract based on concentrations present and AHR activation potential. This is the first study to identify the importance of microbially produced 2-oxindole and KA to AHR activation within the gut. In addition, these tryptophan metabolites exhibit far greater Cyp1a1 activation potency for the human AHR when compared to the mouse AHR. These results need to be taken into account when considering the limitations of the mouse model and its extrapolation to humans. Interestingly, the human AHR has a key amino acid residue, valine 381, in the ligand-binding pocket that is an alanine in other primates; this results in the human AHR having lower affinity for exogenous ligands, such as PAHs.Citation52 However, the amino acid change does not affect the affinity for indole, which suggests that the ability of endogenous tryptophan metabolites to activate the AHR is conserved across all primates.
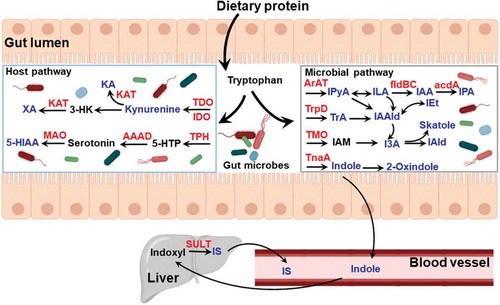
Figure 3. Summary of tryptophan and indole metabolites generated by host and gut microbiota metabolism. Tryptophan is acquired from dietary protein digested in the small intestine and converted to various catabolites. The key metabolites (in blue) have been identified to be AHR ligands and enzymes involved in metabolism are represented in red. AAAD, aromatic amino acid decarboxylase; TDO, tryptophan 2,3-dioxygenase; IDO, indoleamine 2,3-dioxygenase; KA, kynurenic acid; KAT, kynurenine aminotransferase; MAO, monoamine oxidase; TPH, tryptophan hydroxylase; 5-HIAA, 5-hydroxyindole-3-acetic acid; 3-HK, 3-hydroxykynurenine; 5-HTP, 5-hydroxytryptophan; ArAT, aromatic amino acid transaminase; fldBC, phenyllactate dehydratase; IAA, indole acrylic acid; I3A, indole-3-acetic acid; IAld, indole-3-aldehyde; IAAld, indole-3-acetaldehyde; IAM, indole-3-acetamide; ILA, indole-3-lactic acid; IEt, indole-3-ethanol; IPyA, indole-3-pyruvate; IPA, indolic-3-propionic acid; TMO, tryptophan-2-monooxygenase; TnaA, tryptophanase; TrpD, tryptophan decarboxylase; IS, indoxyl sulfate; TrA, tryptamine; SULT, sulfotransferase
AHR ligands and host metabolism in tryptophan catabolism
Tryptophan is an essential amino acid that can only be acquired through dietary protein. Dietary tryptophan is liberated into the small intestine through proteolytic digestion. The majority of free tryptophan is absorbed through intestinal epithelium, principally along the jejunal-ileal axis, and is utilized for protein synthesis by the host; while ~10-20% of tryptophan is further metabolized, the kynurenine and serotonin pathways or by gut microbes ().Citation53 Generation of endogenous AHR ligands, such as kynurenine and KA, is largely achieved through tryptophan metabolism in host cells via the enzymes indoleamine 2,3-dioxygenase (IDO) and tryptophan 2,3-dioxygenase (TDO).Citation54 Kynurenine is a low-affinity AHR ligand that has been linked to regulatory T (Treg) cell functional maturation and suppression of inflammatory cytokine production in dendritic cells (DCs), altering the progression of inflammation leading to immune dysfunction.Citation55 KA, generated by transamination of kynurenine via kynurenine aminotransferases (KAT) activity, is found to be a relatively potent ligand for human AHR and co-mediates the induction of IL6 expression coupled with inflammatory signaling.Citation56 Also, this study found that XA generated by transamination of the kynurenine metabolite 3-hydroxykynurenine (3-HK), activated AHR in a DRE-driven luciferase reporter in human hepatoma cells. Serotonin (5-hydroxytryptamine, 5-HT) known as a brain neurotransmitter, is largely produced in the gut from the amino acid tryptophan via hydroxylation and decarboxylation. Over 90% of serotonin in the body is synthesized by colonic enterochromaffin cells (ECs).Citation57 Gut-derived serotonin is regulated by the gut microbiota, particularly spore-forming bacteria.Citation58 Yano et al. showed that 60% less serotonin were produced in the ECs from germ-free mice compared to specific pathogen-free mice. In a recent study, Manzella et al. found serotonin is an endogenous activator of AHR.Citation59 While it is not a ligand for AHR, serotonin acting as a CYP1A1 substrate, interferes with metabolic clearance of AHR ligands through inhibition of CYP1A1 activity leading to a decrease in ligand degradation and promotion of AHR activation in intestinal epithelial cells.Citation60,Citation61 Clearly, whether other microbial metabolites are capable of modulating AHR activity through inhibition of CYP1A1 should be explored. In a recent study, a novel AHR ligand of 5-HIAA derived from serotonin by monoamine oxidase (MAO) activity was discovered to be a transcriptional marker for regulatory B cells function via AHR activation.Citation42
AHR ligands and yeast infections
Providing vital protection from physical or chemical harm and from infection, the skin serves as the barrier against the environment. A significant amount of AHR ligands can be produced by microorganisms of the skin microbiome, e.g., Malassezia. Malassezia yeast are commensal microbes that, under certain host pathological conditions, can result in disease. Culturing a number of specific strains of Malassezia in tryptophan-rich media results in the production of a number of tryptophan metabolites with potent AHR agonist activity, such as indole[3,2b]carbazole (ICZ), 6-formylindolo[3,2b]carbazole (FICZ), and malassezin.Citation62,Citation63 Extracts isolated from skin scale from patients with Malassezia-associated diseases revealed an increase of 10–100-fold increase in AHR activation potential compared to control skin extracts. A number of potent AHR ligands were identified in these extracts that are seen in Malassezia yeast cultures. Such observations were among the first to establish a direct relationship between the microbiota, its associated metabolites, AHR, and pathology. It would be expected that potent activation of the AHR in the host epithelium, coupled with the presence of the yeast, would result in a heightened inflammatory response. Extra-mammary Paget’s disease (EMPD) is a skin cancer and the possible role of Malassezia-derived AHR ligands in the progression of the disease has been investigated.Citation64 In lesioned tissue, EMPD keratinocytes expression of the AHR target genes CYP1A1 and CCL20 is enhanced. These results are consistent with the assertion that AHR activation participates in inflammatory signaling in human disease. In fact, a previous study by Zelante et al. has revealed the activation of AHR by tryptophan metabolites provides antifungal resistance to Candida albicans via interleukin-22 and mucosal protection from inflammation.Citation50 In another yeast-mediated disease, oropharyngeal candidiasis, which is most often associated with Candida albicans infection of the oral epithelium in AIDS patients, the AHR may be involved in the progression of the disease.Citation65 Kynurenine activated AHR in oral epithelial cells increases activation of Src family kinases that enhances fungal endocytosis. These studies reinforce the concept that microbes that produce AHR ligands can modulate disease status and offers the potential of therapeutic intervention with AHR modulators.
AHR ligands and gut microbiota in tryptophan catabolism
A number of studies have revealed the microbiota as a driving force mediating the degradation of tryptophan in the large intestine. Interestingly, a number of endogenous AHR ligands are produced by metabolic activity of microbiota that inhabit the GI tract. For instance, TrA, produced by tryptophan decarboxylases (TrpDs) from C. sporogenes and Ruminococcus gnavus,Citation66 exhibit AHR-dependent anti-inflammatory activities in Caco2 intestinal cells.Citation67 In addition, TrA is the precursor of IAAld, which also manifests as an AHR ligand.Citation43 Indole-3-ethanol (IEt), known as tryptophol, is formed from IAAld by alcohol dehydrogenase. A recent study determined that IEt, IPyA and IAld derived from tryptophan by gut microbiota could regulate gut barrier function via AHR activation.Citation68 IPyA, a transamination product of dietary tryptophan by microbiota, such as Clostridium sporogenes,Citation69 is a precursor of several microbiota-derived AHR agonists, such as I3A, IAld, and IAAld, and can form various AHR agonists in the reaction of aqueous solution.Citation70 Several intestinal bacteria, including Bacteroides fragilis, Bacteroides thetaiotaomicron and Citrobacter sp.Citation71 and clostridia,Citation72 Clostridioides difficile,Citation73 Clostridium sticklandii, Clostridium lituseburense, Clostridium subterminale and Clostridium putrefaciens, can produce I3A from tryptophan. One study revealed that I3A regulated proinflammatory and oxidative effects in endothelial cells, and activated a signaling pathway related to AHR, resulting in cyclooxygenase-2 (COX2) expression.Citation74 Levels of I3A in serum have been associated with high rates of mortality and cardiovascular events in patients with chronic kidney disease (CKD), thus I3A is suggested to be a predictor of mortality and major cardiovascular events in CKD.Citation74 Also, a study reported that I3A, which was reduced in mice on a high-fat diet versus a low-fat diet, attenuated cytokine-mediated lipogenesis in hepatocytes, playing a potential protective role in lipid metabolism.Citation43 Some strains of Lactobacillus reuteri and Lactobacillus johnsonii can induce the generation of IAld from tryptophan via several pathways ().Citation50 In mice, AHR activation by IAld induced IL22 production resulting in enhanced restoration of mucosal immune homeostasis.Citation50 3-Methylindole (skatole) is formed by the decarboxylation of I3A by Lactobacillus sp. in the large intestine of humans.Citation75 In a study by Hubbard et al., in primary human bronchial epithelial and Caco2 cells, skatole can activate AHR at physiologically attainable concentrations to promote the expression of AHR target genes, such as CYP1A1. In addition, skatole acts as a competitive antagonist of TCDD-mediated AHR activation, indicating that skatole is a weak AHR agonist.Citation76 Through aromatic amino acid aminotransferase (ArAT) and indolelactic acid dehydrogenase (ILDH) activity,Citation77,Citation78 tryptophan can be converted to ILA. It has been reported that ILA could be produced by Bifidobacterium spp and Clostridium Sporogenes.Citation69,Citation79 Meng et al. reported ILA suppressed the transcription of inflammatory cytokine IL8 by interacting with AHR activation.Citation80 Recently, a study using untargeted metabolomics analyses found two tryptophan microbial metabolites, IPA and IAA, are produced by Peptostreptococcus. russellii and Peptostreptococcus. stomatis in a phenyllactate dehydratase fldABC-dependent manner; In addition, metagenomic analysis of stool samples from IBD patients revealed that production of IAA is associated with anti-inflammatory activity.Citation81
Indole metabolism has also been revealed as a rich reservoir of AHR ligands. Indole is synthesized from tryptophan by microbial tryptophanase (TnaA) activity, first recognized in the late 1800s. Many bacterial species encoding the TnaA gene can produce indole, consisting of Gram-positive bacteria such as, Bacillus alveiCitation82 and Clostridium tetani,Citation72 and Gram-negative bacteria such as E. coli,Citation83 Bacteroides thetaiotaomicron,Citation84 Pasturella multocidaCitation85 and Vibrio cholerae (previous Asiatic cholera).Citation86 Consequently, indole is present within the intestinal lumen and human feces at micromolar concentrations.Citation87 Oxidation of indole to 2-oxindole is putatively involved in the pathophysiology of hepatic encephalopathy.Citation88 A study by Hubbard et al. suggests indole and 2-oxindole at physiological concentrations can activate the human AHR within the GI tract.Citation89 Oxidation and sulfation of indole by hepatic oxidases, including cytochrome P450 2E1 (CYP2E1) and sulfotransferase 1A1 (SULT1A1), generate the systemic AHR ligand IS.Citation90,Citation91 IS is typically subject to nephrotic elimination but reaches high serum levels during kidney failure and chronic kidney disease and manifests as an uremic toxin. As a potent endogenous AHR agonist, IS selectively activates the human AHR at nanomolar concentrations in primary human hepatocytes and regulates transcription of multiple genes.Citation92 Studies have revealed that the formation of serum levels of IS in mice is dependent upon the presence of gut flora and suggests a linkage between the gut microbiota and kidney disease.Citation93,Citation94 In CKD patients, serum concentrations of IS are greater than 50-fold higher than those in healthy subjects.Citation95 These studies lead to the conclusion that the bacterial composition of the microbial community, the amount of bacterial metabolic activity, and the amount of available tryptophan in the gut will dictate the level of AHR ligands generated by the microbiota. However, the appropriate level of these AHR ligands to maintain optimal health will need to be further explored.
AHR-dependent regulation in health and diseases via tryptophan and indole metabolites
In the last few decades, it has become apparent that the gut microbiota is an important deterrent or contributor to many noninfectious disease states. In particular, microbial metabolites have been shown to facilitate the interaction between host and gut microbiota involved in systemic homeostasis across the so-called microbiota-gut-brain and microbiota-gut-liver axes.Citation96,Citation97 Although the mechanisms of microbial metabolites mediating signal between host and gut microbiota remain poorly understood, recent studies revealed AHR to be a key regulator in such interaction between host and gut microbiota. Next, we discuss the microbial metabolites that activate the AHR regulating crosstalk between the host and gut microbiota under pathological conditions.
AHR and inflammatory bowel disease
The etiology of IBD consisting of Crohn’s disease (CD) and ulcerative colitis (UC) remains incompletely understood,Citation98 but is believed to be multi-factorial, involving genetic predisposition, lifestyle, diet, and a strong microbiota component.Citation99,Citation100 In a recent study, the association between serum levels of tryptophan and its metabolites in patients with IBD, and clinical and serologic features was systematically evaluated.Citation101 Subsequently, reduced levels of tryptophan were observed in the serum from IBD patients and there was an inverse association between disease activity and serum levels of tryptophan. This negative correlation could also be observed in dextran sulfate sodium-induced colitis in mice.Citation102 Furthermore, the microbiota from mice deficient in caspase recruitment domain 9 (CARD9), a susceptibility gene for colitis,Citation103 failed to produce AHR ligands metabolized from tryptophan.Citation104 Lamas et al. found that the subsequent reduction in AHR activity was associated with reduced IL22 expression and enhanced susceptibility to colitis. Interestingly, not only tryptophan but also microbiota-derived metabolites from tryptophan and indole, could modulate the severity of IBD. Diminished I3A levels and AHR activation are also observed in fecal samples from patients with IBD compared with healthy individuals.Citation104 In addition, in experimental colitis models IPyA is shown to be a potent AHR agonist and exhibits significant anti-inflammatory effects, through increased abundance of colonic IL10 positive T cells combined with a reduction in pro-inflammatory Th1 cells.Citation105 Several studies have revealed that microbiota-dependent tryptophan metabolites, such as indole, serve an important role in maintaining intestinal homeostasis through AHR activation.Citation106,Citation107 These observations suggest tryptophan metabolites and AHR activation potential are involved in the etiology or progression of IBD, thus providing a promising target of therapeutic or nutritional intervention for IBD treatment.
AHR and central nervous system
In recent years, there has been significant interest in the gut–brain axis, and data suggest that AHR agonists provided by the interaction of the gut microbiota and host metabolism, can cross the blood-brain barrier and contribute to control of central nervous system (CNS) function. Emerging evidence suggests that dysregulation of tryptophan metabolism by the kynurenine pathway is associated with neurodegenerative and other neurological disorders, along with psychiatric diseases.Citation108 The production of downstream products of the kynurenine pathway is dependent on expression of IDO and TDO. Considering the expression of IDO/TDO is regulated by activation of AHR;Citation109,Citation110 tryptophan catabolites via kynurenine pathway are involved in immune, metabolic and neural communication mechanisms in gut-brain axis, which is controlled by AHR activation.Citation111 In addition, certain microbiota that produce indole and indole derivatives from dietary tryptophan can also control astrocyte activity to influence CNS inflammation.Citation112 Another study reported AHR agonists in serum are altered during different stages of multiple sclerosis,Citation113 the most common neurological autoimmune disease of CNS. In this cohort study, reduced AHR activation in patients with relapsing-remitting MS was observed when compared to healthy controls. Also, a decrease of AHR transcriptional activity was detected in patients with MS in remission compared to those with active inflammation. Interestingly, there was no difference between patients with benign MS and controls, while an increase in agonist-mediated AHR activity is observed in patients with acute inflammation. These results suggest that deficits in AHR agonists provided by interaction between host metabolism and gut microbiota may contribute to the pathogenesis of MS. These AHR agonists represent a potential biomarker to predict disease stages of MS, including relapse frequency and disability progression, which is often difficult to predict even with a combination of clinical criteria, magnetic resonance imaging, and cerebrospinal fluid findings.
AHR and rheumatoid arthritis
Millions of people worldwide suffer from the autoimmune disorder of RA and its mortality is increasing. Although the mechanisms driving RA progression are not well understood,Citation114 recent data implicate the gut microbiota in the etiology of RA.Citation115 In a recent study by Rosser et al., RA patients exhibited reduced levels of the microbiota-derived SCFA butyrate compared to healthy controls; and butyrate levels showed a significant positive correlation with the frequency of total CD19+ CD24hiCD38hiB cells and with IL10+B cells.Citation42 SCFAs generated by gut microbiota are known to play a vital role in the homeostasis of intestinal epithelium, including providing nutrients for intestinal epithelial cells, supporting barrier function and regulating the production of cytokines and chemokines.Citation2 Notably, studies of activation of the AHR by TCDD in mice have highlighted its role for differentiation and function of adaptive immune cells, such as T cells and DC cells.Citation116–118 Consistent with AHR activation inducing immunosuppression, Piper et al. observed that AHR played a critical role in inducing regulatory B cells differentiation and IL10 transcription to attenuate autoimmunity and inflammation.Citation119 Building on these findings, a significant increase in microbiota-induced AHR ligand 5-hydroxyindole-3-acetic acid (5-HIAA), metabolized from serotonin, was discovered to attenuate arthritis with butyrate supplementation by promoting IL10 transcription in B cells in an AHR-dependent manner.Citation42
AHR role in obesity and gut/liver axis
Obesity is a global health problem that is exacerbated by western diets high in caloric content, fat and carbohydrate and often characterized by the low consumption of fruit and vegetables. Along with obesity, fatty liver and increased visceral fat can lead to an elevation in inflammatory signaling. Accumulating evidence reveals that the AHR plays a key role in the crosstalk between obesity and gut microbiota. The supplementation of the diet in mice with an AHR ligand (e.g. FICZ) or a Lactobacillus strain that produces AHR ligand(s) is capable of improving metabolic syndrome factors, such as hepatic triglyceride levels, fasting glucose levels, and insulin in ob/ob mice.Citation120 Importantly, in humans, a high body mass index correlated with a reduced level of AHR active tryptophan metabolites and overall AHR activation potential in fecal extracts. A similar level of AHR activity in stool was shown in fecal microbiota transplantation experiment. Germ-free mice colonized with fecal microbiota of high-fat diet-fed mice resulted in a reduction of AHR activity compared with mice colonized with microbiota of conventional diet-fed microbiota. These findings indicate a decrease of microbiota-derived AHR ligands was associated with a shift in composition of gut microbiota in obese humans. In fact, a lower abundance of Bacteroidetes was observed in the obese mice compared to lean mice.Citation121 Another possible explanation that can, at least in part, provide a mechanism for this observation is the ability of AHR activation in the GI tract to induce IL22 production and its subsequent ability to attenuate adverse metabolic parameters in ob/ob mice.Citation122 This observation, coupled with results demonstrating that mice deficient in IL22 receptor expression fed a high-fat diet are susceptible to the development of metabolic dysfunction, would suggest that AHR activation in the gut has a protective effect against obesity. Importantly, AHR activation provides protection for the stem cell niche in the intestinal epithelium through restricting cell proliferation and influencing differentiation and enhances overall barrier function.Citation123
A different story emerges from studies designed to examine the influence of AHR activation in the liver on obesity. Transgenic mouse models that express a constitutively activated AHR in the liver have revealed that elevated expression of the CD36/fatty acid translocase results in a fatty liver phenotype through the increased uptake of fatty acids.Citation124 The administration of 3-methyl cholanthrene, a potent AHR ligand, in C57BL6/J mice results in an increase in both liver triglycerides and CD36 expression.Citation125 This would suggest that high-level AHR activation can lead to fatty liver. Another possible mechanism is the ability of CYP1B1 enzyme to influence fatty acid metabolism and prevent obesity, supported by results obtained in Cyp1b1−/- mice.Citation126 The loss of Cyp1b1 expression in mice is correlated with suppression of high-fat diet (HFD)-induced obesity via decreased expression of stearoyl-CoA desaturase 1 (SCD1),Citation127 the rate-limiting enzyme synthesizing monounsaturated fatty acids from saturated fatty acids.Citation128 Moreover, expression of PPARγ and target genes regulated by PPARα are also reduced in Cyp1b1 knockout mouse.Citation129 Activation of PPARγ promotes induction of adipocytes from preadipocytes and storage of triglyceride, while genes stimulated by PPARα plays a critical role in fatty acid transport and mitochondrial fatty acid β-oxidation.Citation130 Taken together, these results suggest the involvement of AHR in regulation of CYP1B1 expression, which promotes fatty acid synthesis, and inhibition of CYP1B1 could be a potential target for clinic treatment of obesity. Indeed, dietary exposure to AHR antagonist α-naphthoflavone is capable of inhibiting fatty liver and obesity in mice fed a western diet.Citation131,Citation132
Therapeutic potential of AHR in clinical applications
Recent technical progress in the field of global untargeted metabolomics, which focuses on identifying low molecular weight metabolites that impact the host, have facilitated the screening of microbial metabolites that may induce AHR activity and subsequently influence host-gut microbiota homeostasis. Such analyses performed on cecal contents from conventional and germ-free mice with subsequent activity assays have identified 2,8-dihydroxyquinoline (2,8-DHQ) as a microbial-derived AHR ligand.Citation133 In addition, accumulating evidence suggests that AHR plays a vital role in the regulation of inflammatory signaling leading to changes in the abundance and function of various immune cells involved in both the innate and adaptive immune systems, such as Th17, Treg and DC.Citation134 Taken together, progress in the identification and function of microbiota-derived AHR ligands may lead to novel therapeutic options for multiple inflammatory diseases. Indeed, the therapeutic modulation of the gut microbiota and consequently microbial AHR ligands is being investigated for improving autoimmune diseases, such as IBD, CNS disease, and metabolic syndrome ().Citation112,Citation135 For example, probiotics containing L. reuteri, which have been demonstrated to generate AHR agonists, have been successfully utilized to alleviate inflammation.Citation136 Not only the use of AHR agonist-producing probiotics, but also direct oral administration of microbial AHR agonists, such as IPA, indole, and IPyA, have been shown to reduce experimental intestinal inflammation in an experimental colitis model.Citation105,Citation137,Citation138 A recent study demonstrated that a diet supplemented with the tryptophan metabolites indole, IS, IPA and IAld limited CNS inflammation in antibiotic-treated mice.Citation112 Furthermore, in order to minimize off-target AHR effects in non-targeted tissues, nanoparticles have been used for the targeted delivery of AHR agonists to the desired tissues and cells.Citation139 Current research has shown that, antigen delivered by nanoparticle can be used to promote protective immunity against microorganisms and cancer.Citation134
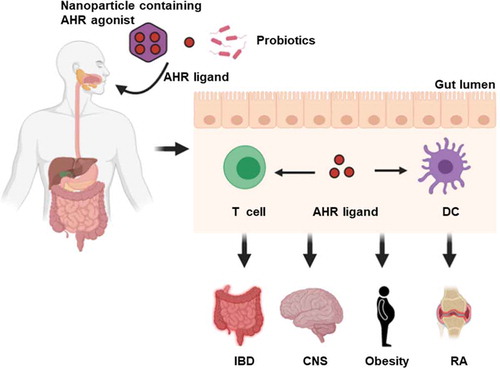
Figure 4. AHR as a therapeutic target. Novel strategies such as oral administration of nanoparticle-mediated cell-type-specific delivery of AHR agonist, AHR ligands and specific probiotics may serve to influence autoimmune inflammatory response. DC, dendritic cell; IBD, inflammatory bowel disease; CNS, central nervous system; RA, rheumatoid arthritis
Conclusions
Numerous studies now support the conclusion that the AHR plays an important role in maintaining gut homeostasis both under homeostatic conditions and during chemical or bacterial challenge. However, there is a need to further expand our understanding of the role for the AHR in multiple cell types within the intestinal tract, including various immune cell types, in the gut epithelium, in the underlying lamina propria, and in the enteric nervous system. This information will be critical to understanding what level of pseudo-endogenous ligands (e.g. tryptophan metabolites) or dietary ligands (e.g. indolo[3,2b]carbazole) is optimal in maintaining healthy gut health. Whether these microbial or dietary AHR ligands could be used for therapeutic purposes also warrant further investigation. This concept is supported by the observation that indole-3-carbinol can attenuate the impact of C. difficile infection in mice.Citation140 Yet another area of investigation involves understanding the impact of interspecies difference in AHR activation. In particular, it is important to note that there are several important structural differences in the human versus mouse AHR, thus raising concern about the use of the mouse models. Nevertheless, most studies on microbial AHR ligand intervention have been evaluated in mouse model. A number of studies establish that the affinity of the AHR for TCDD in mice is 10-fold higher than in humans.Citation141 In contrast, IS is a more potent ligand for the human AHR and has a 500-fold higher potency for human versus the mouse AHR.Citation92 In addition, another study revealed that different levels of species-dependent AHR activation were observed for the indole derivatives; indole, 2-oxindole and skatole, which potently activate the human AHR in a dose-dependent manner, whereas only low concentrations of IPA could induce activity.Citation89 Yet another area to explore is the exact function of each AHR ligand in the different intestinal segments and cell types. Importantly, these microbial AHR ligands from tryptophan and indole metabolism exhibited markedly different concentrations in the luminal contents. For example, Karlin et al. identified the quantity of indole in human feces as a major microbial metabolite, at concentration of ~58.0 µg/g.Citation87,Citation142 Skatole in human fecal matter varies considerably, with an average quantity of 34.5 µg/g.Citation87 A recent study reported 2,8-DHQ, a human AHR-selective ligand, was quantified in human feces with the concentrations between 0 and 3.4 pmol/mg.Citation133 These observations bring up the question whether the microbiomes in individual humans differ in their capacity to produce tryptophan metabolites. Thus, a more comprehensive understanding of the complex effects of microbiota derived AHR ligands, capable of modulating overall gut health and subsequent resistance to disease, is needed. Given the important role of AHR as a mediator in host-gut microbiota communication, this review highlights the utility of the AHR as a promising therapeutic target in human disease, which can be altered through changes in the microbiota, modifying diet, or the administration of drugs.
Disclosure of potential conflicts of interest
The authors declare no conflict of interest.
Additional information
Funding
References
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi:10.1038/nature08821.
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. doi:10.1126/science.1223813.
- Dorrestein PC, Mazmanian SK, Knight R. Finding the missing links among metabolites, microbes, and the host. Immunity. 2014;40:824–832.
- Blacher E, Levy M, Tatirovsky E, Elinav E. Microbiome-modulated metabolites at the interface of host immunity. J Immunol 2017;198(2):572–580. doi:10.4049/jimmunol.1601247.
- Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018;9:3294.
- Muku GE, Murray IA, Perdew GH. Activation of the ah receptor modulates gastrointestinal homeostasis and the intestinal microbiome. Curr Pharmacol Rep 2019;5(5):319–331. doi:10.1007/s40495-019-00197-2.
- Gutierrez-Vazquez C, Quintana FJ. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity. 2018;48(1):19–33. doi:10.1016/j.immuni.2017.12.012.
- Qiu J, Heller JJ, Guo X, Chen Z-M, Fish K, Fu Y-X, Zhou L. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36(1):92–104. doi:10.1016/j.immuni.2011.11.011.
- Murray IA, Patterson AD, Perdew GH. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat Rev Cancer. 2014;14(12):801–814. doi:10.1038/nrc3846.
- Esser C, Rannug A, Ma Q. The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol Rev 2015;67(2):259–279. doi:10.1124/pr.114.009001.
- Petrulis JR, Perdew GH. The role of chaperone proteins in the aryl hydrocarbon receptor core complex. Chem Biol Interact 2002;141(1–2):25–40. doi:10.1016/S0009-2797(02)00064-9.
- Bersten DC, Sullivan AE, Peet DJ, Whitelaw ML. bHLH–PAS proteins in cancer. Nat Rev Cancer. 2013;13(12):827–841. doi:10.1038/nrc3621.
- Kumar MB, Perdew GH. Nuclear receptor coactivator SRC-1 interacts with the Q-rich subdomain of the AhR and modulates its transactivation potential. Gene Expr 1999;8:273–286.
- Kobayashi A, Numayama-Tsuruta K, Sogawa K, Fujii-Kuriyama Y. CBP/p300 functions as a possible transcriptional coactivator of Ah receptor nuclear translocator (Arnt)1. J Biochem 1997;122(4):703–710. doi:10.1093/oxfordjournals.jbchem.a021812.
- Beischlag TV, Wang S, Rose DW, Torchia J, Reisz-Porszasz S, Muhammad K, Nelson WE, Probst MR, Rosenfeld MG, Hankinson O, et al. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol Cell Biol 2002;22(12):4319–4333. doi:10.1128/MCB.22.12.4319-4333.2002.
- Kumar MB, Tarpey RW, Perdew GH. Differential recruitment of coactivator RIP140 by Ah and estrogen receptors absence of a role for LXXLL motifs. the J. Biol. Chem. 1999;274:22155–22164.
- Kim JH, Stallcup MR. Role of the coiled-coil coactivator (CoCoA) in aryl hydrocarbon receptor-mediated transcription. J Biol Chem 2004;279(48):49842–49848. doi:10.1074/jbc.M408535200.
- Chen Y-H, Beischlag TV, Kim JH, Perdew GH, Stallcup MR. Role of GAC63 in transcriptional activation mediated by the aryl hydrocarbon receptor. J Biol Chem 2006;281(18):12242–12247. doi:10.1074/jbc.M512537200.
- Kollara A, Brown TJ. Functional interaction of nuclear receptor coactivator 4 with aryl hydrocarbon receptor. Biochem Biophys Res Commun 2006;346(2):526–534. doi:10.1016/j.bbrc.2006.05.148.
- Beischlag TV, Taylor RT, Rose DW, Yoon D, Chen Y, Lee W-H, Rosenfeld MG, Hankinson O. Recruitment of thyroid hormone receptor/retinoblastoma-interacting protein 230 by the aryl hydrocarbon receptor nuclear translocator is required for the transcriptional response to both dioxin and hypoxia. J Biol Chem 2004;279(52):54620–54628. doi:10.1074/jbc.M410456200.
- Ohtake F, Takeyama K-I, Matsumoto T, Kitagawa H, Yamamoto Y, Nohara K, Tohyama C, Krust A, Mimura J, Chambon P, et al. Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature. 2003;423(6939):545–550. doi:10.1038/nature01606.
- Tian Y, Rabson AB, Gallo MA. Ah receptor and NF-κB interactions: mechanisms and physiological implications. Chem Biol Interact 2002;141(1–2):97–115. doi:10.1016/S0009-2797(02)00068-6.
- Vogel CF, Matsumura F. A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NF-κB family. Biochem Pharmacol 2009;77(4):734–745. doi:10.1016/j.bcp.2008.09.036.
- Karin M, Yamamoto Y, Wang QM. The IKK NF-κB system: a treasure trove for drug development. Nat Rev Drug Discov 2004;3(1):17–26. doi:10.1038/nrd1279.
- Lawrence T. The nuclear factor NF- B pathway in inflammation. Cold Spring Harb Perspect Biol 2009;1(6):a001651. doi:10.1101/cshperspect.a001651.
- Patel RD, Murray IA, Flaveny CA, Kusnadi A, Perdew GH. Ah receptor represses acute-phase response gene expression without binding to its cognate response element. Lab Invest 2009;89(6):695–707. doi:10.1038/labinvest.2009.24.
- Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA. Ah receptor and NF-κB interactions, a potential mechanism for dioxin toxicity. J Biol Chem 1999;274(1):510–515. doi:10.1074/jbc.274.1.510.
- Vogel CF, Sciullo E, Li W, Wong P, Lazennec G, Matsumura F. RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol Endocrinol 2007;21(12):2941–2955. doi:10.1210/me.2007-0211.
- Xiao G, Harhaj EW, Sun S-C. NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol Cell. 2001;7(2):401–409. doi:10.1016/S1097-2765(01)00187-3.
- Sun S-C. The noncanonical NF-κB pathway. Immunol Rev 2012;246(1):125–140. doi:10.1111/j.1600-065X.2011.01088.x.
- Vogel CF, Li W, Wu D, Miller JK, Sweeney C, Lazennec G, Fujisawa Y, Matsumura F. Interaction of aryl hydrocarbon receptor and NF-κB subunit RelB in breast cancer is associated with interleukin-8 overexpression. Arch Biochem Biophys 2011;512(1):78–86. doi:10.1016/j.abb.2011.05.011.
- Vogel CF, Sciullo E, Matsumura F. Involvement of RelB in aryl hydrocarbon receptor-mediated induction of chemokines. Biochem Biophys Res Commun 2007;363(3):722–726. doi:10.1016/j.bbrc.2007.09.032.
- Vogel CF, Nishimura N, Sciullo E, Wong P, Li W, Matsumura F. Modulation of the chemokines KC and MCP-1 by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice. Arch Biochem Biophys 2007;461(2):169–175. doi:10.1016/j.abb.2007.01.015.
- N’Diaye M, Le Ferrec E, Lagadic-Gossmann D, Corre S, Gilot D, Lecureur V, Monteiro P, Rauch C, Galibert M-D, Fardel O, et al. Aryl hydrocarbon receptor- and calcium-dependent induction of the chemokine CCL1 by the environmental contaminant Benzo[a]pyrene. J Biol Chem 2006;281(29):19906–19915. doi:10.1074/jbc.M601192200.
- Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit Rev Eukaryot Gene Expr 2008;18(3):207–250. doi:10.1615/CritRevEukarGeneExpr.v18.i3.20.
- Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat Rev Cancer. 2006;6:947–960.
- Oshima M, Mimura J, Yamamoto M, Fujii-Kuriyama Y. Molecular mechanism of transcriptional repression of AhR repressor involving ANKRA2, HDAC4, and HDAC5. Biochem Bioph Res Co 2007;364(2):276–282. doi:10.1016/j.bbrc.2007.09.131.
- Pollenz RS. The aryl-hydrocarbon receptor, but not the aryl-hydrocarbon receptor nuclear translocator protein, is rapidly depleted in hepatic and nonhepatic culture cells exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Mol Pharmacol 1996;49:391–398.
- Xing X, Bi H, Chang AK, Zang M-X, Wang M, Ao X, Li S, Pan H, Guo Q, Wu H, et al. SUMOylation of AhR modulates its activity and stability through inhibiting its ubiquitination. J Cell Physiol 2012;227(12):3812–3819. doi:10.1002/jcp.24092.
- Durrin LK, Jones PB, Fisher JM, Galeazzi DR, Whitlock JP Jr. 2,3,7,8-Tetrachlorodibenzo-p-dioxin receptors regulate transcription of the cytochrome P1-450 gene. J Cell Biochem 1987;35(2):153–160. doi:10.1002/jcb.240350208.
- Benson JM, Shepherd DM. Aryl hydrocarbon receptor activation by TCDD reduces inflammation associated with Crohn’s disease. Toxicol Sci 2011;120(1):68–78. doi:10.1093/toxsci/kfq360.
- Rosser EC, Piper CJM, Matei DE, Blair PA, Rendeiro AF, Orford M, Alber DG, Krausgruber T, Catalan D, Klein N, et al. Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory B cells. Cell Metab 2020;31(4):837–51 e10. doi:10.1016/j.cmet.2020.03.003.
- Krishnan S, Ding Y, Saedi N, Choi M, Sridharan GV, Sherr DH, Yarmush ML, Alaniz RC, Jayaraman A, Lee K, et al. Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep 2018;23(4):1099–1111. doi:10.1016/j.celrep.2018.03.109.
- Kerley-Hamilton JS, Trask HW, Ridley CJ, Dufour E, Ringelberg CS, Nurinova N, Wong D, Moodie KL, Shipman SL, Moore JH, et al. Obesity is mediated by differential aryl hydrocarbon receptor signaling in mice fed a Western diet. Environ Health Perspect 2012;120(9):1252–1259. doi:10.1289/ehp.1205003.
- Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol 2003;43(1):309–334. doi:10.1146/annurev.pharmtox.43.100901.135828.
- Hubbard TD, Murray IA, Perdew GH. Indole and tryptophan metabolism: endogenous and dietary routes to Ah receptor activation. Drug Metab Dispos 2015;43(10):1522–1535. doi:10.1124/dmd.115.064246.
- Fernandez-Salguero PM, Ward JM, Sundberg JP, Gonzalez FJ. Lesions of Aryl-hydrocarbon Receptor–deficient Mice. Vet Pathol 1997;34(6):605–614. doi:10.1177/030098589703400609.
- Fernandez-Salguero P, Pineau T, Hilbert DM, McPhail T, Lee SS, Kimura S, Nebert D, Rudikoff S, Ward J, Gonzalez F, et al. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science. 1995;268(5211):722–726. doi:10.1126/science.7732381.
- Takamura T, Harama D, Fukumoto S, Nakamura Y, Shimokawa N, Ishimaru K, Ikegami S, Makino S, Kitamura M, Nakao A, et al. Lactobacillus bulgaricus OLL1181 activates the aryl hydrocarbon receptor pathway and inhibits colitis. Immunol Cell Biol 2011;89(7):817–822. doi:10.1038/icb.2010.165.
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372–385. doi:10.1016/j.immuni.2013.08.003.
- Dong F, Hao F, Murray IA, Smith PB, Koo I, Tindall AM, Kris-Etherton PM, Gowda K, Amin SG, Patterson AD, et al. Intestinal microbiota-derived tryptophan metabolites are predictive of Ah receptor activity. Gut Microbes. 2020;12(1):1–24. doi:10.1080/19490976.2020.1788899.
- Hubbard TD, Murray IA, Bisson WH, Sullivan AP, Sebastian A, Perry GH, Jablonski NG, Perdew GH. Divergent Ah receptor ligand selectivity during hominin evolution. Mol Biol Evol 2016;33(10):2648–2658. doi:10.1093/molbev/msw143.
- Le Floc’h N, Otten W, Merlot E. Tryptophan metabolism, from nutrition to potential therapeutic applications. Amino Acids. 2011;41:1195–1205.
- Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478(7368):197–203. doi:10.1038/nature10491.
- Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol 2010;185:3190–3198.
- DiNatale BC, Murray IA, Schroeder JC, Flaveny CA, Lahoti TS, Laurenzana EM, Omiecinski CJ, Perdew GH. Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicol Sci: Official J Soc Toxicol. 2010;115(1):89–97. doi:10.1093/toxsci/kfq024.
- Hata T, Asano Y, Yoshihara K, Kimura-Todani T, Miyata N, Zhang X-T, Takakura S, Aiba Y, Koga Y, Sudo N, et al. Regulation of gut luminal serotonin by commensal microbiota in mice. Plos One. 2017;12(7):e0180745. doi:10.1371/journal.pone.0180745.
- Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler C, Ismagilov R, Mazmanian S, Hsiao E, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161(2):264–276. doi:10.1016/j.cell.2015.02.047.
- Manzella C, Singhal M, Alrefai WA, Saksena S, Dudeja PK, Gill RK. Serotonin is an endogenous regulator of intestinal CYP1A1 via AhR. Sci Rep 2018;8(1):6103. doi:10.1038/s41598-018-24213-5.
- Wincent E, Bengtsson J, Mohammadi Bardbori A, Alsberg T, Luecke S, Rannug U, Rannug A. Inhibition of cytochrome P4501-dependent clearance of the endogenous agonist FICZ as a mechanism for activation of the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A. 2012;109(12):4479–4484. doi:10.1073/pnas.1118467109.
- Manzella CR, Ackerman M, Singhal M, Ticho AL, Ceh J, Alrefai WA, et al. Serotonin modulates AhR activation by interfering with CYP1A1-mediated clearance of AhR ligands. Cell Physiol Biochem 2020;54:126–141.
- Gaitanis G, Magiatis P, Stathopoulou K, Bassukas ID, Alexopoulos EC, Velegraki A, Skaltsounis A-L. AhR ligands, malassezin, and indolo[3,2-b]carbazole are selectively produced by Malassezia furfur strains isolated from seborrheic dermatitis. J Invest Dermatol 2008;128(7):1620–1625. doi:10.1038/sj.jid.5701252.
- Magiatis P, Pappas P, Gaitanis G, Mexia N, Melliou E, Galanou M, Vlachos C, Stathopoulou K, Skaltsounis AL, Marselos M, et al. Malassezia yeasts produce a collection of exceptionally potent activators of the Ah (dioxin) receptor detected in diseased human skin. J Invest Dermatol 2013;133(8):2023–2030. doi:10.1038/jid.2013.92.
- Sato Y, Fujimura T, Tanita K, Chunbing L, Matsushita S, Fujisawa Y, Otsuka A, Yamamoto Y, Hidaka T, Aiba S, et al. Malassezia -derived aryl hydrocarbon receptor ligands enhance the CCL20/Th17/soluble CD163 pathogenic axis in extra-mammary Paget’s disease. Exp Dermatol 2019;28(8):933–939. doi:10.1111/exd.13944.
- Solis NV, Swidergall M, Bruno VM, Gaffen SL, Filler SG, Alspaugh JA. The aryl hydrocarbon receptor governs epithelial cell invasion during oropharyngeal candidiasis. Mbio. 2017;8(2). doi:10.1128/mBio.00025-17.
- Williams BB, Van Benschoten AH, Cimermancic P, Donia MS, Zimmermann M, Taketani M, Ishihara A, Kashyap P, Fraser J, Fischbach M, et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe. 2014;16(4):495–503. doi:10.1016/j.chom.2014.09.001.
- Jin U-H, Lee S-O, Sridharan G, Lee K, Davidson LA, Jayaraman A, Chapkin RS, Alaniz R, Safe S. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol Pharmacol 2014;85(5):777–788. doi:10.1124/mol.113.091165.
- Scott SA, Fu J, Chang PV. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A. 2020;117(32):19376–19387. doi:10.1073/pnas.2000047117.
- Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. 2017;551(7682):648–652. doi:10.1038/nature24661.
- Bittinger MA, Nguyen LP, Bradfield CA. Aspartate aminotransferase generates proagonists of the aryl hydrocarbon receptor. Mol Pharmacol 2003;64(3):550–556. doi:10.1124/mol.64.3.550.
- Chung KT, Anderson GM, Fulk GE. Formation of indoleacetic acid by intestinal anaerobes. J Bacteriol 1975;124(1):573–575. doi:10.1128/JB.124.1.573-575.1975.
- Elsden SR, Hilton MG, Waller JM. The end products of the metabolism of aromatic amino acids by clostridia. Arch Microbiol 1976;107(3):283–288. doi:10.1007/BF00425340.
- Lawson PA, Citron DM, Tyrrell KL, Finegold SM. Reclassification of clostridium difficile as clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe. 2016;40:95–99. doi:10.1016/j.anaerobe.2016.06.008.
- Dou L, Sallee M, Cerini C, Poitevin S, Gondouin B, Jourde-Chiche N, Fallague K, Brunet P, Calaf R, Dussol B, et al. The cardiovascular effect of the uremic solute indole-3 acetic acid. J Am Soc Nephrol 2015;26(4):876–887. doi:10.1681/ASN.2013121283.
- Yokoyama MT, Carlson JR. Production of Skatole and para-Cresol by a Rumen Lactobacillus sp. †. Appl Environ Microbiol 1981;41(1):71–76. doi:10.1128/AEM.41.1.71-76.1981.
- Rasmussen MK, Balaguer P, Ekstrand B, Daujat-Chavanieu M, Gerbal-Chaloin G-CS, Ryffel B. Skatole (3-Methylindole) is a partial aryl hydrocarbon receptor agonist and induces CYP1A1/2 and CYP1B1 expression in primary human hepatocytes. Plos One. 2016;11(5):e0154629. doi:10.1371/journal.pone.0154629.
- O’Neil SR, DeMoss RD. Tryptophan transaminase from Clostridium sporogenes. Arch Biochem Biophys 1968;127:361–369. doi:10.1016/0003-9861(68)90237-3.
- Jean M, DeMoss RD. Indolelactate dehydrogenase from Clostridium sporogenes. Can J Microbiol 1968;14(4):429–435. doi:10.1139/m68-068.
- Aragozzini F, Ferrari A, Pacini N, Gualandris R. Indole-3-lactic acid as a tryptophan metabolite produced by Bifidobacterium spp. Appl Environ Microbiol 1979;38(3):544–546. doi:10.1128/AEM.38.3.544-546.1979.
- Meng D, Sommella E, Salviati E, Campiglia P, Ganguli K, Djebali K, et al. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr Res 2020.
- Wlodarska M, Luo C, Kolde R, d’Hennezel E, Annand JW, Heim CE, Krastel P, Schmitt EK, Omar AS, Creasey EA, et al. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe. 2017;22(1):25–37 e6. doi:10.1016/j.chom.2017.06.007.
- Hoch JA, Demoss RD. Physiological effects of a constitutive tryptophanase in bacillus alvei. J Bacteriol. 1965;90(3):604–610. doi:10.1128/JB.90.3.604-610.1965.
- Dawes EA. Production of indole by Escherichia coli. Nature. 1948;162(4110):229–231. doi:10.1038/162229a0.
- Tannock GW. Characteristics of Bacteroides isolates from the cecum of conventional mice. Appl Environ Microbiol 1977;33(4):745–750. doi:10.1128/AEM.33.4.745-750.1977.
- Clemons KV, Gadberry JL. Increased indole detection for Pasteurella multocida. J Clin Microbiol 1982;15(4):731–732. doi:10.1128/JCM.15.4.731-732.1982.
- Smith T. A modification of the method for determining the production of indol by bacteria. J Exp Med 1897;2(5):543–547. doi:10.1084/jem.2.5.543.
- Karlin DA, Mastromarino AJ, Jones RD, Stroehlein JR, Lorentz O. Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J Cancer Res Clin Oncol 1985;109(2):135–141. doi:10.1007/BF00391888.
- Riggio O, Mannaioni G, Ridola L, Angeloni S, Merli M, Carla V, Salvatori FM, Moroni F. Peripheral and splanchnic indole and oxindole levels in cirrhotic patients: a study on the pathophysiology of hepatic encephalopathy. Am J Gastroenterol 2010;105(6):1374–1381. doi:10.1038/ajg.2009.738.
- Hubbard TD, Murray IA, Bisson WH, Lahoti TS, Gowda K, Amin SG, Patterson AD, Perdew GH. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep 2015;5(1):12689. doi:10.1038/srep12689.
- Niwa T, Nomura T, Sugiyama S, Miyazaki T, Tsukushi S, Tsutsui S. The protein metabolite hypothesis, a model for the progression of renal failure: an oral adsorbent lowers indoxyl sulfate levels in undialyzed uremic patients. Kidney Int Suppl 1997;62:S23–8.
- Patel KP, Luo FJ, Plummer NS, Hostetter TH, Meyer TW. The production of p-cresol sulfate and indoxyl sulfate in vegetarians versus omnivores. Clin J Am Soc Nephrol 2012;7(6):982–988. doi:10.2215/CJN.12491211.
- Schroeder JC, Dinatale BC, Murray IA, Flaveny CA, Liu Q, Laurenzana EM, Lin JM, Strom SC, Omiecinski CJ, Amin S, et al. The uremic toxin 3-indoxyl sulfate is a potent endogenous agonist for the human aryl hydrocarbon receptor. Biochemistry. 2010;49(2):393–400. doi:10.1021/bi901786x.
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106(10):3698–3703. doi:10.1073/pnas.0812874106.
- Barreto FC, Barreto DV, Liabeuf S, Meert N, Glorieux G, Temmar M, Choukroun G, Vanholder R, Massy ZA. Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin J Am Soc Nephrol 2009;4(10):1551–1558. doi:10.2215/CJN.03980609.
- Itoh Y, Ezawa A, Kikuchi K, Tsuruta Y, Niwa T. Protein-bound uremic toxins in hemodialysis patients measured by liquid chromatography/tandem mass spectrometry and their effects on endothelial ROS production. Anal Bioanal Chem 2012;403(7):1841–1850. doi:10.1007/s00216-012-5929-3.
- Jameson KG, Olson CA, Kazmi SA, Hsiao EY Toward understanding microbiome-neuronal signaling. Mol Cell 2020.
- Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol 2018;15:397–411.
- Fiocchi C. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology. 1998;115(1):182–205. doi:10.1016/S0016-5085(98)70381-6.
- Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol 2017;14(10):573–584. doi:10.1038/nrgastro.2017.88.
- Weingarden AR, Vaughn BP. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes. 2017;8(3):238–252. doi:10.1080/19490976.2017.1290757.
- Nikolaus S, Schulte B, Al-Massad N, Thieme F, Schulte DM, Bethge J, Rehman A, Tran F, Aden K, Häsler R, et al. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology. 2017;153(6):1504–16 e2. doi:10.1053/j.gastro.2017.08.028.
- Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487(7408):477–481. doi:10.1038/nature11228.
- Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet 2011;43(11):1066–1073. doi:10.1038/ng.952.
- Lamas B, Richard ML, Leducq V, Pham H-P, Michel M-L, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 2016;22(6):598–605. doi:10.1038/nm.4102.
- Aoki R, Aoki-Yoshida A, Suzuki C, Takayama Y. Indole-3-pyruvic acid, an aryl hydrocarbon receptor activator, suppresses experimental colitis in mice. J Immunol 2018;201(12):3683–3693. doi:10.4049/jimmunol.1701734.
- Liang H, Dai Z, Liu N, Ji Y, Chen J, Zhang Y, Yang Y, Li J, Wu Z, Wu G, et al. Dietary L-tryptophan modulates the structural and functional composition of the intestinal microbiome in weaned piglets. Front Microbiol 2018;9:1736. doi:10.3389/fmicb.2018.01736.
- Kurata K, Kawahara H, Nishimura K, Jisaka M, Yokota K, Shimizu H. Skatole regulates intestinal epithelial cellular functions through activating aryl hydrocarbon receptors and p38. Biochem Biophys Res Commun 2019;510(4):649–655. doi:10.1016/j.bbrc.2019.01.122.
- Schwarcz R, Bruno JP, Muchowski PJ, Wu HQ. Kynurenines in the mammalian brain: when physiology meets pathology. Nat Rev Neurosci 2012;13:465–477.
- Vogel CF, Goth SR, Dong B, Pessah IN, Matsumura F. Aryl hydrocarbon receptor signaling mediates expression of indoleamine 2,3-dioxygenase. Biochem Biophys Res Commun 2008;375(3):331–335. doi:10.1016/j.bbrc.2008.07.156.
- Novikov O, Wang Z, Stanford EA, Parks AJ, Ramirez-Cardenas A, Landesman E, Laklouk I, Sarita-Reyes C, Gusenleitner D, Li A, et al. An aryl hydrocarbon receptor-mediated amplification loop that enforces cell migration in ER − /PR − /Her2 − human breast cancer cells. Mol Pharmacol 2016;90(5):674–688. doi:10.1124/mol.116.105361.
- Ma N, He T, Johnston LJ, Ma X. Host–microbiome interactions: the aryl hydrocarbon receptor as a critical node in tryptophan metabolites to brain signaling. Gut Microbes. 2020;11(5):1203–1219. doi:10.1080/19490976.2020.1758008.
- Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, Chao -C-C, Patel B, Yan R, Blain M, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med 2016;22(6):586–597. doi:10.1038/nm.4106.
- Rothhammer V, Borucki DM, Garcia Sanchez MI, Mazzola MA, Hemond CC, Regev K, Paul A, Kivisäkk P, Bakshi R, Izquierdo G, et al. Dynamic regulation of serum aryl hydrocarbon receptor agonists in MS. Neurol Neuroimmunol Neuroinflamm 2017;4(4):e359. doi:10.1212/NXI.0000000000000359.
- McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med 2011;365(23):2205–2219. doi:10.1056/NEJMra1004965.
- Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 2015;21(8):895–905. doi:10.1038/nm.3914.
- Marshall NB, Vorachek WR, Steppan LB, Mourich DV, Kerkvliet NI. Functional characterization and gene expression analysis of CD4 + CD25 + regulatory T cells generated in mice treated with 2,3,7,8-Tetrachlorodibenzo- p -Dioxin. J Immunol 2008;181(4):2382–2391. doi:10.4049/jimmunol.181.4.2382.
- Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, Weiner HL. Control of Treg and TH17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453(7191):65–71. doi:10.1038/nature06880.
- Vaidyanathan B, Chaudhry A, Yewdell WT, Angeletti D, Yen W-F, Wheatley AK, Bradfield CA, McDermott AB, Yewdell JW, Rudensky AY, et al. The aryl hydrocarbon receptor controls cell-fate decisions in B cells. J Exp Med 2017;214(1):197–208. doi:10.1084/jem.20160789.
- Piper CJM, Rosser EC, Oleinika K, Nistala K, Krausgruber T, Rendeiro AF, Banos A, Drozdov I, Villa M, Thomson S, et al. Aryl hydrocarbon receptor contributes to the transcriptional program of IL-10-producing regulatory B cells. Cell Rep. 2019;29(7):1878–92 e7. doi:10.1016/j.celrep.2019.10.018.
- Natividad JM, Agus A, Planchais J, Lamas B, Jarry AC, Martin R, Michel M-L, Chong-Nguyen C, Roussel R, Straube M, et al. Impaired aryl hydrocarbon receptor ligand production by the gut microbiota is a key factor in metabolic syndrome. Cell Metab 2018;28(5):737–49.e4. doi:10.1016/j.cmet.2018.07.001.
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–11075. doi:10.1073/pnas.0504978102.
- Wang X, Ota N, Manzanillo P, Kates L, Zavala-Solorio J, Eidenschenk C, Zhang J, Lesch J, Lee WP, Ross J, et al. Interleukin-22 alleviates metabolic disorders and restores mucosal immunity in diabetes. Nature. 2014;514(7521):237–241. doi:10.1038/nature13564.
- Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E, Li V, Maradana MR, Schiering C, Stockinger B, et al. The environmental sensor AHR protects from inflammatory damage by maintaining intestinal stem cell homeostasis and barrier integrity. Immunity. 2018;49(2):353–62.e5. doi:10.1016/j.immuni.2018.07.010.
- Lee JH, Wada T, Febbraio M, He J, Matsubara T, Lee MJ, Gonzalez FJ, Xie W. A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology. 2010;139(2):653–663. doi:10.1053/j.gastro.2010.03.033.
- Kawano Y, Nishiumi S, Tanaka S, Nobutani K, Miki A, Yano Y, Seo Y, Kutsumi H, Ashida H, Azuma T, et al. Activation of the aryl hydrocarbon receptor induces hepatic steatosis via the upregulation of fatty acid transport. Arch Biochem Biophys 2010;504(2):221–227. doi:10.1016/j.abb.2010.09.001.
- Liu X, Huang T, Li L, Tang Y, Tian Y, Wang S, et al. CYP1B1 deficiency ameliorates obesity and glucose intolerance induced by high fat diet in adult C57BL/6J mice. Am J Transl Res 2015;7:761–771.
- Li F, Jiang C, Larsen MC, Bushkofsky J, Krausz KW, Wang T, Jefcoate CR, Gonzalez FJ. Lipidomics reveals a link between CYP1B1 and SCD1 in promoting obesity. J Proteome Res 2014;13(5):2679–2687. doi:10.1021/pr500145n.
- Flowers MT, Ntambi JM. Role of stearoyl-coenzyme A desaturase in regulating lipid metabolism. Curr Opin Lipidol 2008;19(3):248–256. doi:10.1097/MOL.0b013e3282f9b54d.
- Larsen MC, Bushkofsky JR, Gorman T, Adhami V, Mukhtar H, Wang S, Reeder SB, Sheibani N, Jefcoate CR. Cytochrome P450 1B1: an unexpected modulator of liver fatty acid homeostasis. Arch Biochem Biophys 2015;571:21–39. doi:10.1016/j.abb.2015.02.010.
- Ferré P. The biology of peroxisome proliferator-activated receptors: relationship with lipid metabolism and insulin sensitivity. Diabetes. 2004;53(Suppl Supplement 1):S43–50. doi:10.2337/diabetes.53.2007.S43.
- Moyer BJ, Rojas IY, Kerley-Hamilton JS, Nemani KV, Trask HW, Ringelberg CS, Gimi B, Demidenko E, Tomlinson CR. Obesity and fatty liver are prevented by inhibition of the aryl hydrocarbon receptor in both female and male mice. Nutr Res (New York, NY). 2017;44:38–50. doi:10.1016/j.nutres.2017.06.002.
- Moyer BJ, Rojas IY, Kerley-Hamilton JS, Hazlett HF, Nemani KV, Trask HW, West RJ, Lupien LE, Collins AJ, Ringelberg CS, et al. Inhibition of the aryl hydrocarbon receptor prevents Western diet-induced obesity. Model for AHR activation by kynurenine via oxidized-LDL, TLR2/4, TGFβ, and IDO1. Toxicol Appl Pharmacol 2016;300:13–24. doi:10.1016/j.taap.2016.03.011.
- Hubbard TD, Liu Q, Murray IA, Dong F, Miller C 3rd, Smith PB, Gowda K, Lin JM, Amin S, Patterson AD, et al. Microbiota metabolism promotes synthesis of the human Ah receptor agonist 2,8-Dihydroxyquinoline. J Proteome Res 2019;18(4):1715–1724. doi:10.1021/acs.jproteome.8b00946.
- Rothhammer V, Quintana FJ. The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat. Rev. Immunol. 2019;19:184–197.
- Miani M, Le Naour J, Waeckel-Enee E, Verma SC, Straube M, Emond P, Ryffel B, van Endert P, Sokol H, Diana J, et al. Gut microbiota-stimulated innate lymphoid cells support β-defensin 14 expression in pancreatic endocrine cells, preventing autoimmune diabetes. Cell Metab 2018;28(4):557–72.e6. doi:10.1016/j.cmet.2018.06.012.
- Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, Cortez VS, Caparon MG, Donia MS, Gilfillan S, et al. Lactobacillus reuteri induces gut intraepithelial CD4 + CD8αα + T cells. Science. 2017;357(6353):806–810. doi:10.1126/science.aah5825.
- Alexeev EE, Lanis JM, Kao DJ, Campbell EL, Kelly CJ, Battista KD, Gerich ME, Jenkins BR, Walk ST, Kominsky DJ, et al. Microbiota-derived indole metabolites promote human and murine intestinal homeostasis through regulation of interleukin-10 receptor. Am J Pathol 2018;188(5):1183–1194. doi:10.1016/j.ajpath.2018.01.011.
- Whitfield-Cargile CM, Cohen ND, Chapkin RS, Weeks BR, Davidson LA, Goldsby JS, Hunt CL, Steinmeyer SH, Menon R, Suchodolski JS, et al. The microbiota-derived metabolite indole decreases mucosal inflammation and injury in a murine model of NSAID enteropathy. Gut Microbes. 2016;7(3):246–261. doi:10.1080/19490976.2016.1156827.
- Quintana FJ. Nanoparticles for the induction of antigen-specific Tregs. Immunotherapy. 2013;5(5):437–440. doi:10.2217/imt.13.25.
- Julliard W, De Wolfe TJ, Fechner JH, Safdar N, Agni R, Mezrich JD. Amelioration of clostridium difficile infection in mice by dietary supplementation with indole-3-carbinol. Ann Surg 2017;265(6):1183–1191. doi:10.1097/SLA.0000000000001830.
- Flaveny CA. Transgenic humanized AHR mouse reveals differences between human and mouse AHR ligand selectivity. Mol Cell Pharmacol 2009;1(3):119–123. doi:10.4255/mcpharmacol.09.15.
- Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci U S A. 2010;107(1):228–233. doi:10.1073/pnas.0906112107.
