ABSTRACT
Accumulating evidence indicates an alarming increase in the incidence of early-onset cancers, which are diagnosed among adults under 50 years of age, in the colorectum, esophagus, extrahepatic bile duct, gallbladder, liver, stomach, pancreas, as well as the bone marrow (multiple myeloma), breast, head and neck, kidney, prostate, thyroid, and uterine corpus (endometrium). While the early-onset cancer studies have encompassed research on the wide variety of organs, this article focuses on research on digestive system cancers. While a minority of early-onset cancers in the digestive system are associated with cancer-predisposing high penetrance germline genetic variants, the majority of those cancers are sporadic and multifactorial. Although potential etiological roles of diets, lifestyle, environment, and the microbiome from early life to adulthood (i.e. in one’s life course) have been hypothesized, exact contribution of each of these factors remains uncertain. Diets, lifestyle patterns, and environmental exposures have been shown to alter the oral and intestinal microbiome. To address the rising trend of early-onset cancers, transdisciplinary research approaches including lifecourse epidemiology and molecular pathological epidemiology frameworks, nutritional and environmental sciences, multi-omics technologies, etc. are needed. We review current evidence and discuss emerging research opportunities, which can improve our understanding of their etiologies and help us design better strategies for prevention and treatment to reduce the cancer burden in populations.
Introduction
Carcinomas arising in the digestive system are leading causes of cancer death worldwide.Citation1 Accumulating evidence indicates that the incidence and mortality of digestive system cancers diagnosed in patients younger than 50 years of age has dramatically increased in many parts for the world.Citation2–4 The term “early-onset cancer” is generally used for adulthood cancer diagnosed under 50 years of age. We used this term and definition for consistency throughout this article. We also use the contrasting term “later-onset cancer” for cancer diagnosed in patients aged 50 years or older. However, it should be noted that clinical, pathological, and molecular features of cancer may not sharply change at the age of 50 years. Early-onset digestive system cancers may be associated with familial clustering or cancer-predisposing germline genetic variants. However, the majority of them appear to be sporadic and multifactorial in their origins.Citation5–8 Because younger individuals have longer life expectancies compared to older individuals, developments of strategies for early-onset cancer prevention may have a substantial impact on public health and be cost-effective in a long run. However, there is the scarcity of population-based evidence on risk factors for early-onset cancers necessary to design and implement prevention programs.
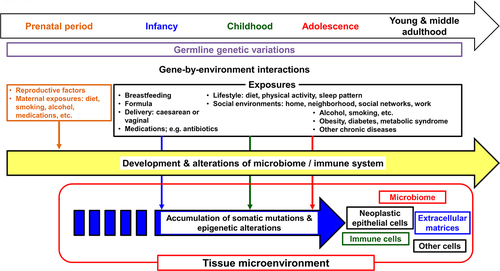
Commensal microorganisms in the human body include bacteria, viruses, fungi, and parasites. A tremendous number of bacteria reside in the gastrointestinal tract, especially in the colon and rectum.Citation9 The intestinal microbiome has been shown to develop throughout early life and influence the systemic metabolic status and immune system.Citation10–12 Maternal exposures, including diet, smoking, alcohol consumption, and medication, as well as infant feeding (breastfeeding vs. formula feeding) have been shown to influence the infant gut microbiome.Citation13–16 As digestive system cancers are heterogeneous diseases influenced by exogenous and endogenous factors such as diets, lifestyle, the microbiome, and immune system (), an integrative approach is required to elucidate their etiology and pathogenesis.Citation17–19 The integration of molecular pathology and epidemiology has generated the transdisciplinary field of molecular pathological epidemiology (MPE),Citation20 which aims to link the exposome with specific pathogenic cellular and molecular signatures.Citation21–23 The integration of microbiology into the MPE approach can provide a better understanding of the interactions between environment exposures, neoplastic cells, immune cells, and the microbiome during the development and progression of early-onset digestive system cancer.Citation24,Citation25 In this review, we summarize current evidence on early-onset digestive system cancers and discuss emerging research opportunities to address the alarming increase in the incidence of early-onset digestive system cancers worldwide.
Figure 1. Life course perspective on factors related to early-onset cancers. Early-onset digestive system cancers develop through the accumulation of somatic mutations and epigenetic alterations in tumor cells under interactive influences of germline genetic variations and various exposures, i.e., gene-by-environmental interactions. An interplay between epithelial, microbial, immune, other cells, and extracellular matrices plays a pivotal role in the tumorigenic (and/or anti-tumorigenic) processes. Theoretically, these processes may start from the prenatal period and remain variably operative throughout one’s life course.
Current evidence of early-onset digestive system cancers
Esophageal cancer
There are two main histological subtypes of esophageal cancer. Esophageal squamous cell carcinoma (ESCC) predominates in certain parts of Asia, such as China and Japan, whereas esophageal adenocarcinoma (EAC) is prevalent in European and North American countries.Citation26,Citation27 EAC typically evolves based on the metaplasia-dysplasia-carcinoma sequence due to chronic gastroesophageal reflux disease and resultant Barrett’s metaplasia. The incidence of early-onset EAC has been increasing in the U.S.Citation28 Studies have shown differences in clinical and molecular features between early-onset and later-onset EAC ().Citation29,Citation30
Table 1. Clinical, pathological, and tumor molecular features of early-onset gastrointestinal tract cancers.
Using data from the Surveillance, Epidemiology, and End Results (SEER) program and the National Cancer Database (NCDB) in the U.S., Kolb et al. demonstrated clinical features and survival outcomes of early-onset EAC.Citation29 Early-onset EAC was associated with stage IV disease.Citation29 However, median overall survival was longest for early-onset EAC (15.2 months) followed by middle age EAC (age 50 to 69 years), and was shortest for old age EAC (age ≥70 years, 10.4 months) (P <.001).Citation29 In multivariate analyses, early-onset EAC was associated with better overall survival (hazard ratio [HR], 0.94; 95% confidence interval [CI], 0.92–0.96) compared to later-onset EAC.Citation29
The increasing incidence of EAC in developed countries may be attributable to the westernized diets and lifestyle. However, the issue of early-onset esophageal cancer has also been recognized in developing countries.Citation55 A study of ESCC patients aged 30–44 years vs. ≥45 years in Tanzania identified risk factors specifically for the younger group, including infrequent teeth cleaning, secondhand smoking, and pest infestation of grains and nuts. Of note, all these factors may affect the microbial composition of the host.Citation56
Gastric cancer
Gastric cancer is the fifth most common and one of the most deadly cancers worldwide.Citation57 Adenocarcinoma (including signet-ring cell carcinoma) is the most common histopathologic type, while less common types include gastrointestinal stromal tumor, MALT (standing for mucosa-associated lymphoid tissue) lymphoma, etc.Citation58 In this section, gastric cancer in most studies refers to adenocarcinoma; however, histological types may not be clearly determined in some studies. Gastric adenocarcinomas can be also divided into tumors in the cardia (the upper part of the stomach adjoining the esophagus) and non-cardia (the middle and distal part of the stomach including the fundus, body, antrum, and pylorus) according to anatomic subsite. An increasing incidence of adenocarcinoma in the gastric cardia has been observed in North America and Western Europe. Gastric non-cardia cancer has been common in Eastern and Central Asia and Eastern Europe.Citation58 The incidence of gastric adenocarcinoma has steadily declined worldwide over the past 50 years due to the increasing availability of the eradication therapy of Helicobacter pylori.Citation57 Paradoxically, the incidence of early-onset gastric cancer has increased in Western countries.Citation32,Citation59,Citation60 Studies have shown differences in clinical and molecular features between early-onset and later-onset gastric cancers ().Citation31–37,Citation61,Citation62
According to data from the SEER program and the NCDB in the U.S., early-onset gastric cancer is associated with Asian/Pacific Islander ancestry, poorly differentiated histologic grade, signet-ring morphology, Lauren diffuse-type histology, and stage IV disease at diagnosis.Citation31,Citation32 Data from Eastern Asia have shown an association between a family history of gastric cancer and early-onset gastric cancer.Citation33,Citation61,Citation62
Using data from next-generation sequencing of tumors in early-onset gastric cancer patients, Setia et al. have shown a higher rate of CDH1 mutations (22% vs. 11%; P =.004) but a similar rate of TP53 mutations in early-onset gastric carcinomas compared to later-onset carcinomas.Citation34 Studies of genomic analyses in early-onset diffuse-type gastric carcinomas have shown significant mutated genes, such as BANP, MUC5B, RHOA, ARID1A, and TGFBR1.Citation35,Citation36 Using clinical and genomic data from the Cancer Genome Atlas (TCGA), Bergquist et al. found that the early-onset gastric cancer was associated with Epstein-Barr virus infection and a genomically stable subtype.Citation32 Ge et al. performed comparative genome-wide analysis of DNA methylation between early-onset and later-onset gastric cancers, and found that the hypermethylation of EIF4E promoter was associated with early-onset gastric cancer.Citation37 These findings suggest that early-onset gastric cancers show different genomic and epigenomic alterations from later-onset gastric cancers.
Colorectal cancer
Colorectal cancer is the second most deadly cancer and third most common malignancy worldwide.Citation63 By far the most common histopathological type is adenocarcinoma (including mucinous and signet-ring cell carcinomas).Citation64 The incidence of colorectal cancer has increased in patients younger than 50 years of age in high-income countries across North America, Europe, and Oceania where colorectal cancer incidence in older adults has been stable or declining.Citation65,Citation66 Accumulating evidence indicates differences in clinical and molecular features between early-onset and later-onset colorectal cancer ().Citation38–50,Citation52–54,Citation67–71
Studies have shown that early-onset colorectal cancer may be associated with a family history of colorectal cancer, advanced disease stage (i.e., stage III or IV) at diagnosis, distal colon and rectal localization, and longer time to diagnosis compared to later-onset colorectal cancer.Citation38–40,Citation45,Citation46 Early-onset colorectal cancer has been pathologically associated with poor differentiation, signet-ring cell morphology, lymphovascular invasion, and a lower degree of antitumor immune response.Citation38,Citation41–43
Studies have shown inconsistent data on prognosis in early-onset vs. later-onset colorectal cancer patients. Using data on patients with stage II or III colon cancer from 6 clinical trials of adjuvant chemotherapy, Fontana et al. showed that, compared to later-onset patients, early-onset colon cancer patients were more likely to experience recurrence and cancer-specific mortality (despite receiving a higher adjuvant treatment intensity).Citation67 In pooled data on 35,713 patients with stage III colon cancer from 25 clinical trials, no significant differences in overall or disease-free survival were observed between early-onset and later-onset patients in multivariable analyses adjusting for tumor molecular features including mismatch repair deficiency, and KRAS and BRAF mutations.Citation48 In metastatic colorectal cancer, there was no statistically significant difference in overall survival between early-onset and later-onset patients although early-onset patients were more likely to receive surgical resection or intensive chemotherapy.Citation47,Citation49 In a cohort study of 769,871 cases with primary colorectal cancer from the NCDB in the U.S., early-onset colorectal cancer was associated with better overall survival compared with later-onset disease after adjusting for disease stage.Citation51
Colorectal cancers consist of heterogeneous tumors with differences in genomic and epigenomic alterations, and the tumor microenvironment through interactions between exposures including diets and lifestyle, microorganisms, and immune cells.Citation72,Citation73 The prevalence of high-level microsatellite instability (MSI) in early-onset colorectal cancer is reported to be 8% to 26%.Citation68–71 Lieu et al. performed next-generation sequencing of 18,218 colorectal cancer specimens and compared genomic landscapes between early-onset and late-onset colorectal cancers.Citation52 In non-MSI colorectal cancers, TP53 and CTNNB1 mutations were associated with early-onset colorectal cancer.Citation52 In MSI-high colorectal tumors, early-onset colorectal cancer was more likely to harbor mutations in APC and KRAS and less likely to harbor mutations in BRAF compared to later-onset cancer.Citation52 In pooled data from 2 clinical trials of metastatic colorectal cancer, FBXW7 and POLE mutations were associated with early-onset disease.Citation50
Several studies demonstrate epigenomic alterations in early-onset colorectal cancer. Akimoto et al. examined methylation levels of long interspersed nucleotide element-1 (LINE-1), which has been well correlated with the global DNA methylation status,Citation74–77 and found that early-onset colorectal cancer was associated with LINE-1 hypomethylation.Citation44 A consortium study of 14,004 cases with colorectal cancer, including 3,089 early-onset cases, found that the proportions of MSI-high, CpG island methylator phenotype (CIMP)-high, BRAF mutated early-onset colorectal cancers gradually increased from the rectum (8.8%, 3.4%, and 3.5%, respectively) to ascending colon (46% MSI-high; 15% CIMP-high) or transverse colon (8.6% BRAF mutated) (all Ptrend <.001 across the rectum to ascending colon), and that later-onset MSI-high colorectal cancers showed a continuous decrease in KRAS mutation prevalence from the rectum (36%) to ascending colon (9%; Ptrend <.001), followed by an increase in the cecum (14%).Citation53 These gradual changes in molecular features of early-onset and later-onset MSI-high colorectal cancers suggest the potential role of the microbiome in the pathogenesis of both early-onset and later-onset colorectal cancer. Overall evidence suggests that differential exposure to the microbiota and their metabolites across different colorectal subsites may lead to differential tumor-microbe-immune interactions and varying developmental processes of colorectal tumor molecular subtypes along the colorectal length in various age groups. Nakamura et al. found that 7 microRNAs (MIR4304, MIR513A, MIR628, MIR194, MIR193A, MIR210, and MIR4453) were significantly upregulated in early-onset colorectal carcinomas compared with late-onset carcinomas, and that 4 of the 7 microRNAs (MIR513A, MIR628, MIR193A, and MIR210) were detectable in plasma specimens from the early-onset colorectal cancer patients.Citation54
Hepatocellular carcinoma (HCC)
Primary liver cancer remains a global health problem and its incidence is growing worldwide. The number of new cases of liver cancer is predicted to increase by 55% in the next two decades, with 1.4 million new diagnoses forecasted for 2040.Citation78
HCC, the most common type of primary liver cancer, is the sixth most common cancer and the third leading cause of cancer-related mortality.Citation79 Although there is limited data published on early-onset HCC, several studies suggest differences in clinical and molecular features between early-onset and later-onset HCC ().Citation80–82
Table 2. Clinical, pathological, and tumor molecular features of early-onset hepatobiliary-pancreatic cancers.
Observational studies have shown that a family history of HCC or hepatitis B virus (HBV) infection is associated with early-onset HCC.Citation80,Citation81 Although further studies are needed, nonalcoholic fatty liver disease and nonalcoholic steatohepatitis have substantially risen in prevalence among adolescents and young adults,Citation93 which may lead to further rise of early-onset HCC in the future. Lessel et al. reported germline mutations in SPRTN among 3 patients from 2 unrelated families presenting early-onset HCC, and demonstrated that SPRTN dysfunction leads to sustained DNA replication stress and consequent replication-related DNA damage in vitro and in vivo.Citation82
Biliary tract cancer
Biliary tract cancer is a relatively rare malignancy arising from epithelial cells in the intrahepatic or extrahepatic bile ducts, or gallbladder.Citation94 Primary sclerosing cholangitis, inflammatory bowel diseases, gallstones, liver fluke infections, biliary malformations, hepatolithiasis, hepatitis C, and liver cirrhosis have been associated with increased incidence of biliary tract cancer.Citation95 Gallbladder carcinoma is the most common cancer of the biliary tract, characterized by a very poor prognosis when diagnosed at advanced stages owing to its aggressive behavior and limited therapeutic options.Citation96 Studies from the U.S., Europe, and Japan have identified an increase in the incidence of biliary tract cancers in younger adults.Citation97–99 Several studies have studied molecular features of early-onset biliary tract cancer ().Citation83,Citation84
A study of clinical and genomic features of cholangiocarcinoma in adolescents and younger adults by Feng et al. has shown that compared to older patients (>45 years old), younger patients with cholangiocarcinoma (≤45 years old) were more likely to have extrahepatic cholangiocarcinoma (29% vs. 17%), poorly differentiated histology, stage III or IV disease, and mutations in the ASXL1 (11% vs. 1%) and KMT2C (19% vs. 4.7%) genes.Citation83 In patients with stage IV cholangiocarcinomas who underwent systemic chemotherapy, the younger group was associated with worse overall survival than the older group (HR, 3.01; 95% CI, 1.14–4.91; P = .03).Citation83 A study of gallbladder cancer by Kumari et al. suggested that a minor allele G and homozygous genotype GG of PARP1 rs1136410 (A/G) are significantly associated with the development of gallbladder carcinoma in younger adults.Citation84
Pancreatic cancer
Pancreatic cancer is the 12th most common cancer worldwide,Citation2 and the fourth leading cause of cancer-related death in both men and women in the U.S.Citation100 Pancreatic cancer has a 5-year survival rate of 6% to 10% and is projected to become the second leading cause of cancer-related mortality in the U.S. by 2040.Citation101 Although early-onset pancreatic cancer accounts for only 5% to 12% of all pancreatic cancer cases, an increase in the incidence of early-onset pancreatic cancer has occurred in the U.S. and other high-income countries over the past few decades.Citation7,Citation8,Citation102 Accumulating evidence indicates differences in clinical and molecular features between early-onset and later-onset pancreatic cancer ().Citation85–89,Citation103,Citation104
Epidemiological studies have shown that potential risk factors for early-onset pancreatic cancer include a family history of pancreatic cancer, smoking, alcohol consumption, obesity, and diabetes.Citation85–87,Citation103 A case-control study in the U.S. reported that individuals who were overweight or obese at the ages of 20 to 49 years were associated with an earlier onset of pancreatic cancer by 2 to 6 years compared to individuals with normal weight.Citation105 Pathological features of early-onset pancreatic cancer have been reported to be poorly differentiated histology and perineural invasion.Citation88,Citation89
Inconsistent data on prognosis of patients with early-onset pancreatic cancer have been reported. Using data on 72,906 patients with pancreatic ductal adenocarcinoma from the SEER database in the U.S., Ansari et al. have shown that early-onset pancreatic ductal adenocarcinoma is associated with worse 5-year overall survival among all patients (6.1% vs. 8.6%; P = .003) and operated patients (17.7% vs. 26.9%; P < .001) compared to those with later-onset disease.Citation86 Another study using data on 248,634 patients with pancreatic cancer from the SEER database has shown that early-onset pancreatic ductal adenocarcinoma is associated with better overall survival compared to those with later-onset disease across all disease stages.Citation104 Some single-institution studies have shown no significant survival difference between early-onset and later-onset pancreatic cancers.Citation85,Citation87,Citation88
The development and progression of pancreatic cancer are affected by somatic mutations and epigenomic regulations in neoplastic cells, and interactions between neoplastic cells, immune cells, and the microbiome in the tumor microenvironment.Citation106 Ben-Aharon et al. performed next-generation sequencing of 293 pancreatic ductal adenocarcinoma specimens and found that mutations in SMAD4 were significantly more prevalent in early-onset pancreatic cancer than in late-onset cancer.Citation90 Bannon et al. examined germline DNA sequencing on pancreatic cancer susceptibility genes and found that germline mutations in BRCA1, BRCA2, or mismatch-repair genes were significantly more prevalent in early-onset pancreatic cancer compared with later-onset disease.Citation91 A genome-wide association study by Campa et al. suggested a SNP at 13q22.3 as a risk allele for early-onset pancreatic cancer.Citation92
Microbiome and digestive system tumor development
Esophageal cancer
Dysbiosis of the oral microbiotaCitation107–109 and Fusobacterium nucleatum (now termed Fusobacterium animalis) in tumor tissuesCitation110 have been associated with ESCC or EAC. These findings have been supported by the recent study by Nomburg et al., which have reported the oral and tumor microbiome from 299 patients with ESCC in five different countries (Tanzania, Malawi, Kenya, China, and Iran) with high incidence of ESCC.Citation111 They found that cancer-associated, traditionally oral bacteria including the genera Fusobacterium, Selenomonas, Prevotella, Streptococcus, Porphyromonas, Veillonella, and Campylobacter, were highly enriched in ESCC tissues. In addition, they also demonstrated that there was a significant correlation between the compositions of the saliva microbiome and ESCC tumor microbiome.Citation111 Hao et al. have reported that the tissue-adherent microbiome in the mouth, esophagus, stomach, and rectum from 37 patients with gastroesophageal reflux, 32 with Barrett’s esophagus, 25 with EAC, and 27 healthy individuals.Citation112 They found that Burkholderia, Lautropia, Ralstonia, and Veillonella in the esophagus positively correlated with disease progression along a spectrum from gastroesophageal reflux to Barrett’s esophagus to EAC.Citation112 These clinical studies suggest dysbiosis of the oral or gut microbiome may influence the development of ESCC and EAC, although exact mechanisms underlying influences of the microbiome on esophageal carcinogenesis remain to be elucidated.
Gastric cancer
Metagenomic analyses of gastric mucosal microbiota showed that a lower microbial diversity, a lower amount of Helicobacter, and higher amounts of certain members of the oral microbiota, including Parvimonas micra, Peptostreptococcus stomatis, and Fusobacterium nucleatum, were observed in both cardia and non-cardia gastric cancer tissues, compared with nontumor tissues.Citation113,Citation114 A recent study has shown that the dysbiosis of the gastric microbiome is associated with the development of gastric cancer after Helicobacter pylori eradication.Citation115 Zhou et al. have examined the fecal and tumor microbiome using 16S ribosomal RNA gene analysis from 1,043 gastric neoplasia patients in China, and found significant enrichments of Streptococcus anginosus and Streptococcus constellatus in gastric carcinoma tissue and stool specimens from patients with intraepithelial neoplasia, early and advanced gastric carcinoma.Citation116 Emerging evidence indicates that dysbiosis of the fungal microbiome (mycobiome) is associated with human digestive system cancers.Citation117 Candida species have been detected in gastric adenocarcinoma tissues and high abundance of Candida species has been associated with early-stage gastric adenocarcinoma.Citation117 The presence of Candida species was associated with the expression of genes involved in cytosolic DNA sensing, Toll-like receptor signaling, and Nod-like receptor signaling in gastric adenocarcinomas.Citation117 These findings suggest potential roles of the oral and gastric microbiome inclusive of mycobiome during gastric carcinogenesis.
Colorectal cancer
Accumulating evidence suggests pathogenic roles of colorectal cancer-associated microbes, such as Fusobacterium nucleatum (now termed Fusobacterium animalis),Citation17,Citation118–129 Peptostreptococcus anaerobius,Citation130–134 Campylobacter jejuni,Citation135 polyketide synthase (pks) positive Escherichia coli,Citation136 enterotoxigenic Bacteroides fragilis,Citation137–145 Enterococcus faecalis,Citation146–149 Streptococcus gallolyticcus,Citation150–152 and Clostridioides difficile.Citation153
Emerging evidence demonstrates interactions between the microbiome and host immunity. Galeano Niño et al. used spatial profiling technologies and single-cell RNA sequencing to examine the spatial distribution and localized effects of the microbiota in human colorectal cancer.Citation154 They found Fusobacterium and Bacteroides to be the most dominant genera in colorectal tumors, and that bacterium-positive areas of colorectal tumors were associated with a downregulation of T cell markers, enrichment of myeloid cells, and upregulations of the immunosuppressive molecule ARG1 and the immune checkpoint protein CTLA4.Citation154 Roelands et al. have reported a large multi-omic analyses of fresh-frozen specimens from 348 patients with primary colon cancer using RNA, whole-exome, deep T cell receptor, bacterial 16S rRNA gene, and whole-genome sequencing.Citation155 They identified a tumor microbiome signature, driven by Ruminococcus bromii, associated with better prognosis. They also developed a composite score (mICRoScore) by the combining tumor microbiome signature and immunological constant of rejection signature, which correlates with the presence of intratumoral T helper 1 cells and cytotoxic immune responses.Citation155 The mICRoScore was associated with patient survival in patients with colorectal cancer.
Several studies have explored the interplay between microorganisms and genetic and/or epigenetic alterations that promote colorectal tumorigenesis. Mouradov and colleagues performed bacterial 16S rRNA gene sequencing in tumor and normal mucosa specimens from 423 patients with stage I to IV colorectal cancer.Citation156 They identified three oncomicrobial community subtypes (OCSs) with distinguishing features. OCS1 (Fusobacterium/oral pathogens, proteolytic, 21%) was associated with proximal location, poor differentiation, high-level MSI, high-level CIMP, consensus molecular subtype (CMS) 1, and BRAF and FBXW7 mutations, whereas both OCS2 (Firmicutes/Bacteroidetes, saccharolytic, 44%) and OCS3 (Escherichia/Pseudescherichia/Shigella, fatty acid β-oxidation, 35%) were associated with distal location and chromosome instability.Citation156 The abundance of certain microorganisms has been associated with oncogenic epigenetic alterations and upregulations of specific non-coding RNAs in colorectal cancer.Citation157 Fusobacterium nucleatum may influence tumor overexpression of MIR21 and MIR1322, which has been shown to potentiate the development and progression of colorectal tumors in mouse models.Citation134,Citation158 DeStefano Shields et al. have shown that colonization by enterotoxigenic Bacteroides fragilis in Apc/Braf-mutant mice developed tumors, which were infiltrated by CD8 T cells and sensitive to anti-CD274 (PD-L1) treatment.Citation159 These findings suggest interactive influences of gut microorganisms and host genetic and epigenetic alterations.
High-throughput sequencing analyses have detected fungi and viruses in colorectal mucosal tissue and fecal specimens, and revealed that dysbiosis of the viral microbiome (virome) or the fungal microbiome (mycobiome) is associated with colorectal cancer.Citation160,Citation161 Some viral taxa, such as Orthobunyavirus, Inovirus, or Tunalikevirus, have been enriched in fecal specimens from patients with colorectal cancer, and dysbiosis of the enteric virome has been associated with worse clinical outcomes in colorectal cancer.Citation160 The role of the gut fungal microbiota in the etiology and progression of colorectal cancer remains uncertain due to the low abundance of fungi in the human gut (≤0.1%–1% of total microorganisms) as well as lack of well-characterized reference genomes for aligning sequencing reads. Coker et al. found that an altered fecal mycobiome composition was associated with colorectal cancer, and that fecal specimens from colorectal cancer patients had higher amounts of Malasseziomycetes and lower amounts of Saccharomycetes and Pneumocystidomycetes compared to cancer-free individuals.Citation161 Lin and colleagues have performed meta-analyses to pool studies with shotgun metagenomic sequencing data from 1,329 individuals, and revealed that several fungal species, including Aspergillus rambellii, Cordyceps sp. RAO-2017, Erysiphe pulchra, Moniliophthora perniciosa, Sphaerulina musiva, and Phytophthora capsica, are enriched and Aspergillus kawachii is depleted in fecal specimens from colorectal cancer patients.Citation162 Multi-kingdom analyses of the presence of bacteria, fungi, archaea, and viruses in fecal specimens from colorecta cancer patients have revealed transkingdom (fungi-bacteria) interactions.Citation162,Citation163 Fecal specimens enriched with Aspergillus rambellii, also harbor high amounts of Fusobacterium and P. micra in patients with colorectal cancer, suggesting the transkingdom interactions between enteric fungi and bacteria during colorectal tumor progression,Citation162 although the underlying mechanisms for this interactions between certain fungi and bacteria in colorectal carcinogenesis are unknown. Pan-cancer analyses of multiple body sites by Dohlman et al. have identified tumor-associated fungi including an enrichment of Candida in colon tumor.Citation117
Accumulating evidence supports the role of microbes in mediating a pathogenic link between environmental factors, including diet and lifestyle, and colorectal carcinogenesis.Citation12,Citation164 Experimental evidence suggests that high-fat diet and cigarette smoking promote colorectal carcinogenesis through the modulation of gut microbiota and metabolites.Citation165 Analyses of two longitudinal prospective cohort studies by Arima et al. have shown that the association of a Western-style diet with colorectal cancer incidence is stronger for tumors containing higher amounts of pks+ Escherichia coli.Citation166 Gao et al. have identified metagenomic signatures of colorectal cancer and validated the signatures in seven published metagenomic datasets of colorectal cancer.Citation167 Lee et al. have shown that there are significant differences in fecal microbial signatures between tubular adenomas and sessile serrated adenomas, and that Flavonifractor plautii and Bacteroides stercoris may influence effects of diet and medications on the development of these premalignant colon lesions.Citation168 Han et al. have demonstrated that the gut commensal Lactobacillus reuteri can potentiate the protective effect of statins on colorectal tumorigenesis via the production of tryptophan catabolite, indole-3-lactic acid.Citation169
HCC
Accumulating evidence indicates that the gut microbiome can influence host immunity and the development of HCC.Citation170 Ma et al. demonstrated that in multiple mouse models, Clostridium species could inhibit the accumulation of hepatic natural killer T cells, and suppress antitumor immune response against both primary and secondary liver tumors.Citation171 A colonization with commensal Clostridium species, gram-positive bacteria involved in the conversion of primary to secondary bile acids, decreased hepatic natural killer T cells and increased liver tumor metastases. Hu et al. found that, after transplantation of the gut microbiota from healthy mice or Lactobacillus reuteri to carcinogen-induced primary HCC-bearing mice, the proportion of IL17A-producing group 3 innate lymphoid cells in the liver was decreased, leading to the reduced IL17A production and suppressed HCC development.Citation172 Li et al. examined the intratumoral microbiome in human HBV-related HCC and identified two microbiome-based HCC subtypes, defined as bacteria-dominant and virus-dominant subtype.Citation173 Higher infiltration of M2 macrophage was associated with bacteria-dominant subtype. Schneider et al. have shown that dysbiosis of the gut microbiome can recruit myeloid-derived suppressor cells and impair T-cell proliferation in the liver, and that Akkermansia muciniphila administration reduced myeloid-derived suppressor cells, liver injury, and liver fibrosis in mice.Citation174
Biliary tract cancer
Intrahepatic cholangiocarcinoma is the second most common primary liver cancer. Its incidence remains low in the Western world but is rising globally.Citation175 Emerging evidence suggests potential interactions between the microbiome, tumor genomic alterations, host immunity, and biliary tract carcinogenesis. In a mouse model of primary sclerosing cholangitis, a decrease in gut barrier function can lead to an increased gut-derived bacteria and lipopolysaccharide in the liver and upregulation of CXCL1 expression in hepatocytes, which results in the recruitment of myeloid-derived suppressor cells in the liver and the development of cholangiocarcinoma.Citation176 Dong et al. performed proteogenomic analyses of paired tumor and adjacent liver tissues from 262 patients with intrahepatic cholangiocarcinoma and identified four subgroups (S1-S4), which had diverse clinical, genomic, immunologic, and microenvironmental features; S1 showed the most abundant expression of inflammatory proteins, such as CD14, MPO, and C5AR1. S2 had the highest level of proteins related to cancer-associated fibroblasts and extracellular matrix, including FAP, POSTN, and FLT1. S3 was characterized by elevated MAPK and metabolic proteins, such as ACAT1, FASN, and IDH1. S4 retained maximum expression of adhesion and biliary-specific proteins, such as ANXA4, KRT18, and EPCAM.Citation177 In addition, they have revealed significant differences in the intratumoral microbiome between these 4 subgroups. Chai and colleagues have found enrichment of Paraburkholderia fungorum in adjacent tissues of intrahepatic cholangiocarcinoma. Paraburkholderia fungorum can inhibit bile duct cancer cell migration and proliferation in in vitro and in vivo experiments.Citation178
Pancreatic cancer
Accumulating evidence indicates potential roles of the microbiome in host immunity and pancreatic tumor development. Pushalkar et al. have demonstrated that gut microbes can migrate to the pancreas and inhibit T-cell-mediated immunity against pancreatic tumors through the recruitment of myeloid-derived suppressor cells into the tumor microenvironment in a mouse model.Citation179 Metagenomic analyses by Riquelme et al. found that high amounts of Pseudoxanthomonas, Streptomyces, or Saccharopolyspora in pancreatic cancer tissue specimens were associated with high density of CD8+ T cells in tumor tissues and better overall survival.Citation180 Nagata et al. performed shotgun metagenomic analysis of fecal and salivary specimens from pancreatic ductal adenocarcinoma patients and cancer-free controls in Japan, Spain, and Germany.Citation181 They have found significant enrichments of Streptococcus and Veillonella spp and a depletion of Faecalibacterium prausnitzii in the gut microbiome of pancreatic cancer patients.Citation181 Ghaddar et al. have developed a computational pipeline of single-cell analysis of host-microbiome interactions in pancreatic cancer and identified a subset of pancreatic tumors harboring intracellular bacteria, which is associated with worse prognosis.Citation182 Chen et al. have shown that deletion of type I collagen produced by pancreatic cancer cells can inhibit pancreatic tumor progression, enhance T cell infiltration, change tumor microbiome with increased Camplyobacterales, and potentiate efficacy of anti-PDCD1 (PD-1) immunotherapy in mice.Citation183 Analyses by Aykut et al. revealed that Malassezia was enriched in human pancreatic cancer tissue specimens, and that Malassezia could potentiate the development of pancreatic tumors in a mouse model.Citation184 Aftab et al. demonstrated that intratumoral mycobiome enhanced cancer cell secretion of pro-inflammatory cytokine IL33 as a chemoattractant for CD4+ type 2 helper T cells cells and group 2 innate lymphoid cells in the pancreatic tumor microenvironment.Citation185 Depletion of fungi or deletion of IL33 in cancer cells was shown to significantly inhibit the progression of pancreatic ductal adenocarcinoma.Citation185
Current evidence for links between the microbiome and early-onset digestive system cancers
Potential risk (or protective) factors for early-onset digestive system cancers include lifestyle and environmental exposures, including obesity, physical inactivity, diet, antibiotics, and aspirin and non-steroidal anti-inflammatory drugs (NSAIDs), all of which have been shown to alter the human microbiome.Citation19,Citation25,Citation122,Citation123,Citation164,Citation186–188 Although there are limited data published on the relationship between the microbiome and early-onset digestive system cancers, studies have suggested differences in the microbiome of stool specimens or tumor tissue between early-onset and later-onset gastrointestinal cancers ().Citation189–191,Citation193
Table 3. Associations between the microbiome and early-onset gastrointestinal cancers.
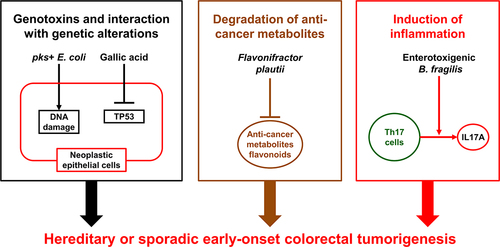
Kadosh et al. have demonstrated a relationship between the gut microbiome and host genetics during colorectal carcinogenesis. They have shown that the gut microbiota-derived gallic acid can inhibit the tumor-suppressive function of mutant TP53 and can promote the development of distal colon tumors in mice harboring the Tp53 p.R270H mutation via hyperactivation of WNT signaling.Citation194 The prevalence of TP53 and CTNNB1 mutations has been shown to be higher in early-onset colorectal cancer than in later-onset colorectal cancer.Citation52 Familial adenomatous polyposis is caused by germline APC mutations.Citation195,Citation196 If left untreated, approximately half of the individuals with familial adenomatous polyposis have been shown to diagnose with colorectal cancer at an age younger than 50 years.Citation71 Dejea et al. have identified that pks-positive Escherichia coli and enterotoxigenic Bacteroides fragilis were more commonly found in colorectal tissues from patients with familial adenomatous polyposis (68% and 60%, respectively), compared to those from healthy individuals (22% and 30%, respectively). In mouse models, co-colonization with these two microbes can potentiate intestinal carcinogenesis through increased DNA damage in colonic epithelium and IL17 induction in the colon.Citation197 Metagenomic analyses of 1,038 fecal specimens from patients with colorectal cancer and age-matched healthy individuals have demonstrated that diversity and composition of the gut microbiome were significantly different between early-onset and later-onset colorectal cancers, and that amounts of some specific bacterial genera, such as Veillonella, Odoribacter, Flavonifractor, Actinomyces, Porphyromonas, and Fusobacterium, were higher in fecal specimens from early-onset colorectal cancer patients than those from age-matched healthy individuals.Citation190 Especially, bacterial species of Flavonifractor plautii were enriched in fecal specimens from early-onset colorectal cancer patients. Another study of integrated metagenomic and metabolomic analyses of fecal specimens from patients with colorectal cancer by Kong et al. have demonstrated that amounts of Flavonifractor plautii were higher in fecal specimens from early-onset colorectal cancer patients than those from age-matched healthy individuals.Citation198 Flavonifractor plautii has been shown to be enriched in stools from Indian patients with colorectal cancer.Citation199 Flavonifractor plautii can degrade flavonoids that have been shown to possess a wide variety of anti-cancer effects by modulating reactive oxygen species-scavenging enzyme activities, arresting the cell cycle, inducing apoptosis and autophagy, and suppressing cancer cell proliferation and invasiveness.Citation200,Citation201 These findings suggest that Flavonifractor plautii may potentiate the development of early-onset colorectal cancer through the degradation of flavonoids, underscoring potential influence of the gut microbiome on host genetics, metabolism, and immunity during tumor development ().
Figure 2. Potential interactions of the gut microbiome with host genetics, metabolism, and immunity during the development of early-onset colorectal cancer. The gut microbiota-derived gallic acid can inhibit the tumor-suppressive function of mutant TP53, which is found more frequently in early-onset colorectal cancer than in later-onset colorectal cancer. pks+ Escherichia coli and enterotoxigenic Bacteroides fragilis can increase DNA damage in colonic epithelium and induce IL17A and colonic inflammation. Flavonifractor plautii can degrade flavonoids, which have been shown to possess a wide variety of anti-cancer effects by modulating reactive oxygen species-scavenging enzyme activities, arresting the cell cycle, inducing apoptosis and autophagy, and suppressing cancer cell proliferation and invasiveness.
The enrichment of Bifidobacterium genus in colorectal cancer tissue has been associated with signet-ring cell morphology,Citation191 which has been a pathological feature of early-onset colorectal cancer.Citation42,Citation202 Bifidobacteria can modulate intestinal epithelial cell differentiation factors in in vitro and in vivo experiments.Citation203 Bifidobacterium genus might influence early-onset colorectal tumorigenesis by modulating epithelial cell differentiation, although further studies are needed to clarify exact mechanisms.
Comparative studies of the gut microbiome in digestive system cancers have found significant differences in the gut microbiome or blood virome between early-onset gastric cancer or HCC and age-matched healthy individuals. Chen et al. have performed 16S rRNA gene sequencing of fecal specimens from 196 patients with gastric cancer and 30 healthy individuals in China, and showed that the amounts of Streptococcus, Veillonella, Tyzzerella, and Aggregatibacter were increased, while those of Odoribacter and Eubacterium ventriosum were lower in stools of early-onset gastric cancer patients than those of age-matched healthy individuals.Citation189 Virome analyses by Liu et al. have demonstrated a blood-based viral exposure signature to predict the occurrence of HCC, and suggested pathogenic roles of viruses other than hepatitis B virus and hepatitis C virus in the development of HCC.Citation193 Further studies are needed to clarify exact mechanisms underlying these associations of the microbiome with early-onset digestive system cancers.
Challenges and emerging opportunities
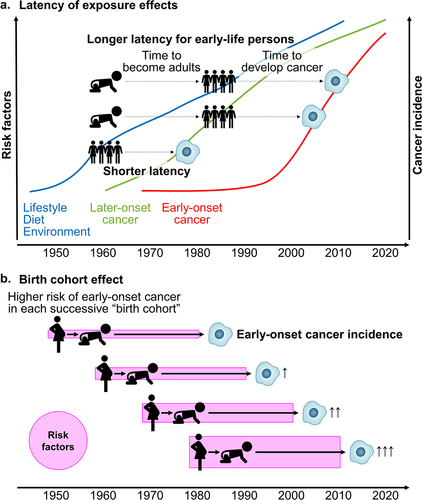
There remain substantial knowledge gaps in the etiology and pathogenesis of early-onset digestive system cancers. It is speculated that dietary, lifestyle, and environmental exposures in early life (from conception to adolescence) may be potential risk factors for the early-onset digestive system neoplasms.Citation3,Citation5,Citation6,Citation204 However, comprehensive analyses of risk factors in early life and young adulthood are difficult without proper study design and data collection. Compared to the prospective design of data and biospecimen collections, it would be much easier to design a case-control study with data and biospecimen collections from cases as well as controls. However, many pitfalls such as differential recall bias and reserve causation exist.Citation25 In the context of microbiome research, reverse causation imply that the presence of disease such as a gastrointestinal disease in cases may influence measured analytes in biospecimens such as stool.Citation25 Utilizing data from existing cohort studies that enrolled early-life participants combined with collections of biospecimens (including blood, stool, saliva, urine, tissue, etc.) enables us to examine early-life exposures in relation to the development of early-onset digestive system cancers.Citation205–207 The conceptualization of combining the molecular pathological epidemiology (MPE) analytical framework into lifecourse epidemiology designs to assess pathogenic effects of early-life factors on diseases later in life has also been developed.Citation208
Reproducibility is another challenge in microbiome research. It has been shown that findings from different studies may be quite disparate. This appears to be due to multiple factors including complex microbial communities, preanalytical and analytical factors, measurement errors, chance variation, among others. Internal and external validations are essential in any given study. It is important to develop rigorous, standardized approaches in microbiome research.
Utilizing the microbiology-MPE research, we can investigate exposures, including diets, lifestyle, and the microbiome in relation to specific cancer subtypes defined by tissue microbial profiling.Citation24 A previous microbiology-MPE study has shown that a so-called prudent diet rich in whole grains and dietary fiber is associated with low incidence of colorectal carcinoma having tissue Fusobacterium nucleatum but not carcinoma without F. nucleatum.Citation188 A further study has revealed a link between inflammatory diets and colorectal carcinomas with Fusobacterium nucleatum, especially in the proximal colon, suggesting that the intestinal inflammatory processes may facilitate colorectal carcinogenesis via altering mucosal barrier functions or augmenting tumorigenic effects of Fusobacterium nucleatum.Citation209 Experimental and observational studies have shown that Fusobacterium nucleatum may contribute carcinogenesis via promoting proliferations of neoplastic clones or suggesting anti-tumor immune reactions.Citation122,Citation123,Citation131,Citation133,Citation210,Citation211 Another putative pathogenic bacterium for colorectal carcinomas is colibactin-producing pks+ Escherichia coli. Colibactin has been shown to cause a specific pattern of genomic mutational changes in colorectal epithelial cells.Citation212 A western-style diet rich in red and processed meat, sugar, and refined grains is another major dietary patterns in the U.S. A microbiology-MPE study has shown that the western dietary pattern is associated with increased incidence of colorectal carcinoma with high-level tissue pks+ Escherichia coli.Citation166 These findings from microbiology-MPE studies support a potential role of bacteria in the relationship of dietary factors with colorectal carcinogenesis. If microbial data are collected from children and/or young adults in a prospective study, we can link microbial profiles with the incidence of specific early-onset cancer subtypes determined by tumor molecular characteristics (e.g. somatic mutations and epigenetic alterations) or tumor microenvironmental features (e.g. pathogenic bacteria). Another knowledge gap is a limited understanding of the pathogenic roles of the microbiome in the development and progression of early-onset digestive system tumors. Hence, further studies are needed to clarify molecular mechanisms by which the microbiome can influence the development and progression of early-onset digestive system tumors.
There is limited evidence for substantial influences of host genetics/epigenetics on the gut microbiome. Existing evidence suggests that maternal exposures, infant feeding, diets, medications, lifestyle patterns, and other environmental exposures can alter the gut microbiome in early life.Citation213 Studies have shown that probiotics intake may generate antiproliferative or pro-apoptotic metabolites in the intestine,Citation214–216 inhibit intestinal inflammation,Citation217,Citation218 reverse gut dysbiosis,Citation219,Citation220 and reactivate antitumor immunity,Citation221,Citation222 thereby leading to the clearance of premalignant cellsCitation223 and prevention of colorectal tumors. Probiotics use, which represents one of the potential prevention strategies of early-onset digestive system tumors, has their relatively safe features and potential health benefits, although further studies are required to assess its short-term and long-term impact on human health and diseases.
Conclusion and future directions
The incidence of early-onset digestive system cancers, diagnosed in individuals under 50 years of age has been increasing worldwide. Although the specific causes remain largely unknown, dietary, lifestyle, and environmental changes that have been occurring across generations since the mid-20th century are considered to play etiological roles (). The gastrointestinal microbiome is a key factor in this emerging trend. Emerging evidence suggests that the gut microbiome may influence host genetics, metabolism, and immunity during early-onset digestive system tumor development. Given potential preventative and therapeutic interventions targeting the microbiome, further research on interactive influences of environmental exposures and the microbiome on digestive system tumor development is needed. Ideally, life-course epidemiological studies combined with prospective collections of biospecimens (stool, blood, saliva, urine, placenta, cord blood, etc.) and tumor tissues will allow for microbiology-MPE research that can study pathogenesis of early-onset cancers. In the meantime, existing cohort studies can be utilized to study risk factor exposures in early life in relation to the development of early-onset cancers. Additionally, we need experimental research encompassing in vitro and in vivo studies to examine effects of the microbiota on tumor development in high-fidelity model systems as well as human intervention trial studies to assess the effects of diets, lifestyle, pre/probiotics, etc. on the intestinal microbiota and/or tumor development. Those collective research efforts will provide evidence required to design and implement intervention programs aimed at young individuals for cancer prevention. A better understanding the pathogenic influences of the gut microbiome on early-onset digestive system cancers will enable tailored screening and preventive strategies, and therapeutic interventions for these cancers.
Figure 3. Simplified schemes illustrating factors related to the rising incidence of early-onset cancers.
Abbreviations
| CI | = | confidence interval |
| CIMP | = | CpG island methylator phenotype |
| CMS | = | consensus molecular subtype |
| EAC | = | esophageal adenocarcinoma |
| EBV | = | Epstein-Barr virus |
| ESCC | = | esophageal squamous cell carcinoma |
| HBV | = | hepatitis B virus |
| HCC | = | hepatocellular carcinoma |
| HGNC | = | HUGO Gene Nomenclature Committee |
| HR | = | hazard ratio |
| HUGO | = | Human Genome Organisation |
| LINE-1 | = | long interspersed nucleotide element-1 |
| MALT | = | mucosa-associated lymphoid tissue |
| MSI | = | microsatellite instability |
| MPE | = | molecular pathological epidemiology |
| NCDB | = | National Cancer Database |
| NSAID | = | non-steroidal anti-inflammatory drug |
| OCS | = | oncomicrobial community subtype |
| PD-1 | = | programmed cell death 1 (PDCD1) |
| PD-L1 | = | programmed cell death 1 ligand 1 (CD274) |
| pks | = | polyketide synthase |
| SEER | = | Surveillance, Epidemiology, and End Results |
| SNP | = | single-nucleotide polymorphism |
| TCGA | = | The Cancer Genome Atlas |
Use of standardized official symbols
We use HUGO (Human Genome Organization) Gene Nomenclature Committee (HGNC)-approved official symbols (or root symbols) for genes and gene products, including ACAT1, ANXA4, APC, ARG1, ARID1A, ASXL1, BANP, BRAF, BRCA1, BRCA2, C5AR1, CD4, CD8, CD14, CD274, CDH1, CTLA4, CTNNB1, CXCL1, EIF4E, EPCAM, FAP, FASN, FBXW7, FLT1, IDH1, IL17A, IL33, KMT2C, KRAS, KRT18, MAPK, MIR21, MIR193A, MIR194, MIR210, MIR513A, MIR628, MIR1322, MIR4304, MIR4453, MPO, MUC5B, PARP1, PDCD1, POLE, POSTN, RHOA, SMAD4, SPRTN, TGFBR1, TP53, WNT; all of which are described at www.genenames.org. The official gene symbols are italicized to differentiate from non-italicized gene product symbols.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–26. doi:10.3322/caac.21763.
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660.
- Ugai T, Sasamoto N, Lee HY, Ando M, Song M, Tamimi RM, Kawachi I, Campbell PT, Giovannucci EL, Weiderpass E, et al. Is early-onset cancer an emerging global epidemic? Current evidence and future implications. Nat Rev Clin Oncol. 2022;19(10):656–673. doi:10.1038/s41571-022-00672-8.
- Zhao J, Xu L, Sun J, Song M, Wang L, Yuan S, Zhu Y, Wan Z, Larsson S, Tsilidis K, et al. Global trends in incidence, death, burden and risk factors of early-onset cancer from 1990 to 2019. BMJ Oncol. 2023;2(1):e000049. doi:10.1136/bmjonc-2023-000049.
- Akimoto N, Ugai T, Zhong R, Hamada T, Fujiyoshi K, Giannakis M, Wu K, Cao Y, Ng K, Ogino S. Rising incidence of early-onset colorectal cancer - a call to action. Nat Rev Clin Oncol. 2021;18(4):230–243. doi:10.1038/s41571-020-00445-1.
- Sinicrope FA, Longo DL. Increasing incidence of early-onset colorectal cancer. N Engl J Med. 2022;386(16):1547–1558. doi:10.1056/NEJMra2200869.
- Huang BZ, Liu L, Zhang J, Team USCPR, Pandol SJ, Grossman SR, Setiawan VW. Rising incidence and racial disparities of early-onset pancreatic cancer in the United States, 1995-2018. Gastroenterology. 2022;163(1):310–312 e1. doi:10.1053/j.gastro.2022.03.011.
- LaPelusa M, Shen C, Arhin ND, Cardin D, Tan M, Idrees K, Geevarghese S, Chakravarthy B, Berlin J, Eng C. Trends in the incidence and treatment of early-onset pancreatic cancer. Cancers Basel. 2022;14(2):283. doi:10.3390/cancers14020283.
- Sorbara MT, Pamer EG. Microbiome-based therapeutics. Nat Rev Microbiol. 2022;20(6):365–380. doi:10.1038/s41579-021-00667-9.
- Derrien M, Alvarez AS, de Vos WM. The gut microbiota in the first decade of life. Trends Microbiol. 2019;27(12):997–1010. doi:10.1016/j.tim.2019.08.001.
- Ronan V, Yeasin R, Claud EC. Childhood development and the microbiome-the intestinal microbiota in maintenance of health and development of disease during childhood development. Gastroenterology. 2021;160(2):495–506. doi:10.1053/j.gastro.2020.08.065.
- Ghosh TS, Shanahan F, O’Toole PW. The gut microbiome as a modulator of healthy ageing. Nat Rev Gastroenterol Hepatol. 2022;19(9):565–584. doi:10.1038/s41575-022-00605-x.
- Baumann-Dudenhoeffer AM, D’Souza AW, Tarr PI, Warner BB, Dantas G. Infant diet and maternal gestational weight gain predict early metabolic maturation of gut microbiomes. Nat Med. 2018;24(12):1822–1829. doi:10.1038/s41591-018-0216-2.
- Maher SE, O’Brien EC, Moore RL, Byrne DF, Geraghty AA, Saldova R, Murphy EF, Van Sinderen D, Cotter PD, McAuliffe FM. The association between the maternal diet and the maternal and infant gut microbiome: a systematic review. Br J Nutr. 2020;129(9):1–29. doi:10.1017/S0007114520000847.
- Garcia-Mantrana I, Selma-Royo M, Gonzalez S, Parra-Llorca A, Martinez-Costa C, Collado MC. Distinct maternal microbiota clusters are associated with diet during pregnancy: impact on neonatal microbiota and infant growth during the first 18 months of life. Gut Microbes. 2020;11(4):962–978. doi:10.1080/19490976.2020.1730294.
- Grech A, Collins CE, Holmes A, Lal R, Duncanson K, Taylor R, Gordon A. Maternal exposures and the infant gut microbiome: a systematic review with meta-analysis. Gut Microbes. 2021;13(1):1–30. doi:10.1080/19490976.2021.1897210.
- Mima K, Ogino S, Nakagawa S, Sawayama H, Kinoshita K, Krashima R, Ishimoto T, Imai K, Iwatsuki M, Hashimoto D, et al. The role of intestinal bacteria in the development and progression of gastrointestinal tract neoplasms. Surg Oncol. 2017;26(4):368–376. doi:10.1016/j.suronc.2017.07.011.
- Mima K, Nakagawa S, Sawayama H, Ishimoto T, Imai K, Iwatsuki M, Hashimoto D, Baba Y, Yamashita YI, Yoshida N, et al. The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett. 2017;402:9–15. doi:10.1016/j.canlet.2017.05.001.
- Mima K, Kosumi K, Baba Y, Hamada T, Baba H, Ogino S. The microbiome, genetics, and gastrointestinal neoplasms: the evolving field of molecular pathological epidemiology to analyze the tumor-immune-microbiome interaction. Hum Genet. 2021;140(5):725–746. doi:10.1007/s00439-020-02235-2.
- Ogino S, Stampfer M. Lifestyle factors and microsatellite instability in colorectal cancer: the evolving field of molecular pathological epidemiology. J Natl Cancer Inst. 2010;102(6):365–367. doi:10.1093/jnci/djq031.
- Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60(3):397–411. doi:10.1136/gut.2010.217182.
- Ogino S, Nowak JA, Hamada T, Phipps AI, Peters U, Milner Jr DA Jr., Giovannucci EL, Nishihara R, Giannakis M, Garrett WS, et al. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut. 2018;67(6):1168–1180. doi:10.1136/gutjnl-2017-315537.
- Ogino S, Nowak JA, Hamada T, Milner DA Jr., Nishihara R. Insights into pathogenic interactions among environment, host, and tumor at the crossroads of molecular Pathology and epidemiology. Annu Rev Pathol. 2019;14:83–103. doi:10.1146/annurev-pathmechdis-012418-012818.
- Hamada T, Nowak JA, Milner DA Jr., Song M, Ogino S. Integration of microbiology, molecular pathology, and epidemiology: a new paradigm to explore the pathogenesis of microbiome-driven neoplasms. J Pathol. 2019;247(5):615–628. doi:10.1002/path.5236.
- Inamura K, Hamada T, Bullman S, Ugai T, Yachida S, Ogino S. Cancer as microenvironmental, systemic and environmental diseases: opportunity for transdisciplinary microbiomics science. Gut. 2022;71(10):2107–2122. doi:10.1136/gutjnl-2022-327209.
- Killcoyne S, Fitzgerald RC. Evolution and progression of Barrett’s oesophagus to oesophageal cancer. Nat Rev Cancer. 2021;21(11):731–741. doi:10.1038/s41568-021-00400-x.
- Thrift AP. Global burden and epidemiology of Barrett oesophagus and oesophageal cancer. Nat Rev Gastroenterol Hepatol. 2021;18(6):432–443. doi:10.1038/s41575-021-00419-3.
- Codipilly DC, Sawas T, Dhaliwal L, Johnson ML, Lansing R, Wang KK, Leggett CL, Katzka DA, Iyer PG. Epidemiology and outcomes of young-onset esophageal adenocarcinoma: an analysis from a population-based Database. Cancer Epidemiol Biomarkers Prev. 2021;30(1):142–149. doi:10.1158/1055-9965.EPI-20-0944.
- Kolb JM, Han S, Scott FI, Murphy CC, Hosokawa P, Wani S. Early onset esophageal adenocarcinoma study G. Early-onset esophageal adenocarcinoma presents with advanced-stage disease but has improved survival compared with older individuals. Gastroenterology. 2020;159(6):2238–2240 e4. doi:10.1053/j.gastro.2020.08.002.
- Wu IC, Zhao Y, Zhai R, Liu G, Ter-Minassian M, Asomaning K, Su L, Liu CY, Chen F, Kulke MH, et al. Association between polymorphisms in cancer-related genes and early onset of esophageal adenocarcinoma. Neoplasia. 2011;13(4):386–392. doi:10.1593/neo.101722.
- LaPelusa M, Shen C, Gillaspie EA, Cann C, Lambright E, Chakravarthy AB, Gibson MK, Eng C. Variation in treatment patterns of patients with early-onset gastric cancer. Cancers Basel. 2022;14(15):3633. doi:10.3390/cancers14153633.
- Bergquist JR, Leiting JL, Habermann EB, Cleary SP, Kendrick ML, Smoot RL, Nagorney DM, Truty MJ, Grotz TE. Early-onset gastric cancer is a distinct disease with worrisome trends and oncogenic features. Surgery. 2019;166(4):547–555. doi:10.1016/j.surg.2019.04.036.
- Pocurull A, Herrera-Pariente C, Carballal S, Llach J, Sanchez A, Carot L, Botargues JM, Cuatrecasas M, Ocana T, Balaguer F, et al. Clinical, molecular and genetic characteristics of early onset gastric cancer: analysis of a large multicenter study. Cancers Basel. 2021;13(13):3132. doi:10.3390/cancers13133132.
- Setia N, Wang CX, Lager A, Maron S, Shroff S, Arndt N, Peterson B, Kupfer SS, Ma C, Misdraji J, et al. Morphologic and molecular analysis of early-onset gastric cancer. Cancer. 2021;127(1):103–114. doi:10.1002/cncr.33213.
- Mun DG, Bhin J, Kim S, Kim H, Jung JH, Jung Y, Jang YE, Park JM, Kim H, Jung Y, et al. Proteogenomic characterization of human early-onset gastric cancer. Cancer Cell. 2019;35(1):111–124 e10. doi:10.1016/j.ccell.2018.12.003.
- Cho SY, Park JW, Liu Y, Park YS, Kim JH, Yang H, Um H, Ko WR, Lee BI, Kwon SY, et al. Sporadic early-onset diffuse gastric cancers have high frequency of somatic CDH1 alterations, but low frequency of somatic RHOA mutations compared with late-onset cancers. Gastroenterology. 2017;153(2):536–549 e26. doi:10.1053/j.gastro.2017.05.012.
- Ge Y, Wu Q, Ma G, Shao W, Liu H, Zhang Q, Xin J, Xue Y, Du M, Zhao Q, et al. Hypermethylation of EIF4E promoter is associated with early onset of gastric cancer. Carcinogenesis. 2018;39(1):66–71. doi:10.1093/carcin/bgx110.
- Chang DT, Pai RK, Rybicki LA, Dimaio MA, Limaye M, Jayachandran P, Koong AC, Kunz PA, Fisher GA, Ford JM, et al. Clinicopathologic and molecular features of sporadic early-onset colorectal adenocarcinoma: an adenocarcinoma with frequent signet ring cell differentiation, rectal and sigmoid involvement, and adverse morphologic features. Mod Pathol. 2012;25(8):1128–1139. doi:10.1038/modpathol.2012.61.
- Chen FW, Sundaram V, Chew TA, Ladabaum U. Advanced-stage colorectal cancer in persons younger than 50 years not associated with longer duration of symptoms or time to diagnosis. Clin Gastroenterol Hepatol. 2017;15(5):728–737 e3. doi:10.1016/j.cgh.2016.10.038.
- O’Sullivan DE, Sutherland RL, Town S, Chow K, Fan J, Forbes N, Heitman SJ, Hilsden RJ, Brenner DR. Risk factors for early-onset colorectal cancer: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2022;20(6):1229–1240 e5. doi:10.1016/j.cgh.2021.01.037.
- Rodriguez L, Brennan K, Karim S, Nanji S, Patel SV, Booth CM. Disease characteristics, clinical management, and outcomes of young patients with colon cancer: a population-based study. Clin Colorectal Cancer. 2018;17(4):e651–e661. doi:10.1016/j.clcc.2018.06.007.
- Willauer AN, Liu Y, Pereira AAL, Lam M, Morris JS, Raghav KPS, Morris VK, Menter D, Broaddus R, Meric-Bernstam F, et al. Clinical and molecular characterization of early-onset colorectal cancer. Cancer. 2019;125(12):2002–2010. doi:10.1002/cncr.31994.
- Ugai T, Vayrynen JP, Lau MC, Borowsky J, Akimoto N, Vayrynen SA, Zhao M, Zhong R, Haruki K, Dias Costa A, et al. Immune cell profiles in the tumor microenvironment of early-onset, intermediate-onset, and later-onset colorectal cancer. Cancer Immunol Immunother. 2022;71(4):933–942. doi:10.1007/s00262-021-03056-6.
- Akimoto N, Zhao M, Ugai T, Zhong R, Lau MC, Fujiyoshi K, Kishikawa J, Haruki K, Arima K, Twombly TS, et al. Tumor long interspersed nucleotide element-1 (LINE-1) hypomethylation in relation to age of colorectal cancer diagnosis and prognosis. Cancers Basel. 2021;13(9):2016. doi:10.3390/cancers13092016.
- Jacome AA, Vreeland TJ, Johnson B, Kawaguchi Y, Wei SH, Nancy You Y, Vilar E, Vauthey JN, Eng C. The prognostic impact of RAS on overall survival following liver resection in early versus late-onset colorectal cancer patients. Br J Cancer. 2021;124(4):797–804. doi:10.1038/s41416-020-01169-w.
- Narayan RR, Aveson VG, Chou JF, Walch HS, Sanchez-Vega F, Santos Fernandes GD, Balachandran VP, D’Angelica MI, Drebin JA, Jarnagin WR, et al. Association of genomic profiles and survival in early onset and screening-age colorectal cancer patients with liver metastases resected over 15 years. J Surg Oncol. 2022;125(5):880–888. doi:10.1002/jso.26797.
- Kanter K, Fish M, Mauri G, Horick NK, Allen JN, Blaszkowsky LS, Clark JW, Ryan DP, Nipp RD, Giantonio BJ, et al. Care patterns and overall survival in patients with early-onset metastatic colorectal cancer. JCO Oncol Pract. 2021;17(12):e1846–e1855. doi:10.1200/OP.20.01010.
- Jin Z, Dixon JG, Fiskum JM, Parekh HD, Sinicrope FA, Yothers G, Allegra CJ, Wolmark N, Haller D, Schmoll HJ, et al. Clinicopathological and molecular characteristics of early-onset stage III colon adenocarcinoma: an analysis of the ACCENT Database. J Natl Cancer Inst. 2021;113(12):1693–1704. doi:10.1093/jnci/djab123.
- Lipsyc-Sharf M, Zhang S, Ou FS, Ma C, McCleary NJ, Niedzwiecki D, Chang IW, Lenz HJ, Blanke CD, Piawah S, et al. Survival in young-onset metastatic colorectal cancer: findings from cancer and Leukemia group B (alliance)/swog 80405. J Natl Cancer Inst. 2022;114(3):427–435. doi:10.1093/jnci/djab200.
- Antoniotti C, Germani MM, Rossini D, Lonardi S, Pietrantonio F, Santini D, Marmorino F, Allegrini G, Daniel F, Raimondi A, et al. FOLFOXIRI and bevacizumab in patients with early-onset metastatic colorectal cancer. A pooled analysis of TRIBE and TRIBE2 studies. Eur J Cancer. 2022;167:23–31. doi:10.1016/j.ejca.2022.02.031.
- Cheng E, Blackburn HN, Ng K, Spiegelman D, Irwin ML, Ma X, Gross CP, Tabung FK, Giovannucci EL, Kunz PL, et al. Analysis of survival among adults with early-onset colorectal cancer in the National cancer Database. JAMA Netw Open. 2021;4(6):e2112539. doi:10.1001/jamanetworkopen.2021.12539.
- Lieu CH, Golemis EA, Serebriiskii IG, Newberg J, Hemmerich A, Connelly C, Messersmith WA, Eng C, Eckhardt SG, Frampton G, et al. Comprehensive genomic landscapes in early and later onset colorectal cancer. Clin Cancer Res. 2019;25(19):5852–5858. doi:10.1158/1078-0432.CCR-19-0899.
- Ugai T, Haruki K, Harrison TA, Cao Y, Qu C, Chan AT, Campbell PT, Akimoto N, Berndt S, Brenner H, et al. Molecular characteristics of early-onset colorectal cancer according to detailed anatomical locations: comparison with later-onset cases. Am J Gastroenterol. 2022;118(4):712–726. doi:10.14309/ajg.0000000000002171.
- Nakamura K, Hernandez G, Sharma GG, Wada Y, Banwait JK, Gonzalez N, Perea J, Balaguer F, Takamaru H, Saito Y, et al. A liquid biopsy signature for the detection of patients with early-onset colorectal cancer. Gastroenterology. 2022;163(5):1242–1251.e2. doi:10.1053/j.gastro.2022.06.089.
- Cheng ML, Zhang L, Borok M, Chokunonga E, Dzamamala C, Korir A, Wabinga HR, Hiatt RA, Parkin DM, Van Loon K. The incidence of oesophageal cancer in Eastern Africa: identification of a new geographic hot spot? Cancer Epidemiol. 2015;39(2):143–149. doi:10.1016/j.canep.2015.01.001.
- Buckle GC, Mmbaga EJ, Paciorek A, Akoko L, Deardorff K, Mgisha W, Mushi BP, Mwaiselage J, Hiatt RA, Zhang L, et al. Risk factors associated with early-onset esophageal cancer in Tanzania. JCO Glob Oncol. 2022;8(8):e2100256. doi:10.1200/GO.21.00256.
- Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71(3):264–279. doi:10.3322/caac.21657.
- Ajani JA, Lee J, Sano T, Janjigian YY, Fan D, Song S. Gastric adenocarcinoma. Nat Rev Dis Primers. 2017;3(1):17036. doi:10.1038/nrdp.2017.36.
- Heer EV, Harper AS, Sung H, Jemal A, Fidler-Benaoudia MM. Emerging cancer incidence trends in Canada: the growing burden of young adult cancers. Cancer. 2020;126(20):4553–4562. doi:10.1002/cncr.33050.
- Arnold M, Park JY, Camargo MC, Lunet N, Forman D, Soerjomataram I. Is gastric cancer becoming a rare disease? A global assessment of predicted incidence trends to 2035. Gut. 2020;69(5):823–829. doi:10.1136/gutjnl-2019-320234.
- Kwak HW, Choi IJ, Kim CG, Lee JY, Cho SJ, Eom BW, Yoon HM, Joo J, Ryu KW, Kim YW. Individual having a parent with early-onset gastric cancer may need screening at younger age. World J Gastroenterol. 2015;21(15):4592–4598. doi:10.3748/wjg.v21.i15.4592.
- Zhou F, Shi J, Fang C, Zou X, Huang Q. Gastric carcinomas in young (younger than 40 years) Chinese patients: clinicopathology, family history, and postresection survival. Med. 2016;95(9):e2873. doi:10.1097/MD.0000000000002873.
- Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14(10):101174. doi:10.1016/j.tranon.2021.101174.
- Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. Colorectal cancer. Nat Rev Dis Primers. 2015;1(1):15065. doi:10.1038/nrdp.2015.65.
- Vuik FE, Nieuwenburg SA, Bardou M, Lansdorp-Vogelaar I, Dinis-Ribeiro M, Bento MJ, Zadnik V, Pellise M, Esteban L, Kaminski MF, et al. Increasing incidence of colorectal cancer in young adults in Europe over the last 25 years. Gut. 2019;68(10):1820–1826. doi:10.1136/gutjnl-2018-317592.
- Siegel RL, Torre LA, Soerjomataram I, Hayes RB, Bray F, Weber TK, Jemal A. Global patterns and trends in colorectal cancer incidence in young adults. Gut. 2019;68(12):2179–2185. doi:10.1136/gutjnl-2019-319511.
- Fontana E, Meyers J, Sobrero A, Iveson T, Shields AF, Taieb J, Yoshino T, Souglakos I, Smyth EC, Lordick F, et al. Early-onset colorectal adenocarcinoma in the IDEA Database: treatment adherence, toxicities, and outcomes with 3 and 6 months of adjuvant fluoropyrimidine and oxaliplatin. J Clin Oncol. 2021;39(36):4009–4019. doi:10.1200/JCO.21.02008.
- Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, Redston M, Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342(2):69–77. doi:10.1056/NEJM200001133420201.
- Mork ME, You YN, Ying J, Bannon SA, Lynch PM, Rodriguez-Bigas MA, Vilar E. High prevalence of hereditary cancer syndromes in adolescents and young adults with colorectal cancer. J Clin Oncol. 2015;33(31):3544–3549. doi:10.1200/JCO.2015.61.4503.
- Pearlman R, Frankel WL, Swanson B, Zhao W, Yilmaz A, Miller K, Bacher J, Bigley C, Nelsen L, Goodfellow PJ, et al. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol. 2017;3(4):464–471. doi:10.1001/jamaoncol.2016.5194.
- Stoffel EM, Koeppe E, Everett J, Ulintz P, Kiel M, Osborne J, Williams L, Hanson K, Gruber SB, Rozek LS. Germline genetic features of young individuals with colorectal cancer. Gastroenterology. 2018;154(4):897–905 e1. doi:10.1053/j.gastro.2017.11.004.
- Schmitt M, Greten FR. The inflammatory pathogenesis of colorectal cancer. Nat Rev Immunol. 2021;21(10):653–667. doi:10.1038/s41577-021-00534-x.
- Bando H, Ohtsu A, Yoshino T. Therapeutic landscape and future direction of metastatic colorectal cancer. Nat Rev Gastroenterol Hepatol. 2023;20(5):306–322. doi:10.1038/s41575-022-00736-1.
- Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122(12):2767–2773. doi:10.1002/ijc.23470.
- Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, Schernhammer ES, Giovannucci EL, Fuchs CS. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100(23):1734–1738. doi:10.1093/jnci/djn359.
- Ogino S, Nishihara R, Lochhead P, Imamura Y, Kuchiba A, Morikawa T, Yamauchi M, Liao X, Qian ZR, Sun R, et al. Prospective study of family history and colorectal cancer risk by tumor LINE-1 methylation level. J Natl Cancer Inst. 2013;105(2):130–140. doi:10.1093/jnci/djs482.
- Inamura K, Yamauchi M, Nishihara R, Lochhead P, Qian ZR, Kuchiba A, Kim SA, Mima K, Sukawa Y, Jung S, et al. Tumor LINE-1 methylation level and microsatellite instability in relation to colorectal cancer prognosis. J Natl Cancer Inst. 2014;106(9):dju195. doi:10.1093/jnci/dju195.
- Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, Laversanne M, McGlynn KA, Soerjomataram I. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol. 2022;77(6):1598–1606. doi:10.1016/j.jhep.2022.08.021.
- Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, Pikarsky E, Zhu AX, Finn RS. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19(3):151–172. doi:10.1038/s41571-021-00573-2.
- Wan DW, Tzimas D, Smith JA, Kim S, Araujo J, David R, Lobach I, Sarpel U. Risk factors for early-onset and late-onset hepatocellular carcinoma in Asian immigrants with hepatitis B in the United States. Am J Gastroenterol. 2011;106(11):1994–2000. doi:10.1038/ajg.2011.302.
- Li Y, Zhang Z, Shi J, Jin L, Wang L, Xu D, Wang FS. Risk factors for naturally-occurring early-onset hepatocellular carcinoma in patients with HBV-associated liver cirrhosis in China. Int J Clin Exp Med. 2015;8:1205–1212.
- Lessel D, Vaz B, Halder S, Lockhart PJ, Marinovic-Terzic I, Lopez-Mosqueda J, Philipp M, Sim JC, Smith KR, Oehler J, et al. Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nat Genet. 2014;46(11):1239–1244. doi:10.1038/ng.3103.
- Feng H, Tong H, Yan J, He M, Chen W, Wang J. Genomic features and clinical characteristics of adolescents and young adults with cholangiocarcinoma. Front Oncol. 2019;9:1439. doi:10.3389/fonc.2019.01439.
- Anjali K, Singh D, Kumar P, Kumar T, Narayan G, Singh S. PARP1 rs1136410 (A/G) polymorphism is associated with early age of onset of gallbladder cancer. Eur J Cancer Prev. 2022;31(4):311–317. doi:10.1097/CEJ.0000000000000708.
- Piciucchi M, Capurso G, Valente R, Larghi A, Archibugi L, Signoretti M, Stigliano S, Zerboni G, Barucca V, La Torre M, et al. Early onset pancreatic cancer: risk factors, presentation and outcome. Pancreatology. 2015;15(2):151–155. doi:10.1016/j.pan.2015.01.013.
- Ansari D, Althini C, Ohlsson H, Andersson R. Early-onset pancreatic cancer: a population-based study using the SEER registry. Langenbecks Arch Surg. 2019;404(5):565–571. doi:10.1007/s00423-019-01810-0.
- Takeda T, Sasaki T, Inoue Y, Okamoto T, Mori C, Mie T, Furukawa T, Yamada Y, Kasuga A, Matsuyama M, et al. Early-onset pancreatic cancer: clinical characteristics and survival outcomes. Pancreatology. 2022;22(4):507–515. doi:10.1016/j.pan.2022.04.003.
- Kang JS, Jang JY, Kwon W, Han Y, Kim SW. Clinicopathologic and survival differences in younger patients with pancreatic ductal adenocarcinoma-A propensity score-matched comparative analysis. Pancreatology. 2017;17(5):827–832. doi:10.1016/j.pan.2017.08.013.
- Ramai D, Lanke G, Lai J, Barakat M, Chandan S, Ofosu A, Dhaliwal A, Adler DG. Early- and late-onset pancreatic adenocarcinoma: a population-based comparative study. Pancreatology. 2021;21(1):124–129. doi:10.1016/j.pan.2020.12.007.
- Ben-Aharon I, Elkabets M, Pelossof R, Yu KH, Iacubuzio-Donahue CA, Leach SD, Lowery MA, Goodman KA, O’Reilly EM. Genomic landscape of pancreatic adenocarcinoma in younger versus older patients: does age matter? Clin Cancer Res. 2019;25(7):2185–2193. doi:10.1158/1078-0432.CCR-18-3042.
- Bannon SA, Montiel MF, Goldstein JB, Dong W, Mork ME, Borras E, Hasanov M, Varadhachary GR, Maitra A, Katz MH, et al. High prevalence of hereditary cancer syndromes and outcomes in adults with early-onset pancreatic cancer. Cancer Prev Res (Phila). 2018;11(11):679–686. doi:10.1158/1940-6207.CAPR-18-0014.
- Campa D, Gentiluomo M, Obazee O, Ballerini A, Vodickova L, Hegyi P, Soucek P, Brenner H, Milanetto AC, Landi S, et al. Genome-wide association study identifies an early onset pancreatic cancer risk locus. Int J Cancer. 2020;147(8):2065–2074. doi:10.1002/ijc.33004.
- Doycheva I, Watt KD, Alkhouri N. Nonalcoholic fatty liver disease in adolescents and young adults: the next frontier in the epidemic. Hepatology. 2017;65(6):2100–2109. doi:10.1002/hep.29068.
- Banales JM, Marin JJG, Lamarca A, Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen JB, Braconi C, et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastro Hepat. 2020;17(9):557–588. doi:10.1038/s41575-020-0310-z.
- Valle JW, Kelley RK, Nervi B, Oh D-Y, Zhu AX. Biliary tract cancer. Lancet. 2021;397(10272):428–444. doi:10.1016/S0140-6736(21)00153-7.
- Roa JC, García P, Kapoor VK, Maithel SK, Javle M, Koshiol J. Gallbladder cancer. Nat Rev Dis Primers. 2022;8(1):69. doi:10.1038/s41572-022-00398-y.
- Utada M, Ohno Y, Tamaki T, Sobue T, Endo G. Long-term trends in incidence and mortality of intrahepatic and extrahepatic bile duct cancer in Japan. J Epidemiol. 2014;24(3):193–199. doi:10.2188/jea.JE20130122.
- Antwi SO, Mousa OY, Patel T. Racial, Ethnic, and age disparities in incidence and survival of intrahepatic cholangiocarcinoma in the United States; 1995-2014. Ann Hepatol. 2018;17(2):274–285. doi:10.5604/01.3001.0010.8659.
- Rahman R, Ludvigsson JF, von Seth E, Lagergren J, Bergquist A, Radkiewicz C. Age trends in biliary tract cancer incidence by anatomical subtype: a Swedish cohort study. Eur J Cancer. 2022;175:291–298. doi:10.1016/j.ejca.2022.08.032.
- Grossberg AJ, Chu LC, Deig CR, Fishman EK, Hwang WL, Maitra A, Marks DL, Mehta A, Nabavizadeh N, Simeone DM, et al. Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA Cancer J Clin. 2020;70(5):375–403. doi:10.3322/caac.21626.
- Rahib L, Wehner MR, Matrisian LM, Nead KT. Estimated projection of US cancer incidence and death to 2040. JAMA Netw Open. 2021;4(4):e214708. doi:10.1001/jamanetworkopen.2021.4708.
- Huang J, Lok V, Ngai CH, Zhang L, Yuan J, Lao XQ, Ng K, Chong C, Zheng ZJ, Wong MCS. Risk worldwide burden of, factors for, and trends in pancreatic cancer. Gastroenterology. 2021;160(3):744–754. doi:10.1053/j.gastro.2020.10.007.
- McWilliams RR, Maisonneuve P, Bamlet WR, Petersen GM, Li D, Risch HA, Yu H, Fontham ET, Luckett B, Bosetti C, et al. Risk factors for early-onset and very-early-onset pancreatic adenocarcinoma: a pancreatic cancer case-control consortium (PanC4) analysis. Pancreas. 2016;45(2):311–316. doi:10.1097/MPA.0000000000000392.
- Saadat LV, Chou JF, Gonen M, Soares KC, Kingham TP, Varghese AM, Jarnagin WR, D’Angelica MI, Drebin JA, O’Reilly EM, et al. Treatment patterns and survival in patients with early-onset pancreatic cancer. Cancer. 2021;127(19):3566–3578. doi:10.1002/cncr.33664.
- Li D, Morris JS, Liu J, Hassan MM, Day RS, Bondy ML, Abbruzzese JL. Body mass index and risk, age of onset, and survival in patients with pancreatic cancer. JAMA. 2009;301(24):2553–2562. doi:10.1001/jama.2009.886.
- Connor AA, Gallinger S. Pancreatic cancer evolution and heterogeneity: integrating omics and clinical data. Nat Rev Cancer. 2022;22(3):131–142. doi:10.1038/s41568-021-00418-1.
- Yu G, Gail MH, Shi J, Klepac-Ceraj V, Paster BJ, Dye BA, Wang GQ, Wei WQ, Fan JH, Qiao YL, et al. Association between upper digestive tract microbiota and cancer-predisposing states in the esophagus and stomach. Cancer Epidemiol Biomarkers Prev. 2014;23(5):735–741. doi:10.1158/1055-9965.EPI-13-0855.
- Chen X, Winckler B, Lu M, Cheng H, Yuan Z, Yang Y, Jin L, Ye W, Wei Q-Y. Oral microbiota and risk for esophageal squamous cell carcinoma in a high-risk area of China. PloS One. 2015;10(12):e0143603. doi:10.1371/journal.pone.0143603.
- Peters BA, Wu J, Pei Z, Yang L, Purdue MP, Freedman ND, Jacobs EJ, Gapstur SM, Hayes RB, Ahn J. Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 2017;77(23):6777–6787. doi:10.1158/0008-5472.CAN-17-1296.
- Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, Sawayama H, Kinoshita K, Ishimoto T, Iwatsuki M, et al. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res. 2016;22(22):5574–5581. doi:10.1158/1078-0432.CCR-16-1786.
- Nomburg J, Bullman S, Nasrollahzadeh D, Collisson EA, Abedi-Ardekani B, Akoko LO, Atkins JR, Buckle GC, Gopal S, Hu N, et al. An international report on bacterial communities in esophageal squamous cell carcinoma. Int J Cancer. 2022;151(11):1947–1959. doi:10.1002/ijc.34212.
- Hao Y, Karaoz U, Yang L, Yachimski PS, Tseng W, Nossa CW, Ye W, Tseng M, Poles M, Francois F, et al. Progressive dysbiosis of human orodigestive microbiota along the sequence of gastroesophageal reflux, Barrett’s esophagus and esophageal adenocarcinoma. Int J Cancer. 2022;151(10):1703–1716. doi:10.1002/ijc.34191.
- Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJY, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67(6):1024–1032. doi:10.1136/gutjnl-2017-314281.
- Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, Figueiredo C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67(2):226–236. doi:10.1136/gutjnl-2017-314205.
- Watanabe T, Nadatani Y, Suda W, Higashimori A, Otani K, Fukunaga S, Hosomi S, Tanaka F, Nagami Y, Taira K, et al. Long-term persistence of gastric dysbiosis after eradication of Helicobacter pylori in patients who underwent endoscopic submucosal dissection for early gastric cancer. Gastric Cancer. 2021;24(3):710–720. doi:10.1007/s10120-020-01141-w.
- Zhou CB, Pan SY, Jin P, Deng JW, Xue JH, Ma XY, Xie YH, Cao H, Liu Q, Xie WF, et al. Fecal signatures of Streptococcus anginosus and Streptococcus constellatus for noninvasive screening and early warning of gastric cancer. Gastroenterology. 2022;162(7):1933–1947 e18. doi:10.1053/j.gastro.2022.02.015.
- Dohlman AB, Klug J, Mesko M, Gao IH, Lipkin SM, Shen X, Iliev ID. A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell. 2022;185(20):3807–3822 e12. doi:10.1016/j.cell.2022.09.015.
- Komiya Y, Shimomura Y, Higurashi T, Sugi Y, Arimoto J, Umezawa S, Uchiyama S, Matsumoto M, Nakajima A. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. 2019;68(7):1335–1337. doi:10.1136/gutjnl-2018-316661.
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298. doi:10.1101/gr.126573.111.
- Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi:10.1101/gr.126516.111.
- Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33(8):1381–1390. doi:10.1007/s10096-014-2081-3.
- Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K, et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 2015;1(5):653–661. doi:10.1001/jamaoncol.2015.1377.
- Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–1980. doi:10.1136/gutjnl-2015-310101.
- Yu J, Chen Y, Fu X, Zhou X, Peng Y, Shi L, Chen T, Wu Y. Invasive fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer. 2016;139(6):1318–1326. doi:10.1002/ijc.30168.
- Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Sci. 2017;358(6369):1443–1448. doi:10.1126/science.aal5240.
- Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, Ng SC, Tsoi H, Dong Y, Zhang N, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6(1):8727. doi:10.1038/ncomms9727.
- Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25(6):968–976. doi:10.1038/s41591-019-0458-7.
- Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25(4):679–689. doi:10.1038/s41591-019-0406-6.
- Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, Beghini F, Manara S, Karcher N, Pozzi C, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. 2019;25(4):667–678. doi:10.1038/s41591-019-0405-7.
- Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, Coker OO, Chan AWH, Chan FKL, Sung JJY, et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat microbiol. 2019;4(12):2319–2330. doi:10.1038/s41564-019-0541-3.
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host & Microbe. 2013;14(2):207–215. doi:10.1016/j.chom.2013.07.007.
- Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi:10.1016/j.immuni.2015.01.010.
- Abed J, Emgard JM, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-GalNAc. Cell Host & Microbe. 2016;20(2):215–225. doi:10.1016/j.chom.2016.07.006.
- Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear Factor−κB, and up-regulating expression of MicroRNA-21. Gastroenterology. 2017;152(4):851–866.e24. doi:10.1053/j.gastro.2016.11.018.
- He Z, Gharaibeh RZ, Newsome RC, Pope JL, Dougherty MW, Tomkovich S, Pons B, Mirey G, Vignard J, Hendrixson DR, et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut. 2019;68(2):289–300. doi:10.1136/gutjnl-2018-317200.
- Pleguezuelos-Manzano C, Puschhof J, Huber AR, van Hoeck A, Wood HM, Nomburg J, Gurjao C, Manders F, Dalmasso G, Stege PB, et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature. 2020;580(7802):269–273. doi:10.1038/s41586-020-2080-8.
- Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology. 2003;124(2):392–400. doi:10.1053/gast.2003.50047.
- Wu S, Rhee KJ, Zhang M, Franco A, Sears CL. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. J Cell Sci. 2007;120(Pt 11):1944–1952. doi:10.1242/jcs.03455.
- Wu S, Powell J, Mathioudakis N, Kane S, Fernandez E, Sears CL. Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-kappaB pathway. Infect Immun. 2004;72(10):5832–5839. doi:10.1128/IAI.72.10.5832-5839.2004.
- Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host & Microbe. 2018;23(2):203–214 e5. doi:10.1016/j.chom.2018.01.007.
- Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol. 2014;14(9):585–600. doi:10.1038/nri3707.
- Chae WJ, Gibson TF, Zelterman D, Hao L, Henegariu O, Bothwell AL. Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proc Natl Acad Sci U S A. 2010;107(12):5540–5544. doi:10.1073/pnas.0912675107.
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491(7423):254–258. doi:10.1038/nature11465.
- Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016–1022. doi:10.1038/nm.2015.
- Wang L, Yi T, Kortylewski M, Pardoll DM, Zeng D, Yu H. IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J Exp Med. 2009;206(7):1457–1464. doi:10.1084/jem.20090207.
- Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23(3):529–536. doi:10.1093/carcin/23.3.529.
- Wang X, Huycke MM. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology. 2007;132(2):551–561. doi:10.1053/j.gastro.2006.11.040.
- Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke MM. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008;68(23):9909–9917. doi:10.1158/0008-5472.CAN-08-1551.
- Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23(8 Pt 1):1298–1303. doi:10.1111/j.1440-1746.2008.05490.x.
- Gupta A, Madani R, Mukhtar H. Streptococcus bovis endocarditis, a silent sign for colonic tumour. Colorectal Dis. 2010;12(3):164–171. doi:10.1111/j.1463-1318.2009.01814.x.
- Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol Cancer. 2010;9(1):249. doi:10.1186/1476-4598-9-249.
- Boleij A, Tjalsma H. The itinerary of Streptococcus gallolyticus infection in patients with colonic malignant disease. Lancet Infect Dis. 2013;13(8):719–724. doi:10.1016/S1473-3099(13)70107-5.
- Drewes JL, Chen J, Markham NO, Knippel RJ, Domingue JC, Tam AJ, Chan JL, Kim L, McMann M, Stevens C, et al. Human colon cancer-derived clostridioides difficile strains drive colonic tumorigenesis in mice. Cancer Discov. 2022;12(8):1873–1885. doi:10.1158/2159-8290.CD-21-1273.
- Galeano Nino JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, Futran N, Houlton J, Sather C, Sicinska E, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. 2022;611(7937):810–817. doi:10.1038/s41586-022-05435-0.
- Roelands J, Kuppen PJK, Ahmed EI, Mall R, Masoodi T, Singh P, Monaco G, Raynaud C, de Miranda N, Ferraro L, et al. An integrated tumor, immune and microbiome atlas of colon cancer. Nat Med. 2023;29(5):1273–1286. doi:10.1038/s41591-023-02324-5.
- Mouradov D, Greenfield P, Li S, In EJ, Storey C, Sakthianandeswaren A, Georgeson P, Buchanan DD, Ward RL, Hawkins NJ, et al. Oncomicrobial community profiling identifies clinicomolecular and prognostic subtypes of colorectal cancer. Gastroenterology. 2023;165(1):104–120. doi:10.1053/j.gastro.2023.03.205.
- Sobhani I, Bergsten E, Couffin S, Amiot A, Nebbad B, Barau C, De’angelis N, Rabot S, Canoui-Poitrine F, Mestivier D, et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc Natl Acad Sci U S A. 2019;116(48):24285–24295. doi:10.1073/pnas.1912129116.
- Xu C, Fan L, Lin Y, Shen W, Qi Y, Zhang Y, Chen Z, Wang L, Long Y, Hou T, et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes. 2021;13(1):1980347. doi:10.1080/19490976.2021.1980347.
- DeStefano Shields CE, White JR, Chung L, Wenzel A, Hicks JL, Tam AJ, Chan JL, Dejea CM, Fan H, Michel J, et al. Bacterial-driven inflammation and mutant BRAF expression Combine to promote Murine colon tumorigenesis that is Sensitive to immune checkpoint therapy. Cancer Discovery. 2021;11(7):1792–1807. doi:10.1158/2159-8290.CD-20-0770.
- Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, Li X, Szeto CH, Sugimura N, Lam TY, et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology. 2018;155(2):529–541.e5. doi:10.1053/j.gastro.2018.04.018.
- Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, Chan FKL, Sung JJY, Yu J. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 2019;68(4):654–662. doi:10.1136/gutjnl-2018-317178.
- Lin Y, Lau HC, Liu Y, Kang X, Wang Y, Ting NL, Kwong TN, Han J, Liu W, Liu C, et al. Altered mycobiota signatures and enriched pathogenic Aspergillus rambellii are associated with colorectal cancer based on multicohort fecal metagenomic analyses. Gastroenterology. 2022;163(4):908–921. doi:10.1053/j.gastro.2022.06.038.
- Liu NN, Jiao N, Tan JC, Wang Z, Wu D, Wang AJ, Chen J, Tao L, Zhou C, Fang W, et al. Multi-kingdom microbiota analyses identify bacterial-fungal interactions and biomarkers of colorectal cancer across cohorts. Nat microbiol. 2022;7(2):238–250. doi:10.1038/s41564-021-01030-7.
- O’Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016;13(12):691–706. doi:10.1038/nrgastro.2016.165.
- Yang J, Wei H, Zhou Y, Szeto CH, Li C, Lin Y, Coker OO, Lau HCH, Chan AWH, Sung JJY, et al. High-fat diet promotes colorectal tumorigenesis through modulating gut microbiota and metabolites. Gastroenterology. 2022;162(1):135–149 e2. doi:10.1053/j.gastro.2021.08.041.
- Arima K, Zhong R, Ugai T, Zhao M, Haruki K, Akimoto N, Lau MC, Okadome K, Mehta RS, Vayrynen JP, et al. Western-style diet, pks island-carrying Escherichia coli, and colorectal cancer: analyses from two large prospective cohort studies. Gastroenterology. 2022;163(4):862–874. doi:10.1053/j.gastro.2022.06.054.
- Gao R, Wu C, Zhu Y, Kong C, Zhu Y, Gao Y, Zhang X, Yang R, Zhong H, Xiong X, et al. Integrated analysis of colorectal cancer reveals cross-cohort gut microbial signatures and associated serum metabolites. Gastroenterology. 2022;163(4):1024–1037 e9. doi:10.1053/j.gastro.2022.06.069.
- Lee JWJ, Plichta DR, Asher S, Delsignore M, Jeong T, McGoldrick J, Staller K, Khalili H, Xavier RJ, Chung DC. Association of distinct microbial signatures with premalignant colorectal adenomas. Cell Host & Microbe. 2023;31(5):827–838 e3. doi:10.1016/j.chom.2023.04.007.
- Han JX, Tao ZH, Wang JL, Zhang L, Yu CY, Kang ZR, Xie Y, Li J, Lu S, Cui Y, et al. Microbiota-derived tryptophan catabolites mediate the chemopreventive effects of statins on colorectal cancer. Nat microbiol. 2023;8(5):919–933. doi:10.1038/s41564-023-01363-5.
- Daniel N, Genua F, Jenab M, Mayen AL, Chrysovalantou Chatziioannou A, Keski-Rahkonen P, Hughes DJ. The role of the gut microbiome in the development of hepatobiliary cancers. Hepatology. 2023;10–97. doi:10.1097/HEP.0000000000000406.
- Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Sci. 2018;360(6391):eaan5931. doi:10.1126/science.aan5931.
- Hu C, Xu B, Wang X, Wan WH, Lu J, Kong D, Jin Y, You W, Sun H, Mu X, et al. Gut microbiota-derived short-chain fatty acids regulate group 3 innate lymphoid cells in HCC. Hepatology. 2023;77(1):48–64. doi:10.1002/hep.32449.
- Li S, Xia H, Wang Z, Zhang X, Song T, Li J, Xu L, Zhang N, Fan S, Li Q, et al. Intratumoural microbial heterogeneity affected tumor immune microenvironment and determined clinical outcome of HBV-related hepatocellular carcinoma. Hepatology. 2023;78(4):1079–91. doi:10.1097/HEP.0000000000000427.
- Schneider KM, Mohs A, Gui W, Galvez EJC, Candels LS, Hoenicke L, Muthukumarasamy U, Holland CH, Elfers C, Kilic K, et al. Imbalanced gut microbiota fuels hepatocellular carcinoma development by shaping the hepatic inflammatory microenvironment. Nat Commun. 2022;13(1):3964. doi:10.1038/s41467-022-31312-5.
- Greten TF, Schwabe R, Bardeesy N, Ma L, Goyal L, Kelley RK, Wang XW. Immunology and immunotherapy of cholangiocarcinoma. Nat Rev Gastroenterol Hepatol. 2023;20(6):349–365. doi:10.1038/s41575-022-00741-4.
- Zhang Q, Ma C, Duan Y, Heinrich B, Rosato U, Diggs LP, Ma L, Roy S, Fu Q, Brown ZJ, et al. Gut microbiome directs hepatocytes to Recruit MDSCs and promote cholangiocarcinoma. Cancer Discov. 2021;11(5):1248–1267. doi:10.1158/2159-8290.CD-20-0304.
- Dong L, Lu D, Chen R, Lin Y, Zhu H, Zhang Z, Cai S, Cui P, Song G, Rao D, et al. Proteogenomic characterization identifies clinically relevant subgroups of intrahepatic cholangiocarcinoma. Cancer Cell. 2022;40(1):70–87 e15. doi:10.1016/j.ccell.2021.12.006.
- Chai X, Wang J, Li H, Gao C, Li S, Wei C, Huang J, Tian Y, Yuan J, Lu J, et al. Intratumor microbiome features reveal antitumor potentials of intrahepatic cholangiocarcinoma. Gut Microbes. 2023;15(1):2156255. doi:10.1080/19490976.2022.2156255.
- Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, Mohan N, Aykut B, Usyk M, Torres LE, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8(4):403–416. doi:10.1158/2159-8290.CD-17-1134.
- Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, Quesada P, Sahin I, Chandra V, San Lucas A, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(4):795–806 e12. doi:10.1016/j.cell.2019.07.008.
- Nagata N, Nishijima S, Kojima Y, Hisada Y, Imbe K, Miyoshi-Akiyama T, Suda W, Kimura M, Aoki R, Sekine K, et al. Metagenomic identification of microbial signatures predicting pancreatic cancer from a multinational study. Gastroenterology. 2022;163(1):222–238. doi:10.1053/j.gastro.2022.03.054.
- Ghaddar B, Biswas A, Harris C, Omary MB, Carpizo DR, Blaser MJ, De S. Tumor microbiome links cellular programs and immunity in pancreatic cancer. Cancer Cell. 2022;40(10):1240–1253 e5. doi:10.1016/j.ccell.2022.09.009.
- Chen Y, Yang S, Tavormina J, Tampe D, Zeisberg M, Wang H, Mahadevan KK, Wu CJ, Sugimoto H, Chang CC, et al. Oncogenic collagen I homotrimers from cancer cells bind to α3β1 integrin and impact tumor microbiome and immunity to promote pancreatic cancer. Cancer Cell. 2022;40(8):818–834.e9. doi:10.1016/j.ccell.2022.06.011.
- Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, Shadaloey SA, Wu D, Preiss P, Verma N, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574(7777):264–267. doi:10.1038/s41586-019-1608-2.
- Alam A, Levanduski E, Denz P, Villavicencio HS, Bhatta M, Alhorebi L, Zhang Y, Gomez EC, Morreale B, Senchanthisai S, et al. Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell. 2022;40(2):153–167 e11. doi:10.1016/j.ccell.2022.01.003.
- Motta JP, Wallace JL, Buret AG, Deraison C, Vergnolle N. Gastrointestinal biofilms in health and disease. Nat Rev Gastroenterol Hepatol. 2021;18(5):314–334. doi:10.1038/s41575-020-00397-y.
- Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y, Shi Y, Song M, da Silva A, Gu M, et al. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin Transl Gastroenterol. 2016;7(11):e200. doi:10.1038/ctg.2016.53.
- Mehta RS, Nishihara R, Cao Y, Song M, Mima K, Qian ZR, Nowak JA, Kosumi K, Hamada T, Masugi Y, et al. Association of dietary patterns with risk of colorectal cancer subtypes classified by Fusobacterium nucleatum in tumor tissue. JAMA Oncol. 2017;3(7):921–927. doi:10.1001/jamaoncol.2016.6374.
- Chen C, Du Y, Liu Y, Shi Y, Niu Y, Jin G, Shen J, Lyu J, Lin L. Characteristics of gastric cancer gut microbiome according to tumor stage and age segmentation. Appl Microbiol Biotechnol. 2022;106(19–20):6671–6687. doi:10.1007/s00253-022-12156-x.
- Yang Y, Du L, Shi D, Kong C, Liu J, Liu G, Li X, Ma Y. Dysbiosis of human gut microbiome in young-onset colorectal cancer. Nat Commun. 2021;12(1):6757. doi:10.1038/s41467-021-27112-y.
- Kosumi K, Hamada T, Koh H, Borowsky J, Bullman S, Twombly TS, Nevo D, Masugi Y, Liu L, da Silva A, et al. The Amount of Bifidobacterium genus in colorectal carcinoma tissue in relation to tumor characteristics and clinical outcome. Am J Pathol. 2018;188(12):2839–2852. doi:10.1016/j.ajpath.2018.08.015.
- Kong C, Liang L, Liu G, Du L, Yang Y, Liu J, Shi D, Li X, Ma Y. Integrated metagenomic and metabolomic analysis reveals distinct gut-microbiome-derived phenotypes in early-onset colorectal cancer. Gut. 2022;72(6):1129–1142. doi:10.1136/gutjnl-2022-327156.
- Liu J, Tang W, Budhu A, Forgues M, Hernandez MO, Candia J, Kim Y, Bowman ED, Ambs S, Zhao Y, et al. A viral exposure signature defines early onset of hepatocellular carcinoma. Cell. 2020;182(2):317–328 e10. doi:10.1016/j.cell.2020.05.038.
- Kadosh E, Snir-Alkalay I, Venkatachalam A, May S, Lasry A, Elyada E, Zinger A, Shaham M, Vaalani G, Mernberger M, et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature. 2020;586(7827):133–138. doi:10.1038/s41586-020-2541-0.
- Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, Koyama K, Utsunomiya J, Baba S, Hedge P. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Sci. 1991;253(5020):665–669. doi:10.1126/science.1651563.
- Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66(3):589–600. doi:10.1016/0092-8674(81)90021-0.
- Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu X, DeStefano Shields CE, Hechenbleikner EM, Huso DL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Sci. 2018;359(6375):592–597. doi:10.1126/science.aah3648.
- Kong C, Liang L, Liu G, Du L, Yang Y, Liu J, Shi D, Li X, Ma Y. Integrated metagenomic and metabolomic analysis reveals distinct gut-microbiome-derived phenotypes in early-onset colorectal cancer. Gut. 2023;72(6):1129–1142. doi:10.1136/gutjnl-2022-327156.
- Gupta A, Dhakan DB, Maji A, Saxena R, Prasoodanan KV, Mahajan S, Pulikkan J, Kurian J, Gomez AM, Scaria J, et al. Association of Flavonifractor plautii, a Flavonoid-Degrading bacterium, with the gut microbiome of colorectal cancer patients in India. mSystems. 2019;4(6):10–128. doi:10.1128/mSystems.00438-19
- Braune A, Blaut M. Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes. 2016;7(3):216–234. doi:10.1080/19490976.2016.1158395.
- Kopustinskiene DM, Jakstas V, Savickas A, Bernatoniene J. Flavonoids as anticancer agents. Nutrients. 2020;12(2):457. doi:10.3390/nu12020457.
- Holowatyj AN, Lewis MA, Pannier ST, Kirchhoff AC, Hardikar S, Figueiredo JC, Huang LC, Shibata D, Schmit SL, Ulrich CM. Clinicopathologic and Racial/Ethnic differences of colorectal cancer among adolescents and young adults. Clin Transl Gastroenterol. 2019;10(7):e00059. doi:10.14309/ctg.0000000000000059.
- Becker S, Oelschlaeger TA, Wullaert A, Vlantis K, Pasparakis M, Wehkamp J, Stange EF, Gersemann M. Bacteria regulate intestinal epithelial cell differentiation factors both in vitro and in vivo. PloS One. 2013;8(2):e55620. doi:10.1371/journal.pone.0055620.
- Liu PH, Wu K, Ng K, Zauber AG, Nguyen LH, Song M, He X, Fuchs CS, Ogino S, Willett WC, et al. Association of obesity with risk of early-onset colorectal cancer among Women. JAMA Oncol. 2019;5(1):37–44. doi:10.1001/jamaoncol.2018.4280.
- Cirillo PM, Cohn BA. Pregnancy complications and cardiovascular disease death: 50-year follow-up of the Child health and development studies pregnancy cohort. Circulation. 2015;132(13):1234–1242. doi:10.1161/CIRCULATIONAHA.113.003901.
- Gupta S, Bharti B, Ahnen DJ, Buchanan DD, Cheng IC, Cotterchio M, Figueiredo JC, Gallinger SJ, Haile RW, Jenkins MA, et al. Potential impact of family history-based screening guidelines on the detection of early-onset colorectal cancer. Cancer. 2020;126(13):3013–3020. doi:10.1002/cncr.32851.
- Murphy CC, Cirillo PM, Krigbaum NY, Singal AG, Lee M, Zaki T, Burstein E, Cohn BA. Maternal obesity, pregnancy weight gain, and birth weight and risk of colorectal cancer. Gut. 2022;71(7):1332–1339. doi:10.1136/gutjnl-2021-325001.
- Nishi A, Kawachi I, Koenen KC, Wu K, Nishihara R, Ogino S. Lifecourse epidemiology and molecular pathological epidemiology. Am J Prev Med. 2015;48(1):116–119. doi:10.1016/j.amepre.2014.09.031.
- Liu L, Tabung FK, Zhang X, Nowak JA, Qian ZR, Hamada T, Nevo D, Bullman S, Mima K, Kosumi K, et al. Diets that promote colon inflammation associate with risk of colorectal carcinomas that contain Fusobacterium nucleatum. Clin Gastroenterol Hepatol. 2018;16(10):1622–1631 e3. doi:10.1016/j.cgh.2018.04.030.
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host & Microbe. 2013;14(2):195–206. doi:10.1016/j.chom.2013.07.012.
- Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17(3):156–166. doi:10.1038/s41579-018-0129-6.
- Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, Gurjao C, Manders F, Dalmasso G, Stege PB, et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature. 2020;580(7802):269–273. doi:10.1038/s41586-020-2080-8.
- Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. 2018;24(4):392–400. doi:10.1038/nm.4517.
- Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40(1):128–139. doi:10.1016/j.immuni.2013.12.007.
- Chen D, Jin D, Huang S, Wu J, Xu M, Liu T, Dong W, Liu X, Wang S, Zhong W, et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating wnt signaling and gut microbiota. Cancer Lett. 2020;469:456–467. doi:10.1016/j.canlet.2019.11.019.
- Sugimura N, Li Q, Chu ESH, Lau HCH, Fong W, Liu W, Liang C, Nakatsu G, Su ACY, Coker OO, et al. Lactobacillus gallinarum modulates the gut microbiota and produces anti-cancer metabolites to protect against colorectal tumourigenesis. Gut. 2021;71(10):2011–2021. doi:10.1136/gutjnl-2020-323951.
- Mohamadzadeh M, Pfeiler EA, Brown JB, Zadeh M, Gramarossa M, Managlia E, Bere P, Sarraj B, Khan MW, Pakanati KC, et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci U S A. 2011;Suppl 108(Suppl supplement_1):4623–4630. doi:10.1073/pnas.1005066107.
- Gamallat Y, Meyiah A, Kuugbee ED, Hago AM, Chiwala G, Awadasseid A, Bamba D, Zhang X, Shang X, Luo F, et al. Lactobacillus rhamnosus induced epithelial cell apoptosis, ameliorates inflammation and prevents colon cancer development in an animal model. Biomed Pharmacother. 2016;83:536–541. doi:10.1016/j.biopha.2016.07.001.
- Li Q, Hu W, Liu WX, Zhao LY, Huang D, Liu XD, Chan H, Zhang Y, Zeng JD, Coker OO, et al. Streptococcus thermophilus inhibits colorectal tumorigenesis through secreting β-galactosidase. Gastroenterology. 2021;160(4):1179–1193.e14. doi:10.1053/j.gastro.2020.09.003.
- Liu X, Jin G, Tang Q, Huang S, Zhang Y, Sun Y, Liu T, Guo Z, Yang C, Wang B, et al. Early life Lactobacillus rhamnosus GG colonisation inhibits intestinal tumour formation. Br J Cancer. 2022;126(10):1421–1431. doi:10.1038/s41416-021-01562-z.
- Wang L, Tang L, Feng Y, Zhao S, Han M, Zhang C, Yuan G, Zhu J, Cao S, Wu Q, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8(+) T cells in mice. Gut. 2020;69(11):1988–1997. doi:10.1136/gutjnl-2019-320105.
- Montalban-Arques A, Katkeviciute E, Busenhart P, Bircher A, Wirbel J, Zeller G, Morsy Y, Borsig L, Glaus Garzon JF, Muller A, et al. Commensal clostridiales strains mediate effective anti-cancer immune response against solid tumors. Cell Host & Microbe. 2021;29(10):1573–1588 e7. doi:10.1016/j.chom.2021.08.001.
- Wong CC, Yu J. Gut microbiota in colorectal cancer development and therapy. Nat Rev Clin Oncol. 2023;20(7):429–452. doi:10.1038/s41571-023-00766-x.
- Popkin BM, Adair LS, Ng SW. Global nutrition transition and the pandemic of obesity in developing countries. Nutr Rev. 2012;70(1):3–21. doi:10.1111/j.1753-4887.2011.00456.x.
- Siegel RL, Fedewa SA, Anderson WF, Miller KD, Ma J, Rosenberg PS, Jemal A. Colorectal cancer incidence patterns in the United States, 1974-2013. J Natl Cancer Inst. 2017;109(8). doi:10.1093/jnci/djw322.
- Stoffel EM, Murphy CC. Epidemiology and mechanisms of the increasing incidence of colon and rectal cancers in young adults. Gastroenterology. 2020;158(2):341–353. doi:10.1053/j.gastro.2019.07.055.
