ABSTRACT
With the increasing of aging population and the consumption of high-fat diets (HFD), the incidence of Alzheimer’s disease (AD) has skyrocketed. Natural antioxidants show promising potential in the prevention of AD, as oxidative stress and neuroinflammation are two hallmarks of AD pathogenesis. Here, we showed that quinic acid (QA), a polyphenol derived from millet, significantly decreased HFD-induced brain oxidative stress and neuroinflammation and the levels of Aβ and p-Tau. Examination of gut microbiota suggested the improvement of the composition of gut microbiota in HFD mice after QA treatment. Metabolomic analysis showed significant increase of gut microbial tryptophan metabolites indole-3-acetic acid (IAA) and kynurenic acid (KYNA) by QA. In addition, IAA and KYNA showed negative correlation with pro-inflammatory factors and AD indicators. Further experiments on HFD mice proved that IAA and KYNA could reproduce the effects of QA that suppress brain oxidative stress and inflammation and decrease the levels of of Aβ and p-Tau. Transcriptomics analysis of brain after IAA administration revealed the inhibition of DR3/IKK/NF-κB signaling pathway by IAA. In conclusion, this study demonstrated that QA could counteract HFD-induced brain oxidative stress and neuroinflammation by regulating inflammatory DR3/IKK/NF-κB signaling pathway via gut microbial tryptophan metabolites.
Introduction
Long-term consumption of high fat-diets (HFDs) is associated with the development of many diseases, such as obesity, dyslipidemia, insulin-dependent diabetes, nonalcoholic fatty liver disease etc.Citation1 HFD is also reported to increase inflammation in the central nervous system and exacerbate the pathologic process of neurodegenerative diseases.Citation2 Neuroinflammation is an innate and adaptive immune response triggered by the release of inflammatory mediators by immune cells, such as microglia and astrocytes.Citation3 Neuroinflammation brings about damage to the blood–brain barrier (BBB) and increases the risk of disease. In addition, neuroinflammation also affects neurons, leading to abnormal neurotransmitter function. While short-term neuroinflammation is thought to be protective and help restore damaged neuronal cells, long-term neuroinflammation is a key factor in the deterioration of many neurological disorders.
Neurodegenerative diseases, including Alzheimer’s disease (AD) and Parkinson’s disease (PD), are increasingly recognized as significant health challenges in aging populations worldwide.Citation4 AD, the most common form of dementia, can be classified into different categories, including familial and sporadic forms, or early and late-onset forms.Citation5 Hyperphosphorylation of Tau protein and excessive deposition of β-amyloid peptides (Aβ) have been widely recognized as the most prominent pathological hallmarks of the disease. Aβ is produced from the cleavage of the β-amyloid precursor protein (APP) by β-secretase (BACE1) and γ-secretase,Citation6 while Presenilin 1 (PS1) and Anterior pharynx-defective 1 (APH1) are the catalytic core of γ-secretase.Citation7
Neuroinflammation have also been implicated as potential pathological features of AD.Citation8 Recent studies have pointed out that microglia will release pro-inflammatory cytokines when stimulated by Aβ, which further stimulate the production of amyloid precursor proteins, leading to increased levels of Aβ and exacerbation of neurodegenerative lesions.Citation9 Moreover, there is a close association between oxidative stress and the inflammatory response. Reactive oxygen species (ROS) could activate the expression of pro-inflammatory factors,Citation10 and the excessive production of ROS by immune cells at the site of inflammation results in oxidative stress and tissue damage.Citation11 As a result, neuroinflammation has been recognized as a target for the prevention and treatment of neurodegenerative diseases,Citation12 and alleviating neuroinflammation is important for regulating brain health.
In recent years, polyphenols have received increasing attention for their potential anti-inflammatory and antioxidant properties. Numerous studies have demonstrated the unrivaled potential of polyphenols in reducing levels of pro-inflammatory mediators and ROS.Citation13 Moreover, polyphenols are also able to regulate neuroinflammation. In clinical studies, it is found that resveratrol reduces MMP9 levels, regulates neuroinflammation and attenuates decline of daily living scores of AD patients.Citation14 Due to low natural absorption rate, the neuro-inflammation regulating effects of polyphenols are always achieved through small-molecule metabolites produced by the gut microbiota.Citation15,Citation16 Quinic acid (QA), which is found in many natural plants, is a cyclohexanecarboxylic acid that exhibits antibacterial, antioxidant, anti-inflammatory, and antiviral activities.Citation17 However, the bioavailability of QA is low, which significantly limits its health effects and clinical values.Citation18 In our previous studies, we found that QA was one of the main constitutes of millet polyphenols which exerted neural protective effects on HFD-induced oxidative stress and neuroinflammation.Citation19
In this study, we aimed to verify the effects of QA on HFD-induced neuroinflammation and its potential in the prevention of AD. The possible mechanism was explored on the perspective of microbiota-brain axis by using metabolomics and transcriptome analysis.
Materials and methods
Materials
Quinic acid (QA) was purchased from Shanghai Yuanye Biotechnology Co., Ltd (Shanghai, China). Kynurenic acid (KYNA) and indole-3-acetic acid (IAA) was purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). QA, KYNA and IAA were analytically pure. Total triglyceride (TG), total cholesterol (TC) and low-density lipoprotein cholesterol (LDL-C) kits were procured from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). Total superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GPx), Malondialdehyde (MDA) and BCA protein assay kits were obtained from Beyotime Biotech Inc (Shanghai, China). All reagents were stored at 4°C.
Animals
Male 6–8-week-old C57BL/6 mice were purchased from Shanghai Jiesijie Laboratory Animal Co., Ltd. (Shanghai, China). All the animals were fed at a temperature of 22 ± 2°C under a 12-h light/12-h dark cycle. All mice were given food and water ad libitum.
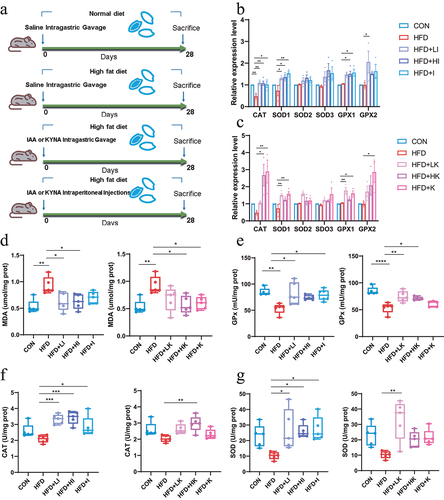
After five days of acclimatization, the first batch of mice were randomly divided into three groups (n = 8): normal diet group (CON), high fat diet (HFD) and high fat diet + quinic acid treated group (30 mg/kg) (HFD+QA). The second batch of mice were randomly divided into eight groups (n = 8). Six gavage groups: normal diet group (CON), high fat diet group (HFD), high fat diet + low-dose (10 mg/kg) IAA group (HFD+LI), high fat diet + high-dose (30 mg/kg) IAA group (HFD+HI), high fat diet + low-dose (10 mg/kg) KYNA group (HFD+LK) and high fat diet + high-dose (30 mg/kg) KYNA group (HFD+HK). Two intraperitoneal injection groups: high fat diet + IAA group (2 mg/kg) (HFD+I) and high fat diet +KYNA group (2 mg/kg) (HFD+K). Meanwhile, the CON and HFD groups were given daily saline gavage. QA stock solution (100 mg/kg) was diluted with water. The stock solution of IAA and KYNA (100 mg/kg) was made up in phosphate-buffered saline (PBS) by the addition of 2 M NaOH to fully resolve it, and then the pH was adjusted to 7.4 with 25% (v/v) HCl. High-fat feed composition: 52.2% basal feed, 20% sucrose, 15% lard, 10% casein, 1.2% cholesterol, 0.6% calcium bicarbonate, 0.4% laboratory animal premix, 0.2% sodium cholate (Jiangsu Xietong Pharmaceutical Bio-engineering Co., Ltd.). The gavage doses of IAA and KYNA were employed in keeping with the dose of QA, and intraperitoneal injection doses of IAA and KYNA was employed based on the determination of IAA in QA treated mice cecum (about 1.4–2.8 mg/kg). After four weeks of treatment, the mice were executed after an overnight fast. Blood, liver, stomach, small intestine, colon, cecum and brain were obtained and stored at −80°C.
Quantification of brain Aβ42 by Enzyme linked immunosorbent assay (ELISA)
Mouse Aβ42 ELISA kit was purchased from Shanghai Enzyme-linked Biotechnology Co., Ltd. (Shanghai China). Frozen brain tissues were thoroughly ground in PBS according to a 1:9 (w/v), while the homogenate was later centrifuged at 5000 × g for 10 min and the supernatant was taken. Collect excess clear supernatant and store at −20°C until next analysis. The level of Aβ42 oligomer in the supernatant was estimated using Aβ42 ELISA kit and the optical densities of the samples and standards were read at 450 nm, standard curves were made and results were calculated.
Immunofluorescence staining
To prepare brains for immunofluorescence imaging, mice were deeply anesthetized and intracardially perfused with saline for 5 min, then with 4% paraformaldehyde (PFA) for 5 min. The brains were removed and stored in PFA at 4°C until the next day. The brain tissue was embedded in paraffin and sliced (4 μm). The brain slices were dewaxed with xylene and rehydrated with graded alcohols. After antigen retrieval in citrate buffer solution, the slices were first treated with 3% H2O2 and then blocked with 2% BSA-PBS. After washing with PBST for three times, the slices were incubated with polyclonal rabbit anti-Iba-1 antibody (1:1000, Abcam Trading Co., Ltd., Shanghai, China) at 4°C overnight. Then, the slices were washed in PBS with 0.05% Tween-20 (PBST) and incubated with the secondary antibody (Goat-anti-rabbit-Rhodamine). The nucleus was stained with DAPI, and the slices were photographed under a fluorescence microscope.
Western blot
Brain tissues were placed in cell lysis buffer for Western and immunoprecipitation (IP) (Beyotime Biotechnology, Co., Ltd. Shanghai, China), well homogenized and then centrifuged at 12,000 × g in a cryo-centrifuge. The supernatant was transferred into a new centrifuge tube, and the protein concentration was determined with BCA method. After added with loading buffer (Beyotime Biotech Inc, Shanghai, China) at a ratio of 1:4 (v/v), the samples were boiled in a metal bath for 5 min and stored at −20°C. The proteins were then separated in a 12% separating gel and transferred from the gel to a 0.45 µm polyvinylidene difluoride (PVDF) membrane by transmembrane. And then the PVDF membrane was placed in a blocking solution (5% skimmed milk for non-phosphorylated proteins and 5% BSA for phosphorylated proteins) for 2 h at room temperature. The primary antibody was then incubated overnight. After the incubation, the PVDF membrane was washed for four times in TBS with 0.05% Tween-20 (TBST) for 5 min each time. Then, horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse IgG (1:1000, Beyotime Biotech Inc, Shanghai, China) was added and incubated for 2 h. The membrane was washed again for four times with TBST and then the protein bands were detected with ECL Substrate Kit under light-avoiding conditions. The ChemiDoc Imaging system (Bio-Rad Co.) was used for imaging and Image Lab software was used for quantitative analysis. Primary antibodies used in this study: anti-β-actin, anti-Gapdh, anti-IκB and anti-p-IκB (1:1000, Beyotime Biotech Inc, Shanghai, China); anti-TNF-α, anti-IL-1β, anti-Tau, anti-p-Tau Thr205, anti-IKK, anti-p-IKK, anti-NF-κB and anti-p-NF-κB (1:1000; Affinity Biosciences Group LTD, Jiangsu, China); anti-DR3 (1:1000, Beijing Biosynthesis Biotechnology Co., Ltd., Beijing, China).
Real-time quantitative polymerase chain reaction
Total RNA from brain tissues was extracted using EZ-10 Total RNA Mini-Preps Kit (Sangon Biotech (Shanghai) Co.,Ltd, Shanghai, China), and then reverse transcribed into cDNA using HiScript III RT SuperMix for qPCR. (+gDNA wiper) (Nanjing Vazyme Biotech Co., Ltd, Nanjing, China) The cDNA was amplified by ChamQ Universal SYBR qPCR Master Mix (Nanjing Vazyme Biotech Co., Ltd, Nanjing, China). The primers used in this study were offered in the Supporting Information.
16S rRNA sequencing
Fecal DNA was extracted using an E.Z.N.A. stool DNA Kit (Omega Bio-tek, Norcross, GA, U.S) according to the manufacturer’s instructions. The region V3-V4 of the bacterial 16S rRNA gene were amplified with primer pairs 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806 R (5′-GGACTACHVGGGTWTCTAAT-3′). 16S rDNA gene was amplified, and the PCR products were extracted from 2% agarose gel, purified by AxyPrep DNA gel extraction kit (Axygen biosciences, union city, Calif., USA), and quantified by Quantus™ fluorometer (Promega, USA). The purified PCR products were sequenced on Illumina MiSeq platform (Illumina, USA) of Majorbio Bio-Pharm Technology Co, Ltd. (Shanghai, China). All data were analyzed on the online platform of Majorbio Cloud Platform (https://cloud.majorbio.com/).
Untargeted metabolomics
Metabolites were extracted from 50 mg of brain tissue in a 2 mL centrifuge tube with a 6 mm diameter grinding bead. 400 μL of extraction solution (methanol:water = 4:1 (v:v)) containing 0.02 mg/mL of internal standard (L-2-chlorophenylalanine) was used for metabolite extraction. The sample solution was ground in a frozen tissue grinder for 6 min (−10°C, 50 kHz), followed by cryosonic extraction for 30 min (5°C, 40 kHz). The samples were allowed to stand at −20°C for 30 min, centrifuged for 15 min (4°C 13,000 g), and the supernatant was pipetted into an injection vial with an internal cannula for analysis. After on-boarding, the LC-MS raw data were imported into the metabolomics processing software Progenesis QI (Waters Corporation, Milford, USA) for baseline filtering, etc. A data matrix of retention times, mass-to-charge ratios, and peak intensities was obtained, and at the same time, the information of the MS and MSMS mass spectra was matched with the metabolism public databases as well as Majorbio’s own libraries. The MS and MSMS mass spectrometry information was matched with the metabolic public database and Majorbio’s own library to obtain metabolite information. The searched data matrix was uploaded and analyzed on the Majorbio’s cloud platform (https://cloud.majorbio.com/).
In vitro anaerobic fermentation and High Performance Liquid Chromatography (HPLC)
Fresh feces were collected from ten healthy mice. The feces were diluted in carbonic acid-phosphate buffer at 1:50 (w/v) and then filtered with four layers of gauze. The carbonic acid-phosphate buffer contained 9.24 g/L of NaHCO3, 2.2824 g/L of Na2HPO4, 0.47 g/L of NaCl, 0.45 g/L of KCl, 0.40 g/L of urea, 0.0728 g/L of CaCl2·2 H2O, 0.1 g/L of Na2SO4, 0.1 g/L of MgCl2·6 H2O, 2 g/L of peptone water, 2 g/L of yeast extract with 10 mL/L trace element containing 3.68 g/L of FeSO4·7 H2O, 1.159 g/L of MnSO4·H2O, 0.44 g/L of ZnSO4·7 H2O, 0.12 g/L of CoCl2·6 H2O, 0.098 g/L of CuSO4·5 H2O, 0.0174 g/L of Mo7(NH4)6O2·4 H2O, and distilled water, and the pH was adjusted to 7.0. The buffer was autoclaved at 121°C for 20 min before use. Sixteen milliliters of carbonic acid-phosphate buffer and 4 mL of fecal slurry were added to a 50 mL penicillin vial, and then QA (QA group) or saline (CON group) was added with a final concentration of 1.0 mg/mL. The mixed solution was shaken thoroughly and placed in an anaerobic environment (90% of nitrogen, 5% of carbon oxide, and 5% of hydrogen). The fermentation broth was collected after 72 h of incubation and the supernatant was collected after centrifugation at 10,000 rpm for 15 min. The supernatant was lyophilized and stored at 4°C for measurement.
Quantification of IAA by HPLC: 5 μg of supernatant powder was dissolved in 100 μL ultrapure water. Subsequently, 180 μL ether and 90 μL ethyl acetate were added and mixed for 10 s. After centrifuging at 12,000 rpm for 10 min, 200 μL supernatant was collected and dried using nitrogen blowing, and re-dissolved in 200 μL acetonitrile. The solution was centrifuged again at 12,000 rpm for 10 min. Before detection, the solution was passed through a 0.22 μm filter membrane. The HPLC instrument (Essentia LC-16, Shimadzu, Japan) equipped with a reversed-phase C18 column (25 cm × 4.6 mm, particle size = 5 μm) was used for the detection of IAA, and 20 μL sample was injected to the column. Mobile phase A: 200 mM ammonium acetate buffer (ice acetic acid adjusted to pH 4.5). Mobile phase B: acetonitrile, 70% B-phase isocratic elution. Retention time was 12 min with a flow rate at 0.8 mL/min and column temperature 35°C. The detection wavelength was 270 nm.
Quantification of KYNA by HPLC: 5 μg powder was dissolved in 300 μL ammonium formate solution (20 mM, contained 0.1% formic acid). After sonicating for 20 min and centrifuging at 13,000 rpm for 15 min, 200 μL supernatant was collected and passed through a 0.22 μm filter membrane before detection. KYNA was also detected by a reversed-phase C18 column. Mobile phase: 20 mM ammonium formate solution (containing 0.1% formic acid), acetonitrile, ethanol (95:2:3). Retention time was 35 min with a flow rate 1.0 mL/min. The column temperature was 30°C, and the detection wavelength was 344 nm.
Reference-based transcriptome analysis
Total RNA was extracted from the tissue using Trizol Reagent. Then RNA quality was determined by 5300 Bioanalyser (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). RNA purification, reverse transcription, library construction and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China). Shortly, messenger RNA was isolated according to polyA selection method by oligo (dT) beads and then fragmented by fragmentation buffer firstly. Secondly, double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen, CA) with random hexamer primers (Illumina). Then the synthesized cDNA was subjected to end-repair, phosphorylation and ‘A’ base addition according to Illumina’s library construction protocol. Libraries were size selected for cDNA target fragments of 300 bp on 2% Low Range Ultra Agarose followed by PCR amplification using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantified by Qubit 4.0, paired-end RNA-seq sequencing library was sequenced with the NovaSeq Xplus sequencer (2 × 150 bp read length). The raw paired end reads were trimmed and quality controlled by fastp with default parameters. Then clean reads were separately aligned to reference genome with orientation mode using HISAT2 software. The mapped reads of each sample were assembled by StringTie in a reference-based approach. All data were analyzed on the online platform of Majorbio Cloud Platform (https://cloud.majorbio.com/).
Statistical analysis
All data were expressed as mean ± standard error of the mean (SEM). Statistical analysis was performed using GraphPad Prism 8.0.2 (GraphPad Software, San Diego, CA, USA). Differences among the experimental data were assessed using one-way analysis of variance (ANOVA), followed by Tukey’s post hoc multiple comparison test. The untargeted metabolomics and transcriptomics data were analyzed on the online platform of Majorbio Cloud Platform (www.majorbio.com). p < .05 was considered statistically significant.
Results
QA alleviated brain oxidative stress induced by HFD
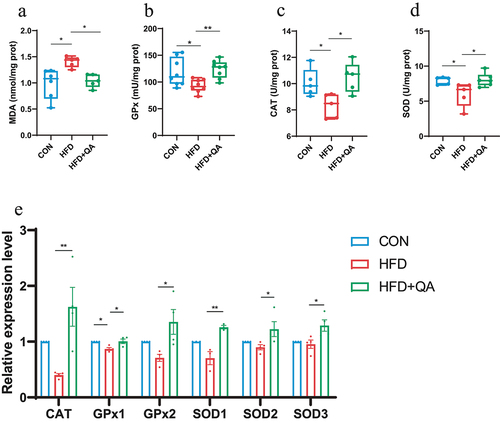
To investigate the impact of QA on high-fat diet-induced mice, the mice were intragastrically administered with 30 mg/kg/d QA during the 4-week high-fat diet induction process (Fig. S1a). At the end of the induction, we found that mice fed with a high-fat diet (HFD) weighed significantly higher than those fed with a normal diet (CON), and intragastric gavage of QA (HFD+QA) reversed the weight of mice to a normal level (Fig. S1b). The serum and liver lipids were determined to verify the establishment of the HFD mice models. From the results, the serum (Fig. S1C) and liver (Fig. S1d) levels of total triglyceride (TG), total cholesterol (TC), and low-density lipoprotein cholesterol (LDL-C) were significantly increased in HFD group compared with CON group, and QA improved the elevation of those lipids caused by HFD. These results indicated a successful establishment of the HFD model.
Malondialdehyde (MDA) is one of the end products of peroxidation of polyunsaturated fatty acids in cells, and therefore it is often considered as a marker of oxidative stress. Total superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (GPx) are the body’s first line of defense against the harmful effects of excess reactive oxygen species (ROS),Citation20 which are also considered as indicators of oxidative stress. From , the level of MDA in brain was significantly increased in HFD group but decreased in HFD+QA group. Whereas the activities of GPx (), CAT (), and SOD () were significantly decreased by HFD but increased by QA. The examination of the expression of those enzymes on mRNA level showed similar results that the expression of those enzymes was decreased in HFD group but increased in HFD+QA group (). These results suggested that QA was able to attenuate brain oxidative stress brought on by HFD.
Figure 1. QA improved brain oxidative stress in mice subjected to HFD.
QA suppressed neuroinflammation and down-regulated the expression of AD-related indicators induced by HFD
The assessment of pro-inflammatory cytokines interleukin-6 (IL-6) and interleukin-1β (IL-1β), as well as the anti-inflammatory cytokine interleukin-10 (IL-10) showed that HFD upregulated the expression of IL-6 and IL-1β and inhibited the expression of IL-10, while QA treatment significantly suppressed the expression of IL-6 and IL-1β and promoted the expression of IL-10 (). The examination of tumor necrosis factor α (TNF-α) and IL-1α on protein level further demonstrated that QA suppressed neuroinflammation induced by HFD ().
Figure 2. QA suppressed neuroinflammation and downregulated AD indicators in HFD mice.
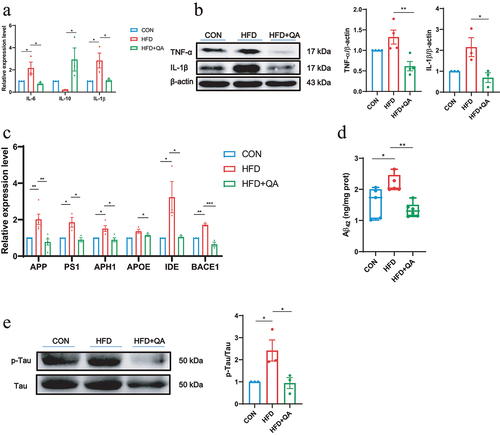
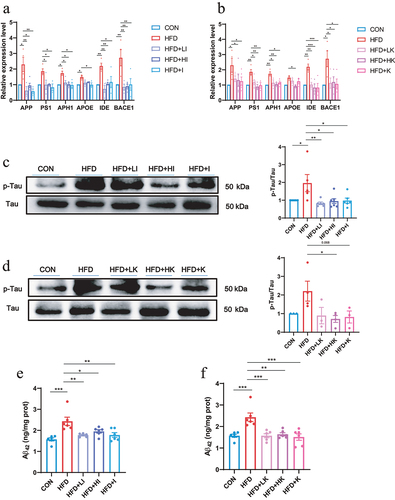
Then, the mRNA levels of AD-related marker genes were evaluated and the results were shown in . These data suggested that HFD induced the up-regulation of AD-related genes, APP, PS1, APH1, Apolipoprotein E (APOE), Insulin degrading enzyme (IDE), BACE1, while QA significantly decreased the expression levels of them. Subsequently, the protein levels of the two markers of AD, phosphorylated Tau and Aβ42 were determined. From the western blot analysis of p-Tau, it was obviously indicated that HFD increased the phosphorylation of Tau and QA suppressed it (). ELISA of Aβ42 showed that HFD led to the increase of Aβ42 in brain while QA reduced its level (). These findings indicated that QA was associated with a reduction in the expression of certain genes linked to AD.
QA improved gut microbiota dysbiosis induced by HFD
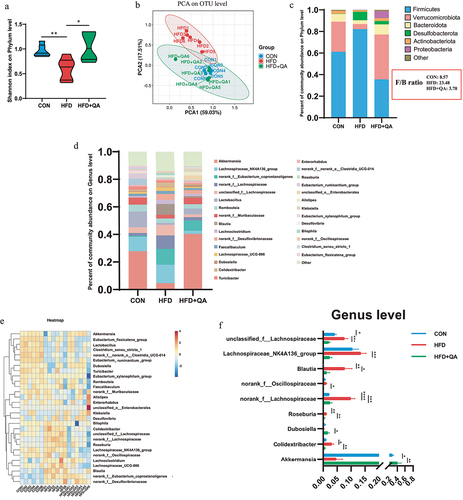
Gut microbial diversity was determined to investigate the alterations of gut microbes after QA treatment. Shannon index which represents the α diversity at the genus level indicated a significant increase in the diversity of intestinal microbiota in HFD+QA group compared with HFD group (). Principal component analysis (PCoA) which represents α diversity showed that the composition of the intestinal microbes of mice in CON group and QA group was significantly different from that of HFD group (). At the phylum level, the analysis of community abundance revealed that HFD increased the abundance of Firmicutes and Desulfobacteria, and decreased the abundance of Verrucomicrobiota and Bacteroidetes, while QA administration reversed the effects of HFD on those phyla. It was also notable that QA treatment also decreased the Firmicutes/Bacteroidetes (F/B) ratio (). As the increasing of F/B ratio is considered as an indicator of gut microbiota dysbiosis, the above results suggested that QA alleviated dysbiosis of HFD mice. The abundance of gut microbes was also examined at the genus level (), from which it could be observed that the dominant species was different among the three groups, where Akkermansia was dominant in CON and QA groups while Lanchnospiraceae was dominant in HFD group. Heatmap clearly showed the relative abundance of different microbes in each mouse (). Among them, the abundance of Akkermansia in the HFD+QA group was significantly increased compared to the HFD group, while the abundance of Colidextribacter, Roseburia, Blautia, Lanchnospiraceae etc. was significantly decreased (). Therefore, the above results suggested that QA improved the gut microbiota composition of HFD mice, which might contribute to the alleviation of brain oxidative stress and neuroinflammation.
Figure 3. QA alleviated HFD-induced gut microbial dysbiosis.
QA altered gut microbe-associated metabolites
As is known that the bioavailability of polyphenols is low and most dietary polyphenols exerts their function through microbial metabolites fermented by gut microorganisms. Therefore, liquid chromatography with mass spectrometry (MS) analysis was used to observe the alterations of microbial metabolites after QA treatment. Partial least squares discriminant analysis showed that the fecal metabolic profiles of mice treated with QA was significantly different from those treated with HFD (). Volcano plots demonstrated the number of metabolites significantly up- or down-regulated in HFD+QA group compared to HFD group (). It suggested that 245 metabolites were up-regulated and 76 metabolites were down-regulated after QA supplementation. The Venn diagram showed the unique or shared metabolites among the three groups (). These results indicated that mice treated with QA significantly altered the composition of metabolites in the feces of HFD mice.
Figure 4. QA altered gut microbe-associated metabolites.
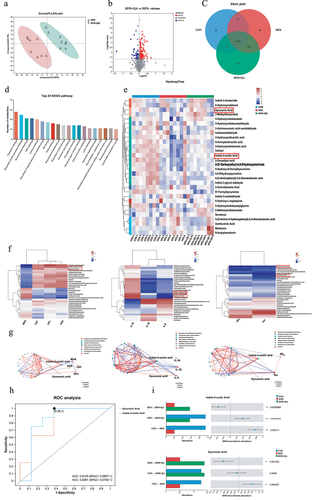
Identified metabolites were then analyzed via KEGG pathway classification, and the top 20 KEGG pathways were shown in . Among them, tryptophan metabolism was the top enriched pathway. The heatmap analysis showed the tryptophan metabolites in each sample (). Among them, the relative abundance of indole-3-acetic acid (IAA) and kynurenic acid (KYNA) were down regulated in HFD group but upregulated in HFD+QA group. Correlation analysis between tryptophan metabolites and the markers of brain oxidative stress, neuroinflammation and AD showed that many tryptophan metabolites including indole-3-acetic acid (IAA) and kynurenic acid (KYNA) exhibited significant positive correlations with CAT, GPx, SOD and IL-10, and negative correlations with MDA, IL-1β, IL-6, Aβ42 and Tau (). The subsequent analysis of multi-factor correlation network maps showed that IAA and KYNA were the key metabolites related to those indicators (). The results of the receiver operating characteristic analysis performed on IAA and KYNA indicated that these two substances were indeed the metabolites that were more critical to the differential effects among the different groups (). The abundance alterations of IAA and KYNA showed that they were decreased in HFD group but increased in HFD+QA group (). In order to verify the results of the non-targeted metabolomics analysis, we quantified the amount of IAA in the feces of the mice cecum with HPLC. The results showed that the content of IAA was reduced in HFD group but increased in HFD+QA group (Fig. S2a), which was consistent with the trend demonstrated in . Subsequently, in vitro anaerobic fermentation was used to figure out if QA altered the tryptophan metabolites of microbes. When the mice feces were fermented with QA for 72 h, the concentration of IAA and KYNA in the bacterial broth was significantly increased (Fig. S2b-c). Therefore, we hypothesized that QA might act through altering the production of IAA and KYNA of the intestinal flora.
QA alleviated oxidative stress and neuroinflammation through microbial metabolites
To verify whether QA reduced brain oxidative stress, neuroinflammation via microbial metabolites, different doses of IAA and KYNA (10 mg/kg/d and 30 mg/kg/d for intragastric treatment, and 2 mg/kg/d for intraperitoneally injection) were employed to verify the function of the two metabolites on HFD-induced brain dysfunction. The design of the experiment was shown in . Data of weight gain suggested that IAA and KYNA treatment could reverse the weight gain caused by HFD (Fig. S3a). The examination of serum and liver lipids indicated that both IAA and KYNA could decrease TC, TG and LDL-C either in serum or in liver (Fig. S3b-e). Interestingly, there was no significant difference between intraperitoneal injection and intragastric gavage.
(A) The diagram of treatment. (B) The relative expression levels of genes related to oxidative stress in the brain after IAA treatment. (C) The relative expression levels of genes related to oxidative stress in the brain after IAA treatment. (D) MDA level in brain. (E) Activity of GPx. (F) Activity of CAT. (G) Activity of SOD. Data are expressed as mean ± SEM, *p < .05, **p < .01, ***p < .001. MDA, malondialdehyde; GPx, glutathione peroxidase; CAT, catalase; SOD, superoxide dismutase.
The expression of antioxidant enzymes indicated that IAA and KYNA administration increased the mRNA levels of CAT, SOD1 and Gpx1 (). The detection of MDA in brain suggested IAA and KYNA could reduce MDA level (). In the meanwhile, both IAA and KYNA could increase the activities of antioxidant enzymes CAT, GPx and SOD (). Concurrently, the mRNA level of IL-6 and IL-1β () and the protein level of TNF-α and IL-1β () after IAA and KYNA treatment decreased significantly, following the same trend as that of QA in HFD mice. These results revealed that brain oxidative stress and inflammatory responses caused by HFD could be mitigated by both IAA and KYNA treatment.
Figure 6. IAA and KYNA ameliorated inflammation of HFD mice.
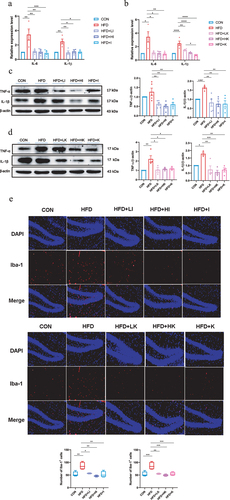
The activation of microglia is regarded as the indicator of neuroinflammation,Citation21 and elevated expression of ionized calcium-binding adaptor molecule 1 (Iba-1) represents microglia activation.Citation22 The immunofluorescence staining of Iba-1 on brain slices showed that the number of Iba-1 positive cells was significantly increased in HFD group, but decreased in both IAA and KYNA groups (). Measurement of AD-related genes indicated that IAA and KYNA could inhibit the up-regulation of APP, PS1, APH1, APOE, IDE, BACE1 induced by HFD (). Furthermore, IAA and KYNA suppressed the phosphorylation of Tau and reduced the level of Aβ42 (). From the above results, it could be concluded that IAA and KYNA reproduced the function of QA on HFD-induced brain oxidative stress, neuroinflammation and expression of AD-related genes.
Figure 7. IAA and KYNA down-regulated AD-related indicators of HFD mice.
QA regulated brain function by inhibiting DR3/IKK/NF-κB signaling pathway through bacterial metabolites
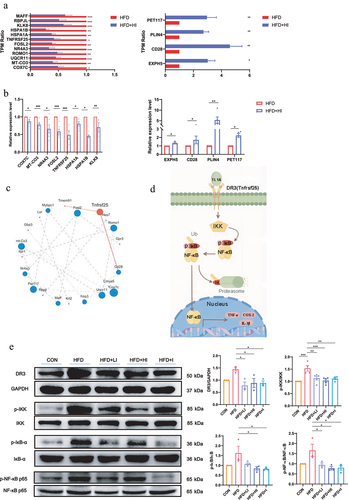
To further investigate the mechanism how QA regulated brain function through microbial metabolites, a reference-based (RB) transcriptome analysis was conducted to determine the impact of IAA on gene expression and the potential regulatory mechanism in brain. The gene expression profile of HFD+HI group was compared with HFD group. Principal component analysis showed significantly different clustering of gene in HFD+HI mice compared with HFD mice (). Volcano plot illustrated 56 up-regulated genes and 58 down-regulated genes in the HFD+HI group compared to the HFD group (). From the Venn diagram, it could be observed that there were 653 genes specifically enriched in the HFD+HI group while 431 genes specifically enriched in HFD group (). Gene Set Enrichment Analysis (GSEA) showed that genes related to oxidative phosphorylation, positive regulation of monocyte chemotactic protein 1 production, organ or tissue specific immune response and amyloid fibril formation were positively correlated with the HFD group and negatively correlated with the HFD+HI group (). From the heatmaps analysis, the gene expression profiles were significantly altered by HI administration (). Among them, Hcrt, Tnfrsf25, Gbp3, etc. were significantly down-regulated in the HFD+HI group while Palb2, Pet117, Plin4, etc. were up-regulated by IAA. The Reactome annotation analysis indicated that these differential genes belong to 12 different functional classes, and genes related to immune system accounted for the largest proportion ().
Figure 8. IAA altered the expression of genes in the brain of HFD mice.
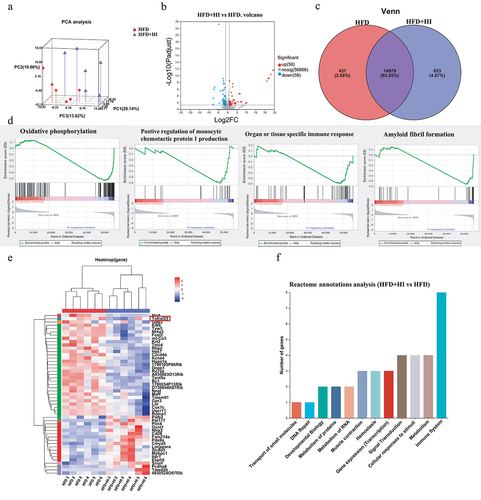
According to the Reactome annotation analysis, the relative abundance of genes (expressed as TPM ratio) related to immune system that were significantly up- and down-regulated was shown in . Those genes were also subjected to QPCR validation, and the results demonstrated the reliability of the RB transcriptome analysis (). showed the interaction of the immune system related genes by protein–protein interaction network analysis. Among those genes, Tnfrsf25 encodes the functional receptor DR3, which has been reported to be important meditators of inflammation.Citation23 It has been established that activation of DR3 by ligand TL1A rapidly initiates downstream signaling cascades involves the phosphorylation of the IκB kinases (IKK) and IκB, and triggers NF-κB signaling pathway,Citation24 leading to the expression of pro-inflammatory factors (). The detection of DR3, p-IKK/IKK, p-IκB/IκB and p-NF-κB/NF-κB () showed that the protein level of DR3 and the phosphorylation level of IKK, IκB and NF-κB was increased by HFD, suggesting the activation of DR3/IKK/NF-κB pathway. Whereas IAA treatment significantly decreased the expression of DR3 and the phosphorylation of IKK, IκB and NF-κB, which indicated the suppression of DR3/IKK/NF-κB pathway. These findings suggested that QA might suppress HFD-induced neuroinflammation by inhibiting DR3/IKK/NF-κB signaling pathway through IAA.
Figure 9. IAA-regulated neuroinflammation via the NF-κB signaling pathway.
Discussion
With the increase of people’s average age, AD has become one of the deadliest diseases today. As oxidative stress and neuroinflammation are major factors in AD, the potential of polyphenols in the treatment of neurodegenerative diseases has attracted more and more attentions.Citation25 In the present study, we demonstrated that QA could significantly alleviate HFD-induced oxidative stress and neuroinflammation, inhibit the phosphorylation of Tau and down-regulate the protein level of Aβ42. We found that QA could improve the dysbiosis of gut microbiota of HFD mice and altered the microbial metabolites, especially the tryptophan-related metabolites, IAA and KYNA, while IAA and KYNA treatment could reproduce the effects of QA on oxidative stress, neuroinflammation and expression of AD related genes in HFD mice. Furthermore, IAA treatment could alter the expression profiles of genes related to immunity and inhibit the activation of DR3/IKK/NF-κB signaling pathway to suppress inflammation in the brain.
Polyphenols have long been recognized to possess numerous biological properties. The potential of polyphenols on the treatment of neuroinflammation has also been discussed before. For example, Epigallocatechin-3-O-gallate protects neurons in the hippocampus of depressed rats by decreasing IL-6 levels and inhibiting neuroinflammation.Citation26 Prolonged neuroinflammation can lead to the exacerbation of neurodegenerative diseases, so polyphenols have also been recognized to have potential in the prevention of AD. Curcumin is reported to inhibit the formation and promote the disaggregation of amyloid-β plaques, attenuate the hyperphosphorylation of Tau and enhance its clearance, and modify microglial activity etc.Citation27 Resveratrol treatment attenuates the decline of cognitive scores, reduces Aβ42 levels during a 52-week trials on AD patients.Citation28 In this study, we observed that daily oral administration of QA to HFD mice alleviated oxidative stress and neuroinflammation as well as decreased the protein level of Aβ and p-Tau (). However, the role of QA in the prevention of AD needs to be further investigated on AD mice models.
It is recognized that large polyphenols are metabolized by intestinal microorganisms into small molecules,Citation15 and the function of dietary polyphenols are always carried out by gut microbiota, especial via gut microbial metabolites. From the results of 16S rRNA sequencing, we found that QA effectively improved the diversity and composition of intestinal microbiota of HFD mice. At the phylum level, the abundance of the two dominate phyla Bacteroidota and Firmicutes was altered by HFD and the F/B ratio was increased in HFD mice. Stability of F/B ratio is one of the most important healthy indicators of gut microbiota. Studies have shown that the F/B ratio is higher in obesity people.Citation29 In our study, we observed that the administration of QA reduced this ratio to a level comparable to that of the CON group (), suggesting the improvement of gut microbiota by QA. The further examination of the relative abundance of microbes at genus level showed that QA significantly increased the abundance of Akkermansia and decreased the abundance of Colidextribacter. As a probiotic, Akkermansia exhibits efficacy in alleviating multiple diseases, mitigating intestinal inflammation and preserving a healthy intestinal barrier.Citation30 Colidextribacter is reported to be negatively correlated with serum SOD activityCitation31 and positively correlated with obesityCitation32 and serum TC levels.Citation33 Thus, our studies indicated that gut microbes might play essential roles during the suppression of neuroinflammation by QA.
From the results of metabolomics, the main metabolic pathways were found to be involved were tryptophan metabolism, biosynthesis of plant secondary metabolites, steroid hormone biosynthesis and ABC transporters (). Tryptophan metabolism is an important metabolic pathway of human. As an essential amino acid, tryptophan could be converted to many bioactive compounds through tryptophan metabolism.Citation34 Tryptophan metabolites have received increasing attention in recent years due to their anti-inflammatory and anti-AD properties.Citation35 Although plant secondary metabolites are substances that occur naturally in plants, studies have shown that they can also have great benefits for human health, such as shikimic acid, which is demonstrated to possess antioxidant and anti-inflammatory properties.Citation36 The alteration of steroid hormone biosynthesis might be due to the alteration of lipid metabolism, as we observed a significant down regulation of blood lipids after QA treatment (Fig. S1). ABC transporters are responsible for ATP-driven translocation of many substrates across membranes,Citation37 and considered as a new line of research for treating AD because some of them can control levels of Aβ in the brain.Citation38 As tryptophan metabolism was the top enriched pathway, we focused on tryptophan metabolites in this study to further investigate the possible mechanisms.
Tryptophan metabolism is considered as a link between gut microbiota and brain.Citation39 Numerous studies have shown that tryptophan metabolites play a key role in maintaining intestinal homeostasis and systemic immunity,Citation40 among other functions. Tryptophan metabolites produced by various ways can cross the BBB and thus modulate the immunoreactivity of astrocytes and microglia.Citation41 Thus, the enzymes involved in tryptophan metabolism have been identified as ideal targets for cancer and neurodegenerative diseases.Citation42 Tryptophan metabolism contains three main pathways: the indole pathway, the kynurenine pathway and serotonin pathway.Citation43 In this study, we found that most of the differential metabolites of tryptophan metabolism belonged to the indole pathway and the kynurenine pathway (). The correlation heatmaps and multivariate correlation network maps analysis on tryptophan metabolites showed that IAA and KYNA were positively correlated with antioxidative enzymes and negatively correlated with pro-inflammatory factors and AD indicators (). ROC analysis of these two substances further indicated the importance of IAA and KYNA, and the abundance of IAA and KYNA were significantly increased in the fecal of the HFD+QA group mice compared with the HFD group mice. In vitro anaerobic fermentation of QA and mice feces confirmed that QA could increase the microbial production of IAA and KYNA indeed (Fig. S2b-c). It is interesting that there seems to be some relationship between Akkermansia and IAA. Studies have shown that Akkermansia improved colitis in mice by increasing the levels of indole-3-acrylic acid and IAA in the gut.Citation44 Furthermore, an anti-obesity study revealed that whole grain Qingke conspicuously augmented the abundance of Akkermansia in HFD mice, concomitant with elevated levels of tryptophan metabolites (Indole and IAA).Citation45 Here in this study, the alteration of Akkermansia was also in keeping with IAA, which is consistent with those studies.
The effects of IAA and KYNA on neuroinflammation has been mentioned by studies. It is reported that IAA could inhibit Aβ aggregation and suppress the NF-κB signaling pathway,Citation46 and KYNA is generally considered to have neuroprotective effects.Citation47 Besides, researchers found that IAA could alleviate nonalcoholic fatty liver disease in mice,Citation48 and attenuate oxidative stress and inflammation. Another study showed that IAA relieved ankylosing spondylitis in mice.Citation49 KYNA is also reported to regulate adipose tissue energy homeostasis and inflammation.Citation50 In the present study, we confirmed that administration of IAA and KYNA could restore the effects of QA on brain oxidative stress, neuroinflammation and AD related markers in HFD mice (). Together, our data suggested that QA alleviated brain oxidative stress and inflammation by regulating tryptophan metabolites.
In the mechanism exploration of this study, we found that the gene expression profiles of brain were significantly influenced by IAA treatment. GSEA analysis showed that genes related to oxidative phosphorylation, positive regulation of monocyte chemotactic protein 1 production, organ or tissue specific immune response and amyloid fibril formation were more highly expressed in the HFD group rather than in the HFD+HI group, which is consistent with the reduction in the expression levels of inflammatory factors and A42 in HFD+HI group. Further categorization of the differential genes revealed that the differential genes were most enriched in the category of the immune system (). These data suggested that QA might suppress neuroinflammation by regulating inflammatory signaling pathways via IAA.
Among the differential genes, Tnfrsf25 could be a candidate target, as it encodes DR3, a member of the TNF super receptor family, that directs the NF-κB signaling pathway thereby causing an inflammatory response in the organism. In addition, the sole ligand of DR3, TL1A, is recognized as a potential therapeutic target for inflammatory conditions, such as intestinal inflammation and tissue fibrosis.Citation23 Activation of the NF-κB signaling pathway is inextricably linked to the development of many diseases such as various types of inflammation and cancer,Citation51 and also leads to activation of microglia. It is reported that NF-κB levels were significantly increased in the cerebral cortex of AD patients, and NF-κB activates BACE1 and promotes the formation of Aβ protofibrils, as well as inhibits the dephosphorylation of Tau.Citation52 It has been shown that TL1A signaling activates NF-κB through DR3Citation53 to promote the expression of a number of target genes, such as IL-6, IL-2, IL-1β and TNF-α, and these released molecules will in turn reactivate NF-κB. In this study, we found the activation of DR3/IKK/NF-κB signaling pathway in HFD group and confirmed the suppression of this pathway by IAA (). Combining with the expression profiles of inflammatory factors TNF-α, IL-1β, IL-6 and IL-10 with or without QA and IAA treatment, our findings demonstrated that QA might suppress HFD-induced neuroinflammation by inhibiting the activation of the DR3/IKK/NF-κB signaling pathway in the brains via microbial metabolites. However, the concrete regulatory mechanism of IAA on the DR3/IKK/NF-κB signaling pathway needs to be investigated further.
It is undeniable that there may be other pathways that contribute to the regulation of QA/IAA on oxidative stress and neuroinflammation, as many other genes were also differentially expressed in HFD and HFD+HI group. GSEA analysis showed that genes related to oxidative phosphorylation were negatively enriched in HI group, and COX7C, a subunit of cytochrome c oxidase, which is involved in oxidative phosphorylation, was significantly downregulated by IAA. Other immune related genes, such as NR4A3 and FOSL2 were also down-regulated by IAA. NR4A3 is defined as an orphan receptor, and is considered as a potential target for the treatment of inflammatory diseases.Citation54 FOSL2 is an important transcriptional regulator that may be associated with many inflammatory diseases.Citation55 HSP family always participate the folding of proteins. HSPA1A belongs to the HSP70 family along with HSPA1B, and their expression was also significantly suppressed by IAA. Several studies have shown that HSP70 has potent neuroprotective properties for the treatment of AD.Citation56 The high expression of HSPA1A and HSPA1B in HFD group might attribute to the up-regulation of Aβ. QA or IAA treatment suppressed the expression of Aβ, so that their expression decreased correspondingly.
Conclusion
In conclusion, this study demonstrated for the first time that QA supplementation improved HFD-induced brain oxidative stress and neuroinflammation by increasing the expression and activity of antioxidant enzymes and inhibiting the expression of pro-inflammatory factors. QA reduced Aβ levels and Tau phosphorylation in HFD mice, and shifted the composition of gut microbiota and relevant metabolites production toward anti-inflammatory. Our results also indicated the involvement of tryptophan metabolites in the regulatory function of QA. Administration of tryptophan metabolites IAA and KYNA reproduced the effects of QA on brain oxidative stress and neuroinflammation. Transcriptome analysis revealed that IAA could suppress the DR3/IKK/NF-κB signaling pathway. Together, these data suggested that QA might suppress HFD-induced brain inflammation by inhibiting DR3/IKK/NF-κB signaling pathway via gut microbial tryptophan metabolites. Our study indicated the potential of QA in the regulation of brain health. However, the exact gut microbiota species responsible for the production of tryptophan metabolites and regulated by QA need further in-depth exploration.
Author contribution
Sen Li and Xiao Guan conceived the concept and designed the experiments. Yuwei Cai and Sen Li performed the experiments and wrote the manuscript. Tong Guan, Ze Zhang and Wanqing Cao contribute to samples collection. Yu Zhang and Kai Huang contribute to the interpretation of the results and the amendments of the manuscript.
Supplementary information.docx
Download MS Word (742 KB)WB.docx
Download MS Word (5.2 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data are available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2024.2374608
Additional information
Funding
References
- Liu P, Wang Z-H, Kang SS, Liu X, Xia Y, Chan C-B, Ye K. High-fat diet-induced diabetes couples to Alzheimer’s disease through inflammation-activated C/EBPβ/AEP pathway. Mol Psychiatry. 2022;27(8):3396–22. doi:10.1038/s41380-022-01600-z.
- Gannon OJ, Robison LS, Salinero AE, Abi-Ghanem C, Mansour FM, Kelly RD, Tyagi A, Brawley RR, Ogg JD, Zuloaga KL. High-fat diet exacerbates cognitive decline in mouse models of Alzheimer’s disease and mixed dementia in a sex-dependent manner. J Neuroinflammation. 2022;19(1):110. doi:10.1186/s12974-022-02466-2.
- Ransohoff RM. How neuroinflammation contributes to neurodegeneration. Science. 2016;353(6301):777–783. doi:10.1126/science.aag2590.
- Gao C, Jiang J, Tan Y, Chen S. Microglia in neurodegenerative diseases: mechanism and potential therapeutic targets. Signal Transduct Target Ther. 2023;8(1):359. doi:10.1038/s41392-023-01588-0.
- Schachter AS, Davis KL. Alzheimer’s disease. Dialogues Clin Neurosci. 2000;2(2):91–100. doi:10.31887/DCNS.2000.2.2/asschachter.
- Zhang T, Chen D, Lee TH. Phosphorylation signaling in APP processing in Alzheimer’s Disease. IJMS. 2020;21(1):209. doi:10.3390/ijms21010209.
- Watanabe H, Yoshida C, Hidaka M, Ogawa T, Tomita T, Futai E. Specific mutations in Aph1 cause γ-Secretase activation. Int J Mol Sci. 2022;23(1):507. doi:10.3390/ijms23010507.
- Heneka MT, Carson MJ, Khoury JE, Landreth GE, Brosseron F, Feinstein DL, Jacobs AH, Wyss-Coray T, Vitorica J, Ransohoff RM, et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015;14(4):388–405. doi:10.1016/S1474-4422(15)70016-5.
- Ozben T, Ozben S. Neuro-inflammation and anti-inflammatory treatment options for Alzheimer’s disease. Clin Biochem. 2019;72:87–89. doi:10.1016/j.clinbiochem.2019.04.001.
- Simpson DSA, Oliver PL. ROS generation in Microglia: understanding oxidative stress and inflammation in neurodegenerative disease. Antioxidants. 2020;9(8):743. doi:10.3390/antiox9080743.
- Aleksandrova K, Koelman L, Rodrigues CE. Dietary patterns and biomarkers of oxidative stress and inflammation: a systematic review of observational and intervention studies. Redox Biol. 2021;42:101869. doi:10.1016/j.redox.2021.101869.
- Zhang W, Xiao D, Mao Q, Xia H. Role of neuroinflammation in neurodegeneration development. Signal Transduct Target Ther. 2023;8:267. doi:10.1038/s41392-023-01486-5.
- Wu M, Luo Q, Nie R, Yang X, Tang Z, Chen H. Potential implications of polyphenols on aging considering oxidative stress, inflammation, autophagy, and gut microbiota. Crit Rev Food Sci Nutr. 2021;61(13):2175–2193. doi:10.1080/10408398.2020.1773390.
- Moussa C, Hebron M, Huang X, Ahn J, Rissman RA, Aisen PS, Turner RS. Resveratrol regulates neuro-inflammation and induces adaptive immunity in Alzheimer’s disease. J Neuroinflammation. 2017a;14(1):1. doi:10.1186/s12974-016-0779-0.
- Carregosa D, Carecho R, Figueira IN, Santos C. Low-molecular weight metabolites from polyphenols as effectors for attenuating neuroinflammation. J Agric Food Chem. 2020;68(7):1790–1807. doi:10.1021/acs.jafc.9b02155.
- Frolinger T, Sims S, Smith C, Wang J, Cheng H, Faith J, Ho L, Hao K, Pasinetti GM. The gut microbiota composition affects dietary polyphenols-mediated cognitive resilience in mice by modulating the bioavailability of phenolic acids. Sci Rep. 2019;9(1):3546. doi:10.1038/s41598-019-39994-6.
- Benali T, Bakrim S, Ghchime R, Benkhaira N, El Omari N, Balahbib A, Taha D, Zengin G, Hasan MM, Bibi S, et al. Pharmacological insights into the multifaceted biological properties of quinic acid. Biotechnol Genet Eng Rev. 2022; 1–30. doi:10.1080/02648725.2022.2122303.
- Inbathamizh L, Padmini E. Quinic acid as a potent drug candidate for prostate cancer - a comparative pharmacokinetic approach. Asian J Pharm Clin Res. 2013;6:106–112.
- Li S, Xian FR, Guan X, Huang K, Yu WW, Liu DD. Neural protective effects of millet and millet polyphenols on high-fat diet-induced oxidative stress in the brain. Plant Food Hum Nutr. 2020;75(2):208–214. doi:10.1007/s11130-020-00802-6.
- Weydert CJ, Cullen JJ. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat Protoc. 2010;5(1):51–66. doi:10.1038/nprot.2009.197.
- Minter MR, Zhang C, Leone V, Ringus DL, Zhang X, Oyler-Castrillo P, Musch MW, Liao F, Ward JF, Holtzman DM, et al. Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer’s disease. Sci Rep. 2016;6:30028. doi:10.1038/srep30028.
- Zhang M, Qian C, Zheng Z, Qian F, Wang Y, Thu P, Zhang X, Zhou Y, Tu L, Liu Q, et al. Jujuboside a promotes Aβ clearance and ameliorates cognitive deficiency in Alzheimer’s disease through activating Axl/HSP90/PPARγ pathway. Theranostics. 2018;8:4262–4278. doi:10.7150/thno.26164.
- Valatas V, Kolios G, Bamias G. TL1A (TNFSF15) and DR3 (TNFRSF25): a co-stimulatory system of cytokines with diverse functions in gut mucosal immunity. Front Immunol. 2019;10:583. doi:10.3389/fimmu.2019.00583.
- Wang J, Al-Lamki RS, Zhu X, Liu H, Pober JS, Bradley JR. TL1-A can engage death receptor-3 and activate NF-kappa B in endothelial cells. BMC Nephrol. 2014;15:178.
- Veurink G, Perry G, Singh S. Role of antioxidants and a nutrient rich diet in Alzheimer’s disease. Open Biol. 2020;10(6):200084. doi:10.1098/rsob.200084.
- Wang J, Xu S, Chen X, Wang L, Li J, Li G, Zhang B. Antidepressant effect of EGCG through the inhibition of hippocampal neuroinflammation in chronic unpredictable mild stress-induced depression rat model. J Funct Foods. 2020;73:104106. doi:10.1016/j.jff.2020.104106.
- Tang M, Taghibiglou C, Liu J. The mechanisms of action of curcumin in Alzheimer’s disease. J Alzheimers Dis. 2017;58(4):1003–1016. doi:10.3233/JAD-170188.
- Moussa C, Hebron M, Huang X, Ahn J, Rissman RA, Aisen PS, Turner RS. Resveratrol regulates neuro-inflammation and induces adaptive immunity in Alzheimer’s disease. J Neuroinflammation. 2017b;14(1):1. doi:10.1186/s12974-016-0779-0.
- Stojanov S, Berlec A, Štrukelj B. The influence of probiotics on the firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms. 2020;8(11):1715. doi:10.3390/microorganisms8111715.
- Cani PD, Depommier C, Derrien M, Everard A, de Vos WM. Akkermansia muciniphila: paradigm for next-generation beneficial microorganisms. Nat Rev Gastroenterol Hepatol. 2022;19(10):625–637. doi:10.1038/s41575-022-00631-9.
- Chen L, Hu T, Wu R, Wang H, Wu H, Wen P. In vivo antioxidant activity of Cinnamomum cassia leaf residues and their effect on gut microbiota of d-galactose-induced aging model mice. J Sci Food Agric. 2023;103(2):590–598. doi:10.1002/jsfa.12170.
- Lu H-Y, Zhao X, Liu T-J, Liang X, Zhao M-Z, Tian X-Y, Yi H-X, Gong P-M, Lin K, Zhang Z, et al. Anti-obesity effect of fucoidan from Laminaria japonica and its hydrothermal degradation product. Food Biosci. 2024;58:103749. doi:10.1016/j.fbio.2024.103749.
- Huang J, Xu Y, Wang M, Yu S, Li Y, Tian H, Zhang C, Li H. Enterococcus faecium R-026 combined with Bacillus subtilis R-179 alleviate hypercholesterolemia and modulate the gut microbiota in C57BL/6 mice. FEMS Microbiology Letters. 2023;370:fnad118. doi:10.1093/femsle/fnad118.
- Xue C, Li G, Zheng Q, Gu X, Shi Q, Su Y, Chu Q, Yuan X, Bao Z, Lu J, et al. Tryptophan metabolism in health and disease. Cell Metab. 2023;35(8):1304–1326. doi:10.1016/j.cmet.2023.06.004.
- Sorgdrager F, Naude P, Kema I, Nollen E, Deyn P. Tryptophan metabolism in inflammaging: from biomarker to therapeutic target. Front Immunol. 2019;10:2565. doi:10.3389/fimmu.2019.02565.
- Gandhi GR, Vasconcelos ABS, Antony PJ, Montalvão MM, de Franca MNF, Hillary VE, Ceasar SA, Liu D. Natural sources, biosynthesis, biological functions, and molecular mechanisms of shikimic acid and its derivatives. Asian Pac J Trop Biomed. 2023;13(4):139–147. doi:10.4103/2221-1691.374230.
- Rees DC, Johnson E, Lewinson O. ABC transporters: the power to change. Nat Rev Mol Cell Biol. 2009;10(3):218–227. doi:10.1038/nrm2646.
- Pereira CD, Martins F, Wiltfang J, da Cruze Silva OAB, Rebelo S. ABC transporters are key players in Alzheimer’s disease. J Alzheimer’s Dis. 2018;61(2):463–485. doi:10.3233/JAD-170639.
- Gao K, Mu C-L, Farzi A, Zhu W-. Tryptophan metabolism: a link between the gut microbiota and brain. Adv Nutr. 2020;11:709–723. doi:10.1093/advances/nmz127.
- Su X, Gao Y, Yang R. Gut microbiota-derived tryptophan metabolites maintain gut and systemic homeostasis. Cells. 2022;11(15):2296. doi:10.3390/cells11152296.
- Zhang Z, Tang H, Chen P, Xie H, Tao Y. Demystifying the manipulation of host immunity, metabolism, and extraintestinal tumors by the gut microbiome. Signal Transduct Target Ther. 2019;4(1):41. doi:10.1038/s41392-019-0074-5.
- Platten M, Nollen EAA, Röhrig UF, Fallarino F, Opitz CA. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov. 2019;18(5):379–401. doi:10.1038/s41573-019-0016-5.
- Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host & Microbe. 2018;23(6):716–724. doi:10.1016/j.chom.2018.05.003.
- Gu Z, Pei W, Shen Y, Wang L, Zhu J, Zhang Y, Fan S, Wu Q, Li L, Zhang Z. Akkermansia muciniphila and its outer protein Amuc_1100 regulates tryptophan metabolism in colitis. Food Funct. 2021;12:10184–10195. doi:10.1039/D1FO02172A.
- Li X, Suo J, Huang X, Dai H, Bian H, Zhu M, Lin W, Han N. Whole grain qingke attenuates high-fat diet-induced obesity in mice with alterations in gut microbiota and metabolite profile. Front Nutr. 2021;8. doi:10.3389/fnut.2021.761727.
- Zhou Y, Chen Y, He H, Peng M, Zeng M, Sun H. The role of the indoles in microbiota-gut-brain axis and potential therapeutic targets: a focus on human neurological and neuropsychiatric diseases. Neuropharmacology. 2023;239:109690. doi:10.1016/j.neuropharm.2023.109690.
- Ostapiuk A, Urbanska EM. Kynurenic acid in neurodegenerative disorders—unique neuroprotection or double-edged sword? CNS Neurosci Ther. 2022;28(1):19–35. doi:10.1111/cns.13768.
- Ji Y, Gao Y, Chen H, Yin Y, Zhang W. Indole-3-acetic acid alleviates nonalcoholic fatty liver disease in mice via attenuation of hepatic lipogenesis, and oxidative and inflammatory stress. Nutrients. 2019;11(9):2062. doi:10.3390/nu11092062.
- Shen J, Yang L, You K, Chen T, Su Z, Cui Z, Wang M, Zhang W, Liu B, Zhou K, et al. Indole-3-Acetic acid alters intestinal microbiota and alleviates ankylosing spondylitis in mice. Front Immunol. 2022;13:762580. doi:10.3389/fimmu.2022.762580.
- Agudelo LZ, Ferreira DMS, Cervenka I, Bryzgalova G, Dadvar S, Jannig PR, Pettersson-Klein AT, Lakshmikanth T, Sustarsic EG, Porsmyr-Palmertz M, et al. Kynurenic acid and Gpr35 regulate adipose tissue energy homeostasis and inflammation. Cell Metabolism. 2018;27(2):378–392.e5. doi:10.1016/j.cmet.2018.01.004.
- Yu H, Lin L, Zhang Z, Zhang H, Hu H. Targeting NF-κB pathway for the therapy of diseases: mechanism and clinical study. Signal Transduct Target Ther. 2020;5(1):209. doi:10.1038/s41392-020-00312-6.
- Sun E, Motolani A, Campos L, Lu T. The pivotal role of NF-kB in the pathogenesis and therapeutics of Alzheimer’s disease. Int J Mol Sci. 2022;23(16):8972. doi:10.3390/ijms23168972.
- Xu W-D, Li R, Huang A-F. Role of TL1A in inflammatory autoimmune diseases: a comprehensive review. Front Immunol. 2022;13:891328. doi:10.3389/fimmu.2022.891328.
- Rodríguez-Calvo R, Tajes M, Vázquez-Carrera M. The NR4A subfamily of nuclear receptors: potential new therapeutic targets for the treatment of inflammatory diseases. Expert Opin Ther Targets. 2017;21(3):291–304. doi:10.1080/14728222.2017.1279146.
- Renoux F, Stellato M, Haftmann C, Vogetseder A, Huang R, Subramaniam A, Becker MO, Blyszczuk P, Becher B, Distler JHW, et al. The AP1 transcription factor Fosl2 promotes systemic autoimmunity and inflammation by repressing treg development. Cell Rep. 2020;31(13):107826. doi:10.1016/j.celrep.2020.107826.
- Zatsepina OG, Evgen’ev MB, Garbuz DG. Role of a heat shock transcription factor and the major heat shock protein Hsp70 in memory formation and neuroprotection. Cells. 2021;10(7):1638. doi:10.3390/cells10071638.