Abstract
The initiation of DNA replication is a highly regulated event in eukaryotic cells to ensure that the entire genome is copied once and only once during S phase. The primary target of cellular regulation of eukaryotic DNA replication initiation is the assembly and activation of the replication fork helicase, the 11-subunit assembly that unwinds DNA at a replication fork. The replication fork helicase, called CMG for Cdc45-Mcm2–7, and GINS, assembles in S phase from the constituent Cdc45, Mcm2–7, and GINS proteins. The assembly and activation of the CMG replication fork helicase during S phase is governed by 2 S-phase specific kinases, CDK and DDK. CDK stimulates the interaction between Sld2, Sld3, and Dpb11, 3 initiation factors that are each required for the initiation of DNA replication. DDK, on the other hand, phosphorylates the Mcm2, Mcm4, and Mcm6 subunits of the Mcm2–7 complex. Sld3 recruits Cdc45 to Mcm2–7 in a manner that depends on DDK, and recent work suggests that Sld3 binds directly to Mcm2–7 and also to single-stranded DNA. Furthermore, recent work demonstrates that Sld3 and its human homolog Treslin substantially stimulate DDK phosphorylation of Mcm2. These data suggest that the initiation factor Sld3/Treslin coordinates the assembly and activation of the eukaryotic replication fork helicase by recruiting Cdc45 to Mcm2–7, stimulating DDK phosphorylation of Mcm2, and binding directly to single-stranded DNA as the origin is melted.
Introduction to DNA Replication and the Cell Cycle
DNA replication is restricted to the S phase of the eukaryotic cell cycle.Citation1 During S phase, the parental genomic DNA is replicated precisely once to provide an additional copy of DNA for each daughter cell.Citation1 It is important that replication is restricted to S phase, and that the parental is only duplicated just once.Citation2 Over-replication of DNA, under-replication of DNA, or replication of DNA outside of S phase can lead to genome instability and possibly cell death or cancer.Citation2
The core enzymes of DNA replication are the DNA polymerases, which copy the parental duplex DNA.Citation3 Pol α-primase supplies the initial nucleic acid primer for subsequent elongation by Pol δ and Pol ε .Citation3 Pol ε is devoted to replication on the leading strand, while Pol δ is devoted to replication on the lagging strand,Citation4 although recently this division of labor has been questioned.Citation5 Importantly, Pol α-primase, Pol δ, and Pol ε each require single-stranded DNA templates (primed for Pol δ and Pol ε, or unprimed for Pol α-primase) for activity. Since the parental duplex is double-stranded, the replication fork helicase is required to melt the parental duplex to form single DNA strands (ssDNA).Citation6 The replication fork helicase therefore functions before the replicative polymerases, and therefore the replication fork helicase is the physiologic target for cell cycle regulation.Citation6
The Replication Fork Helicase is CMG
The replication fork helicase is an 11-subunit enzyme called CMG, composed of Cdc45 (Cell-division cycle 45], Mcm2–7 (Minichromosome maintenance, a heterohexamer of Mcm2, Mcm3, Mcm4, Mcm5, Mcm6, and Mcm7), and GINS (Japanese for Go, Ichi, Nii, Sans, or 5,1,2,3, or Sld5 [synthetic lethal with dpb11-1 (DNA-polymerase B-interacting protein)], Psf1 (partner with Sld5), Psf2, Psf3).Citation7 The Mcm2–7 heterohexameric ring complex is the motor of the CMG helicase.Citation7,8 The 6 homologous Mcm subunits contain an N-terminal DNA-interacting and oligomerization domain and a C-terminal AAA+ motor domain.Citation9 The Mcm2–7 complex has very weak helicase and ATPase activity on its own, but the assembly with Cdc45 and the tetrameric GINS complex stimulates Mcm2–7 activity.Citation7 Together, the CMG is very efficient in hydrolyzing ATP and processively unwinding DNA.Citation7
The Mcm2–7 complex loads as an inactive head to head double hexamer to encircle double-stranded DNA during G1 phase in a reaction known as replication licensing.Citation10,11 This reaction is ATP-dependent and catalyzed by Orc, Cdc6, and Cdt1.Citation10,11 The details of this reaction have recently been elucidated by a number of labs.Citation12-14 This reaction is restricted to G1 phase in the cell cycle.Citation15 Since Mcm2-7 complexes cannot load during S phase, this restriction of Mcm2–7 loading to G1 phase prevents re-replication or over-replication of the parental duplex DNA.Citation15
An excess of Mcm2–7 complexes load onto double-stranded DNA relative to the Mcm2–7 complexes that are activated to unwind DNA at a replication fork.Citation16 This was known as the Mcm paradox, since it was initially unclear why an excess of dormant Mcm2–7 complexes are loaded to encircle double-stranded DNA.Citation16 Later it was discovered that the excess Mcm2–7 complexes are loaded as a “back-up” mechanism if cells are subjected to replication stress.Citation16 This is important because it ensures that there is a sufficient number of loaded Mcm2–7 complexes before S phase begins, which is when licensing becomes inhibited. If cells encounter replication stress, additional loaded Mcm2–7 complexes are activated to help accomplish the complete replication of the genomic DNA.Citation16 This “back-up” mechanism helps ensure that once DNA replication starts, DNA replication will eventually be completed to avoid genome instability.Citation16 Dormant origins have also been shown to be an active tumor suppressor mechanism.Citation17
Although the Mcm2–7 complex loads to encircle double-stranded DNA during G1, the Mcm2–7 complex assembles with Cdc45 and GINS to form the CMG helicase during S phase.Citation7,18-20 Thus, assembly of the CMG from Cdc45, Mcm2–7, and GINS is a critical S phase regulatory mechanism that restricts DNA replication to S phase.Citation7,18-20 Furthermore, whereas there is an excess of loaded Mcm2–7 complexes relative to Mcm2–7 complexes that are subsequently activated, there is no evidence that fully assembled CMGs are present in excess.Citation19,20 Thus, the assembly of Cdc45 and GINS with Mcm2–7 completes the assembly of the active helicase and marks the particular Mcm2–7 complex for activation and unwinding.Citation7,18-21
Role of CDK and DDK in Helicase Assembly and Activation
The assembly of the CMG can only occur during S phase because this step is regulated by 2 S-phase kinases, S-CDK (S-phase cyclin-dependent kinase) and DDK (Dbf4-dependent kinase).Citation6 Moreover, CMG complex formation is also regulated in budding yeast by 4 initiation-specific protein factors, Sld2 (RecQL4 or RecQ4 in humans), Sld3 (Treslin or TICRR in humans), Sld7, and Dpb11 (TopBP1 in humans).Citation6 These factors do not travel as part of the CMG active helicase.Citation22 S-CDK plays also an important role in inhibiting the Mcm2–7 loading reaction during S phase, allowing licensing to take place only during G1.Citation15 However, in addition, S-CDK phosphorylates Sld2 and Sld3 to trigger complex formation with Dpb11.Citation23,24 This reaction is conserved from yeast to humans.Citation25-28 Furthermore, Sld3 and Sld7 form a tetramer containing 2 subunits of Sld3 and 2 subunits of Sld7 in solution.Citation29 Sld2, Sld3, Dpb11, and Dbf4 subunit of DDK are limiting during S phase, and overexpression of these factors causes late origins to fire early.Citation30 Sld3 is also a target of the replication checkpoint, which functions to silence late origins during replication stress.Citation31
The role of DDK during S phase is to phosphorylate subunits of the Mcm2–7 complex.Citation32,33 The Mcm2, Mcm4, and Mcm6 subunits are phosphorylated by DDK during S phase.Citation34,35 Phosphorylation of Mcm4 by DDK is important for the recruitment of Cdc45 to Mcm2–7.Citation35-37 Furthermore, phosphorylation of Mcm4 by DDK alleviates an inhibitor activity present in the N-terminal region of Mcm4.Citation38 The role of DDK phosphorylation of Mcm6 is currently under investigation.Citation35
The role of DDK phosphorylation of Mcm2 has been less clear and controversial. While some studies, including papers from our group, have suggested that DDK phosphorylation of Mcm2 is required for DNA replication under normal growth conditions,Citation39,40 other studies have suggested that DDK phosphorylation of Mcm2 is only important during replication stress.Citation41 Our studies suggesting that DDK phosphorylation of Mcm2 is required for cell growth under normal conditions used a galactose-inducible promoter, wherein galactose levels were varied to achieve wild-type levels of Mcm2 expression under normal conditions.Citation42 Under these conditions, expression of kinase-dead Mcm2 (Mcm2-S164A,S170A) is lethal to these cells, and overexpression of kinase dead Mcm2 results in a dominant negative severe growth defect.Citation42
The studies that conclude that DDK phosphorylation of Mcm2 is only important during replication stress were based upon a genomic insertion of the kinase-dead mutant of mcm2 at the genomic locus, under regulation by native promoter.Citation41 We recently acquired the yeast strain that harbored the genomic insertion of the kinase-dead mutant of mcm2.Citation43 We overexpressed the kinase-dead mutant of mcm2 in the wild-type yeast strain and in the yeast strain harboring the kinase dead mutant.Citation43 Whereas overexpression of the kinase-dead mutant of mcm2 in wild-type cells results in a severe growth defect, overexpression of the same kinase dead mutant of mcm2 resulted in no defect in the cells harboring the genomic copy of kinase-dead mcm2.Citation43 These data suggest that the strain with the genomic copy of kinase-dead mcm2 harbored a suppressor mutation.Citation43 Thus, this suppressor mutation explains the discrepancy between the conclusions from our different laboratories, and the data now point to the conclusion that DDK phosphorylation of Mcm2 is required for DNA replication under normal growth conditions.Citation39,42,43
The interaction between Mcm2 and Mcm5 in the Mcm2–7 ring may dissociate under certain conditions, acting as a ‘gate’ for the movement of dsDNA into the Mcm2–7 ring during G1.Citation8,44 Work from our laboratory showed how DDK phosphorylation of Mcm2 may have a critical role in modulating this gate. The phosphorylation of the N-terminal of Mcm2 by DDK may open the Mcm2–7 ring at the Mcm2-Mcm5 interface, allowing for ssDNA extrusion.Citation42 The generation of ssDNA at origins of replication is critical for the timely assembly of the CMG helicase.Citation42 Furthermore, expression of mcm5-bob1 (mcm5-P83L) bypasses the requirement for DDK in the cell,Citation45 and expression of mcm5-bob1 also partially suppresses the growth inhibition induced by overexpression of kinase-dead mutant of mcm2.Citation42 Mcm5-bob1 is also inhibited from binding Mcm2 relative to wild-type Mcm5, suggesting that the Mcm2-Mcm5 interface is a target of regulation by DDK.Citation42
Roles of Sld2, Sld3, and Dpb11 in Helicase Assembly and Activation
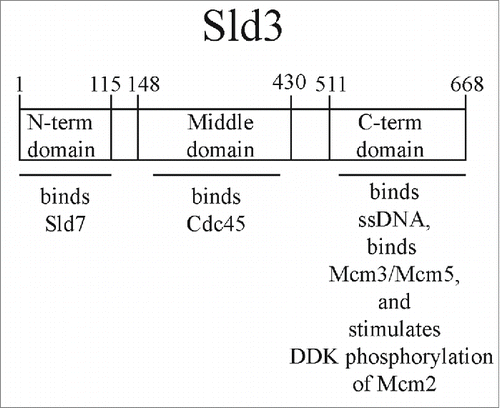
The roles of the essential initiation factors, Sld2, Sld3, and Dpb11, are beginning to be elucidated. For example, recent structural data shows that the N-terminal region of Sld3 binds to Sld7, and Sld3 and Sld7 from a tetramer of 2 subunits each of Sld3 and Sld7 (Fig. 1).Citation29 Furthermore, the middle domain of Sld3 binds to Cdc45, and structural data on this interaction is available as well.Citation46 Moreover, Sld3 recruits Cdc45 to Mcm2–7 during S phase in a manner that is dependent on DDK.Citation32,47 Dpb11 is believed to be a scaffolding protein that attaches to CDK-phosphorylated Sld2 and Sld3.Citation23,24 Each of these proteins, Sld2, Sld3, and Dpb11, bind directly to Mcm2-7 and single-stranded DNA (ssDNA).Citation48-50 These data suggest that Sld2, Sld3, and Dpb11 may play an active role in assembly or activation of the helicase.Citation48-50 Furthermore, the ssDNA-binding properties of Sld2 and Sld3 are conserved in their human counterparts, RecQL4 and Treslin, respectively.Citation50,51 In addition, the region of Sld3 that binds to ssDNA and Mcm3 and Mcm5 subunits is the C-terminal domain of Sld3.Citation50
What could be the functional role of Sld2, Sld3, and Dpb11 binding to ssDNA? Work from other labs suggests that single-stranded DNA is formed at an origin of replication during the process of replication initiation, in a process known as origin melting.Citation52 Furthermore, in one model of replication elongation, the steric exclusion model, the lagging single-stranded DNA template is excluded from the central channel of the CMG replication fork helicase.Citation53 Other models for replication fork helicase mechanism involve a ploughshare at the back of the helicase,Citation54 or an exit channel for single-stranded DNA extrusion through the CMG.Citation55 While details regarding CMG mechanism remain to be resolved, one theme in each of the models is for the separation of the 2 single DNA strands during helicase activation. Thus, during replication initiation, an ssDNA-binding platform is potentially generated for Sld2, Sld3, and Dpb11. Since the Sld2, Sld3, and Dpb11 proteins bind to one another in a CDK-dependent mechanism Citation23,24 and since Sld2, Sld3 or Dpb11 alone has considerable affinity for ssDNA,Citation48-50 the Sld2-Sld3-Dpb11 complex likely has a very tight affinity for ssDNA.
Recent data from our lab suggest that Sld2, Sld3, and Dpb11 attachment to ssDNA at an origin of replication is required for DNA replication.Citation48-50 In addition, we found that Sld2, Sld3, and Dpb11 interaction with ssDNA may be important for GINS attachment to Mcm2–7.Citation48-50 This conclusion follows from the observation that Sld2, Sld3, and Dpb11 compete with GINS for Mcm2–7 in a manner that is influenced by ssDNA addition.Citation48-50 In other words, Sld2, Sld3, and Dpb11 each interact with Mcm5 and Mcm3, the subunits of Mcm2–7 that contact GINS.Citation48-50 Furthermore, addition of Sld2, Sld3, and Dpb11 to Mcm2–7 can dislodge GINS from Mcm2–7.Citation48-50 However, single-stranded DNA also competes with Sld2, Sld3, and Dpb11, but not GINS, for Mcm3/Mcm5 interaction.Citation48-50 In addition, the addition of single-stranded DNA releases Sld2, Sld3, and Dpb11 from Mcm3 and Mcm5, allowing GINS to bind to Mcm2–7 by a passive sequestration mechanism.Citation48-50 There is also a recent manuscript from the Speck lab demonstrating that Sld2 and Sld3 exhibit an anti-cooperative mechanism for recruiting Cdc45 to loaded Mcm2–7 complexes.Citation56 In other words, Sld2 and Sld3 release from Mcm2–7 once Cdc45 is recruited to Mcm2–7.Citation56 Further details about the process of origin melting and helicase activation are not yet resolved, but the development of an in vitro replication initiation assay by the Diffley lab holds promise that many of these details will soon be revealed.Citation32
Dbf4, Sld2, Sld3, and Dpb11 are present in low amounts in the cell.Citation30 Thus, they can only bind to certain Mcm2–7 complexes, marking them for subsequent CMG helicase assembly and activation. But what is to prevent DDK from phosphorylating one Mcm2–7 complex, while Sld3, for example, binds to a different Mcm2–7 complex? A potential answer to this question has recently been revealed by work from our lab.Citation43 DDK phosphorylation of Mcm2 is a very weak reaction in vitro, and only a fraction of Mcm2 proteins are phosphorylated by DDK in a kinase assay using purified DDK and Mcm2.Citation40 In spite of this weak activity in vitro, we demonstrated an in vivo DNA replication function for DDK phosphorylation of Mcm2.Citation42,43 Thus, the data suggested that a critical factor was missing from our in vitro phosphorylation assay.Citation43,44
Is Sld3/Treslin a Conductor of Helicase Assembly and Activation?
We examined Sld2, Sld3, and Dpb11 for their effect on DDK phosphorylation of Mcm2 in vitro, and found that Sld3 exerts an 11-fold stimulation of DDK phosphorylation of Mcm2.Citation43 In contrast, the ability of Sld2 and Dpb11 to stimulate DDK phosphorylation of Mcm2 is very slight.Citation43 Furthermore, Treslin, the human ortholog of Sld3, stimulates human DDK phosphorylation of human Mcm2 by 15-fold.Citation43 Thus, the ability of Sld3/Treslin to substantially stimulate DDK phosphorylation of Mcm2 is conserved from yeast to human.Citation43
This observation may help explain the question asked above, what keeps Sld3 from binding to one Mcm2–7 complex, while DDK phosphorylates a different Mcm2–7 complex? The answer is surprisingly simple. Sld3/Treslin binds to Mcm2–7 and activates DDK phosphorylation of Mcm2 at the same time.Citation43,50 Thus, Sld3/Treslin is a conductor of replication initiation. Sld3/Treslin performs several functions upon binding to Mcm2-7: Citation1 Sld3/Treslin recruits Cdc45 to Mcm2–7 in a manner that depends upon DDK,Citation47,2 Sld3/Treslin stimulates DDK phosphorylation of the Mcm2 subunit of the Mcm2–7 complex,Citation43,3 Sld3/Treslin binds to ssDNA once the origin is melted, dissociating from Mcm2–7 and allowing GINS to bind Mcm2–7.Citation50 By performing these 3 biochemical reactions at a given Mcm2–7 complex, Sld3/Treslin coordinates helicase assembly with helicase activation.Citation43,47,50
The Role of Mcm10 in DNA Replication Initiation
Mcm10 is also required for the initiation of DNA replication.Citation57-59 Some early reports suggest that Mcm10 is involved in the recruitment of Cdc45 to Mcm2–7,Citation60-63 while recent reports suggest that Mcm10 is involved in a step following CMG assembly.Citation64-66 These recent reports suggest that Mcm10 plays a role in activating the fully-assembled CMG. It may be that both models are correct; in other words, Mcm10 may have multiple functions in replication initiation. Future work may help resolve this controversy. There are also reports concluding that Mcm10 is a component of the CMG during replication elongation, providing stability to the Pol α-primase.Citation67,68 Thus, Mcm10 is required for initiation, and it is also required for replication elongation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Funding was provided by Florida State University and the National Science Foundation, grant number 1265431 to D.L.K.
References
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem 2002; 71:333-74; PMID:12045100; http://dx.doi.org/10.1146/annurev.biochem.71.110601.135425
- Green B, Finn K, Li J. Loss of DNA replication control is a potent inducer of gene amplification. Science 2010; 329:943-46; PMID:20724634; http://dx.doi.org/10.1126/science.1190966
- Georgescu R, Schauer G, Yao N, Langston L, Yurieva O, Zhang D, Finkelstein J, O'Donnell, M. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. Elife 2015; 4:e04988; PMID:25871847; http://dx.doi.org/10.7554/eLife.04988
- Pursell Z, Isoz I, Lundström E, Johansson E, Kunkel T. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science 2007; 317:127-30; PMID:17615360; http://dx.doi.org/10.1126/science.1144067
- Johnson R, Klassen R, Prakash L, Prakash S. A Major Role of DNA Polymerase δ in Replication of Both the Leading and Lagging DNA Strands. Mol Cell 2015; 59:163-75; PMID:26145172; http://dx.doi.org/10.1016/j.molcel.2015.05.038
- Tognetti S, Riera A, Speck C. Switch on the engine: how the eukaryotic replicative helicase MCM2-7 becomes activated. Chromosoma 2015; 124:13-26; PMID:25308420; http://dx.doi.org/10.1007/s00412-014-0489-2
- Ilves I, Petojevic T, Pesavento J, Botchan M. Activation of the MCM2-7 Helicase by Association with Cdc45 and GINS Proteins. Mol Cell 2010; 37:247-58; PMID:20122406; http://dx.doi.org/10.1016/j.molcel.2009.12.030
- Bochman M, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell 2008; 31:287-93; PMID:18657510; http://dx.doi.org/10.1016/j.molcel.2008.05.020
- Fletcher R, Shen J, Gomez-Llorente Y, Martin CS, Carazo JM, Chen X. Double hexamer disruption and biochemical activities of Methanobacterium thermoautotrophicum MCM. J Biol Chem 2005; 280:42405-10; PMID:16221679; http://dx.doi.org/10.1074/jbc.M509773200
- Remus D, Beuron F, Tolun G, Griffith J, Morris E, Diffley J. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 2009; 139:719-730; PMID:19896182; http://dx.doi.org/10.1016/j.cell.2009.10.015
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A 2009; 106:20240-20245; PMID:19910535; http://dx.doi.org/10.1073/pnas.0911500106
- Ticau S, Friedman L, Ivica N, Gelles J, Bell S. Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell 2015; 161:513-525; PMID:25892223; http://dx.doi.org/10.1016/j.cell.2015.03.012
- Frigola J, Remus D, Mehanna A, Diffley J. ATPase-dependent quality control of DNA replication origin licensing. Nature 2013; 495:339-343; PMID:23474987; http://dx.doi.org/10.1038/nature11920
- Fernández-Cid A, Riera A, Tognetti S, Herrera M, Samel S, Evrin C, Winkler C, Gardenal E, Uhle S, Speck C. An ORC/Cdc6/MCM2-7 Complex Is Formed in a Multistep Reaction to Serve as a Platform for MCM Double-Hexamer Assembly. Mol Cell 2013; 50:577-588; http://dx.doi.org/10.1016/j.molcel.2013.03.026
- Diffley J. The many faces of redundancy in DNA replication control. Cold Spring Harb Symp Quant Biol 2010; 75:135-142; PMID:21502406; http://dx.doi.org/10.1101/sqb.2010.75.062
- Ge X, Jackson D, Blow J. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev 2007; 21:3331-3341; PMID:18079179; http://dx.doi.org/10.1101/gad.457807
- Kawabata T, Luebben S, Yamaguchi S, Ilves I, Matise I, Buske T, Botchan M, Shima N. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell 2011; 41:543-553; PMID:21362550; http://dx.doi.org/10.1016/j.molcel.2011.02.006
- Moyer S, Lewis P, Botchan M. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A 2006; 103:10236-10241; PMID:16798881; http://dx.doi.org/10.1073/pnas.0602400103
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, Deursen FV, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 2006; 8:358-366; PMID:16531994; http://dx.doi.org/10.1038/ncb1382
- Pacek M, Tutter A, Kubota Y, Takisawa H, Walter J. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 2006; 21:581-587; PMID:16483939; http://dx.doi.org/10.1016/j.molcel.2006.01.030
- Costa A, Renault L, Swuec P, Petojevic T, Pesavento J, Ilves I, MacLellan-Gibson K, Fleck R, Botchan M, Berger J. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. Elife 2014; 3 Aug 12:e03273
- Kanemaki M, Labib K. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J 2006; 25:1753-1763; PMID:16601689; http://dx.doi.org/10.1038/sj.emboj.7601063
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 2007; 445:281-285; PMID:17167417; http://dx.doi.org/10.1038/nature05432
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 2007; 445:328-332; PMID:17167415; http://dx.doi.org/10.1038/nature05465
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy W. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 2010; 140:349-359; PMID:20116089; http://dx.doi.org/10.1016/j.cell.2009.12.049
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy W. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J Cell Biol 2011; 193:995-1007; PMID:21646402; http://dx.doi.org/10.1083/jcb.201102003
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase α in the initiation of DNA replication. Mol Cell Biol 2006; 26:4843-4852; PMID:16782873; http://dx.doi.org/10.1128/MCB.02267-05
- Boos D, Sanchez-Pulido L, Rappas M, Pearl L, Oliver A, Ponting C, Diffley J. Regulation of DNA Replication through Sld3-Dpb11 Interaction Is Conserved from Yeast to Humans. Curr Biol 2011; 21:1152-1157; PMID:21700459; http://dx.doi.org/10.1016/j.cub.2011.05.057
- Itou H, Shirakihara Y, Araki H. The quaternary structure of the eukaryotic DNA replication proteins Sld7 and Sld3. Acta Crystallogr D Biol Crystallogr 2015; 71:1649-1656; PMID:26249346; http://dx.doi.org/10.1107/S1399004715010457
- Mantiero D, Mackenzie A, Donaldson A, Zegerman P. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J 2011; 30:4805-4814; PMID:22081107; http://dx.doi.org/10.1038/emboj.2011.404
- Zegerman P, Diffley JFX. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst) 2009; 8:1077-1088; PMID:19505853; http://dx.doi.org/10.1016/j.dnarep.2009.04.023
- Yeeles J, Deegan T, Janska A, Early A, Diffley J. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015; 519:431-435; PMID:25739503; http://dx.doi.org/10.1038/nature14285
- Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells?s Genes Dev 2010; 24:1208-1219; PMID:20551170; http://dx.doi.org/10.1101/gad.1933010
- Randell J, Fan A, Chan C, Francis L, Heller R, Galani K, Bell S. Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol Cell 2010; 40:353-363; PMID:21070963; http://dx.doi.org/10.1016/j.molcel.2010.10.017
- Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, Kim J, Ishii A, Tanaka T, Kobayashi T, et al. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem 2006; 281:39249-39261; PMID:17046832; http://dx.doi.org/10.1074/jbc.M608935200
- Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docing site-mediated mechanism to promote S phase progression. Mol Cell 2006; 24:101-113; PMID:17018296; http://dx.doi.org/10.1016/j.molcel.2006.07.033
- Sheu Y, Kinney J, Lengronne A, Pasero P, Stillman B. Domain within the helicase subunit Mcm4 integrates multiple kinase signals to control DNA replication initiation and fork progression. Proc Natl Acad Sci U S A 2014; 111:E1899-1908; PMID:24740181; http://dx.doi.org/10.1073/pnas.1404063111
- Sheu Y, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 2010; 463:113-117; PMID:20054399; http://dx.doi.org/10.1038/nature08647
- Lei M, Kawasaki Y, Young M, Kihara M, Sugino A, Tye B. MCM2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev 1997; 11:3365-3374; PMID:9407029; http://dx.doi.org/10.1101/gad.11.24.3365
- Bruck I, Kaplan D. Dbf4-Cdc7 phosphorylation of Mcm2 is required for cell growth. J Biol Chem 2009; 284:28823-28831; PMID:19692334; http://dx.doi.org/10.1074/jbc.M109.039123
- Stead B, Brandl C, Davey M. Phosphorylation of Mcm2 modulates Mcm2-7 activity and affects the cell's response to DNA damage. Nucleic Acids Res 2011; 39:6998-7008; PMID:21596784; http://dx.doi.org/10.1093/nar/gkr371
- Bruck I, Kaplan DL. The Dbf4-Cdc7 kinase promotes Mcm2-7 ring opening to allow for single-stranded DNA extrusion and helicase assembly. J Biol Chem 2015; 290:1210-1221; PMID:25471369; http://dx.doi.org/10.1074/jbc.M114.608232
- Bruck I, Kaplan D. Conserved mechanism for coordinating replication fork helicase assembly with phosphorylation of the helicase. Proc Natl Acad Sci U S A 2015; 112:11223-11228; PMID:26305950; http://dx.doi.org/10.1073/pnas.1509608112
- Samel S, Fernández-Cid A, Sun J, Riera A, Tognetti S, Herrera M, Li H, Speck C. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev 2014; 28:1653-1666; PMID:25085418; http://dx.doi.org/10.1101/gad.242404.114
- Hardy CFJ, Dryga O, Seematter S, Pahl PMB, Sclafani RA. mcm5/cdc46-bob1 bypasses the requirement for the S phase activatorCdc7p. Proc Natl Acad Sci U S A 1997; 94:3151-3155; PMID:9096361; http://dx.doi.org/10.1073/pnas.94.7.3151
- Itou H, Muramatsu S, Shirakihara Y, Araki H. Crystal structure of the homology domain of the eukaryotic DNA replication proteins Sld3 Treslin Structure 2014; 22:1341-1347; PMID:25126958; http://dx.doi.org/10.1016/j.str.2014.07.001
- Kamimura Y, Tak YS, Sugino A, Araki H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J 2001; 20:2097-2107; PMID:11296242; http://dx.doi.org/10.1093/emboj/20.8.2097
- Dhingra N, Bruck I, Smith S, Ning B, Kaplan D. Dpb11 helps control assembly of the Cdc45-Mcm2-7-GINS replication fork helicase. J Biol Chem 2015; 290:7586-7601; PMID:25659432; http://dx.doi.org/10.1074/jbc.M115.640383
- Bruck I, Kaplan D. The replication initiation protein sld2 regulates helicase assembly. J Biol Chem 2014; 289:1948-1959; PMID:24307213; http://dx.doi.org/10.1074/jbc.M113.532085
- Bruck I, Kaplan DL. The replication initiation protein Sld3/Treslin orchestrates the assembly of the replication fork helicase during S phase. J Biol Chem 2015; 290(45):27414-24; PMID: 26405041; http://dx.doi.org/10.1074/jbc.M115.688424
- Ohlenschläger O, Kuhnert A, Schneider A, Haumann S, Bellstedt P, Keller H, Saluz HP, Hortschansky P, Hänel F, Grosse F, et al. The N-terminus of the human RecQL4 helicase is a homeodomain-like DNA interaction motif. Nucleic Acids Res 2012; 40:8309-8324; http://dx.doi.org/10.1093/nar/gks591
- Geraghty D, Ding M, Heintz N, Pederson D. Premature structural changes at replication origins in a yeast minichromosome maintenance (MCM) mutant. J Biol Chem 2000; 275:18011-18021; PMID:10751424; http://dx.doi.org/10.1074/jbc.M909787199
- Fu Y, Yardimci H, Long D, Ho T, Guainazzi A, Bermudez V, Hurwitz J, van Oijen A, Schärer O, Walter J. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 2011; 146:931-941; PMID:21925316; http://dx.doi.org/10.1016/j.cell.2011.07.045
- Takahashi TS, Wigley DB, Walter JC. Pumps, paradoxes, and ploughshares: mechanism of the Mcm2-7 DNA helicase. Trends Biochem Sci 2005; 30:437-444; PMID:16002295; http://dx.doi.org/10.1016/j.tibs.2005.06.007
- Gai D, Chang Y, Chen X. Origin DNA melting and unwinding in DNA replication. Curr Opin Struct Biol 2010; 20:756-762; PMID:20870402; http://dx.doi.org/10.1016/j.sbi.2010.08.009
- Herrera, MC, Tognetti, S, Riera, A, Zech, J, Clarke P, Fernández-Cid A, Speck C. A reconstituted system reveals how activating and inhibitory interactions control DDK dependent assembly of the eukaryotic replicative helicase. Nucleic Acids Res 2015; 43(21):10238-10250; PMID: 26338774; http://dx.doi.org/10.1093/nar/gkv881
- Solomon N, Wright M, Chang S, Buckley A, Dumas L, Gaber R. Genetic and molecular analysis of DNA43 and DNA52: two new cell-cycle genes in Saccharomyces cerevisiae. Yeast 1992; 8:273-289; PMID:1514326; http://dx.doi.org/10.1002/yea.320080405
- Merchant A, Kawasaki Y, Chen Y, Lei M, Tye BK. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in S. cerevisiae. Mol Cell Biol 1997; 17:3261-3271; PMID:9154825; http://dx.doi.org/10.1128/MCB.17.6.3261
- Thu Y, Bielinsky A. Enigmatic rles of Mcm10 in DNA replication. Trends Biochem Sci 2013; 38:184-194; PMID:23332289; http://dx.doi.org/10.1016/j.tibs.2012.12.003
- Wohlschlegel J, Dhar S, Prokhorova T, Dutta, A, Walter J. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol Cell 2002; 9:233-240; PMID:11864598; http://dx.doi.org/10.1016/S1097-2765(02)00456-2
- Gregan J, Lindner K, Brimage L, Franklin R, Namdar M, Hart E, Aves S, Kearsey S. Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding. Mol Biol Cell 2003; 14:3876-3887; PMID:12972571; http://dx.doi.org/10.1091/mbc.E03-02-0090
- Sawyer S, Cheng I, Chai W, Tye B. Mcm10 and Cdc45 cooperate in origin activation in Saccharomyces cerevisiae. J Mol Biol 2004; 340:195-202; PMID:15201046; http://dx.doi.org/10.1016/j.jmb.2004.04.066
- Im JS, Ki SH, Farina A, Jung DS, Hurwitz J, Leea JK. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc Natl Acad Sci U S A 2009; 106:15628-15632; PMID:19805216; http://dx.doi.org/10.1073/pnas.0908039106
- Watase G, Takisawa H, Kanemaki M. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr Biol 2012; 22:343-349; PMID:22285032; http://dx.doi.org/10.1016/j.cub.2012.01.023
- van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J 2012; 31:2195-2206; PMID:22433841; http://dx.doi.org/10.1038/emboj.2012.69
- Kanke M, Kodama Y, Takahashi T, Nakagawa T, Masukata H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J 2012; 31:2182-2194; PMID:22433840; http://dx.doi.org/10.1038/emboj.2012.68
- Ricke R, Bielinsky A. A conserved Hsp10-like domain in Mcm10 is required to stabilize the catalytic subunit of DNA polymerase-α in budding yeast. J Biol Chem 2006; 281:18414-18425; PMID:16675460; http://dx.doi.org/10.1074/jbc.M513551200
- Ricke RM, Bielinsky AK. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol Cell 2004; 16:173-185; PMID:15494305; http://dx.doi.org/10.1016/j.molcel.2004.09.017