ABSTRACT
Background and Objective
The potential of probiotics on the prevention and control of periodontitis and other chronic inflammatory conditions has been suggested. Lactobacillus and Bifidobacterium species influence P. gingivalis interaction with gingival epithelial cells (GECs) but may not act in a unique way. In order to select the most appropriate probiotic against P. gingivalis, we aimed to evaluate the effect of several strains on Porphyromonas gingivalis biofilm formation and transcription virulence-associated factors (PgVAFs).
Methods
Cell-free pH neutralized supernatants (CFS) and living Lactobacillus spp. and Bifidobacterium spp. were tested against P. gingivalis ATCC 33277 and W83, in mono- and multi-species (with Streptococcus oralis and S. gordonii) biofilms. Relative transcription of P. gingivalis genes (fimA, mfa1, kgp, rgp, ftsH and luxS) was determined in biofilms and under GECs co-infection.
Results
Probiotics CFS reduced P. gingivalis ATCC 33277 levels in mono-species biofilms and living probiotics reduced P. gingivalis abundance in multi-species biofilms. L. acidophilus LA5 down-regulated transcription of most PgVAFs in biofilms and GECs.
Conclusions
Probiotics affect P. gingivalis biofilm formation by down-regulating overall PgVAFs with the most pronounced effect observed for L. acidophilus LA5.
Introduction
Periodontitis is a chronic inflammatory disease in response to a polymicrobial dysbiotic biofilm that affects supporting periodontal tissues surrounding the teeth and leads to alveolar bone resorption [Citation1]. Porphyromonas gingivalis is considered a keystone pathogen in periodontitis due to its properties to trigger an exaggerated pro-inflammatory response that dictates the periodontal destruction, and to evade host defense mechanisms [Citation2]. At the same time, P. gingivalis drives a shift in the microbial composition towards a dysbiotic-related state [Citation3].
The current conventional periodontal treatment is based on mechanical debridement of the biofilm plus the use of antimicrobial drugs, aiming to alter the microbial community composition of oral sites, thus leading to control the inflammatory response [Citation4]. However, recent evidences [Citation5] highlight the limitations of this approach and shed light on the search for new therapies. Thus, a proposal to guide periodontal pocket recolonization by the use of probiotics has been raised as a promising strategy [Citation6], as suggested by experimental and clinical studies [Citation5–Citation8]. Meanwhile, despite the evidences showing the benefits of probiotics on the control of the dysbiotic resident microbiota [Citation7,Citation8], little is known on their effects on periodontopathogens.
We have previously shown that probiotic strains of the genera Lactobacillus and Bifidobacterium promote a reduction in P. gingivalis adhesion to and invasion of gingival epithelial cells, and alter the cell response to the pathogen [Citation9]. These beneficial microorganisms may also directly exert an effect on certain pathogens by attenuating their virulence components [Citation10], such as by impairing toxins production [Citation11]. Regarding oral pathogens, lactobacilli may attenuate the virulence of Candida albicans by inhibiting yeast-hypha differentiation [Citation12], and of Aggregatibacter actinomycetemcomitans by down-regulating the expression of exotoxins [Citation13]. Nonetheless, little is known about the effects of interactions between probiotics and the keystone pathogen P. gingivalis.
A successful colonization of the oral cavity by P. gingivalis depends on its ability to interact with early colonizers in the dental biofilm, e.g. Streptococcus oralis and S. gordonii [Citation14,Citation15], to adhere to and invade the epithelial barrier [Citation15–Citation17], and to evade host immunity [Citation18]. Despite intra-species diversity, P. gingivalis pathogenic potential is associated with factors such as the expression of capsule, the main (FIMA) and minor (MFA1) fimbriae [Citation19], lysine-(KGP) and arginine-(RGP) gingipains [Citation20], quorum sensing components (LUXS) [Citation21], and metallopeptidases (FTSH) [Citation22].
Thus, we aimed to investigate whether Lactobacillus and Bifidobacterium probiotics would interfere in P. gingivalis biofilm formation, by using mono- and multi-species in vitro models. Since microbial interactions exert a profound effect on gene expression in a strain-specific fashion [Citation23], we also evaluated the effects of probiotics on the transcription profiles of two P. gingivalis strains in biofilms and after gingival epithelial cells co-infection.
Material and methods
Strains and culture conditions
Six probiotic strains were tested [Lactobacillus acidophilus LA-5™ (CHR Hansen Holding A/S, Hørsholm, Denmark), L. rhamnosus HN001 Howaru™ (Danisco, Madison, WI, USA), L. reuteri DSM 17938 (BioGaia AB, Lund, Sweden), Bifidobacterium breve 1101A, B. pseudolongum 1191A and B. bifidum 1622A (isolated from feces of healthy children, obtained at the Federal University of Minas Gerais) [Citation24]. P. gingivalis (W83 and ATCC 33277) were used as periodontopathogens. S. oralis ATCC10557 and S. gordonii DL1 [Citation25] were used in multi-species biofilm assays.
Bacteria were cultivated from frozen stocks at −80°C. Lactobacilli were grown on Lactobacilli MRS agar (Difco Laboratories, Detroit, MI, USA) and streptococci were cultivated on Tryptic Soy agar [TSA] (Difco Laboratories), both under microaerophilic conditions at 10% CO2, 37°C. Bifidobacteria were grown on BSM agar (Bifidus Selective Medium, Sigma-Aldrich, St. Louis, MO, USA) at 37°C, under anaerobic conditions (90% N2, 5% CO2 and 5% H2) in an anaerobe chamber (Plas-Labs Model 855, Lansing, MI, USA). P. gingivalis were also grown under anaerobic conditions (90% N2, 5% CO2 and 5% H2) at 37°C on blood agar plates [TSA (Difco Laboratories) enriched with 5% defibrinated sheep blood, 0.5 mg/mL hemin (Sigma-Aldrich) and 1 mg/mL menadione (Sigma-Aldrich)].
Cell-free pH neutralized supernatants of probiotic cultures
After growth overnight in Brain-Heart Infusion broth (Neogen, Lansing, MI, USA) supplemented with 5 mg/mL hemin and 10 mg/mL menadione (BHIHM), probiotics cultures were centrifuged at 8,000xg for 10 min, and cells resuspended in BHIHM to an OD600nm equivalent to 108 colony forming units (CFU)/mL, diluted 1:10 and incubated under agitation (80 rpm) for 24 h under anaerobiosis. Then, bacteria were removed by centrifugation and the pH of supernatants adjusted to 7.0 with 0.1 M NaOH. Cell-free pH neutralized supernatants (CFS) were sterilized by filtration (0.22 μm pore, Life Sciences, Ann Arbor, MI, USA) and used in biofilm assays.
Effect of CFS and living cells on biofilm formation
Biofilms were formed in polystyrene 96 wells/plates (Corning Incorporated Costar®, Kennebunk, ME, USA). For mono-species biofilm, aliquots of overnight grown culture in BHIHM of each P. gingivalis strain (1 × 107 CFU/well) were inoculated to a total volume of 100 μL. For multi-species biofilm, P. gingivalis, S. oralis and S. gordonii were inoculated at 1 × 107 CFU/well of each species. In order to evaluate the effect of probiotics CFS on biofilm formation, a final dilution of 1:2.5 of CFS of each probiotic strain was added to BHIHM. In order to evaluate the effect of living probiotics cells on biofilm formation, each probiotic strain was inoculated at 1 × 107 CFU/well. Negative control consisted of non-inoculated medium, and positive controls consisted of mono- or multi-species biofilm without CFS or without living probiotics.
Plates were incubated for 24 h under agitation (80 rpm) on a shaking platform (Sunflower Mini-Shaker, Biosan, Riga, Latvia) in anaerobiosis (90% N2, 5% CO2 and 5% H2) at 37°C using an anaerobe chamber (Plas-Labs). Biofilm biomass was estimated as previously reported [Citation26]. Briefly, non-adherent bacteria were removed by washing with 1 X Phosphate Buffered Saline (PBS, pH 7.4), and biofilms were stained with 0.4% safranin for 15 min. Excess of dye was removed by washing with distilled water, followed by dye extraction with 95% ethanol for 15 min, and absorbance measured at 490 nm, which represented the biomass amount. All assays were performed in triplicate and independently repeated at least twice.
Quantification of P. gingivalis, S. oralis, S. gordonii, Lactobacillus spp. and Bifidobacterium spp. by qPCR
DNA in biofilms was extracted using Master PureTM DNA Purification Kit (Epicentre, Madison, WI, USA), and treated with RNAse. Amplification of P. gingivalis, S. oralis, S. gordonii, lactobacilli, and bifidobacteria 16SrRNA was performed with 0.5 µL of each primer (25 pmol) (Table 1), 10 µL of Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), 1 µL DNA (10 ng/µL), in a final volume of 20 µL and qPCR was performed by StepOne Plus (Applied Biosystems). Temperature profiles consisted of denaturation (95°C for 10 min), amplification and quantification (40 cycles of 95°C for 15 s and 60°C for 1 min) and melting curve (95°C for 15 s, 60°C for 1 min and heating rate of 0.3°C-95°C for 15 s). For absolute quantification and estimation of PCR efficiency, a standard curve was generated using 16SrRNA amplicons. Reactions with standards and tested DNA were performed in duplicate. Ct values were correlated with 16SrRNA copy numbers, and data expressed as the number of each species/well, considering four copies of 16SrRNA gene per chromosome [Citation27].
Interaction of P. gingivalis and probiotics with gingival epithelial cells
The assays were performed according to our previous work [Citation9]. Briefly, immortalized human gingival epithelial cells (OBA-9/GECs) were cultured in Keratinocyte-Serum Free Medium (KSFM) (GIBCO™, Life Technologies, Carlsbad, CA, USA) supplied with human recombinant epidermal growth factor. GECs were seeded in 24-well culture plates at a cell density of 2.0 × 105 cells per well, in KSFM without antibiotic. After 24 h, GECs were challenged with P. gingivalis strains and/or probiotics at a multiplicity of infection (MOI) of 1:1,000. After 2 h incubation, unattached bacteria were removed by washing, and GECs were lysed followed by RNA extraction.
Effect of probiotics on gene expression of Porphyromonas gingivalis VAFs
Total RNA in adherent biofilms and co-infected epithelial cells were extracted using RNeasy KIT (QIAGEN, Valencia, CA, USA). The quality and concentration of the extracted RNA were determined by measurement of absorbance at 260 and 280 nm in a NanoDrop™ One Spectrophotometer (Thermo Scientific, Waltham, MA, USA). After RNase-free DNase I treatment (Invitrogen Life Technologies), cDNA was synthesized, using 5X VILOTM and 10X SuperScriptTM Enzyme (Invitrogen Life Technologies, Waltham, MA, USA) in a GeneAmp PCR System 2400 thermocycler (Applied Biosystems®) set at 25°C for 10 min, 42°C for 60 min and 85°C for 5 min. Control reactions using no enzyme were performed. Gene expression was evaluated with the primers listed in . Reactions consisted of 0.5 µL of each primer (25 pmol), 10 µL of Power SYBR Green PCR Master Mix (Applied Biosystems®), 1 µL of cDNA at a concentration of 40 ng/µL, completed to a final volume of 20 µL. The RT-qPCR reaction consisted of an activation step of denaturation program (95°C for 10 min), and 40 cycles (95°C for 15 s, 60°C for 1 min with a single fluorescence measurement), melting curve program (95°C for 15 s, 60°C for 1 min, with a heating rate of 0.3°C-95°C for 15 s and a continuous fluorescence measurement) in a StepOne Plus thermocycler (Applied Biosystems®). The efficiency for both the internal control (16SrRNA) and gene of interest (GOI) was determined using P. gingivalis ATCC 33277 and W83 cDNA templates dilution series as standard curve. Comparative quantification was determined by calculating the Ct difference between the target gene in the test and calibrator samples (assays without probiotics CFS or living cells), normalized to the reference gene Ct values and adjusted for variations in amplification efficiency [Citation28].
Table 1. Oligonucleotides primers for real-time PCR assays used for bacteria quantification in biofilms and for transcription analysis of P. gingivalis virulence-associated genes.
Statistical analysis
Data were expressed as mean ± standard deviation (SD) from three independent experiments. Statistical analyses were performed using GraphPad Prism version 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). One-way ANOVA with post hoc Tukey’s test was used in all analyses, and a significance level of 0.05 was established.
Results
Probiotic CFS reduced biomass of mono- and multi-species P. gingivalis biofilms
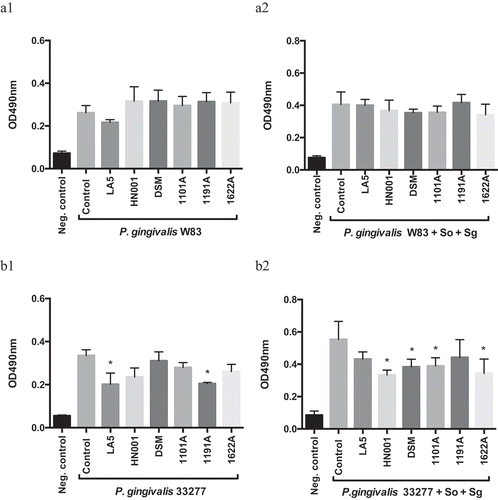
Some probiotic CFS reduced mono- and multi-species biofilm biomass of P. gingivalis ATCC 33277, but not of strain W83 (). However, this reduction was not only dependent on the P. gingivalis and probiotic strain, but on the environmental condition (mono- or multi-species biofilm).
Figure 1. Effect of probiotics cell-free supernatants (CFS) diluted at 1:2.5 on P. gingivalis W83 (a1, mono-species; a2, multi-species) and ATCC 33277 (b1, mono-species; b2, multi-species) biofilm biomass, represented by the OD490nm of the standard biofilm dye. Groups: Neg. control- Negative control represents non-inoculated medium; Control- CFS-free positive controls of P. gingivalis mono- and multi-species biofilms (So – S. oralis and Sg – S. gordonii), and experimental group with CFS of: LA5 – L. acidophilus LA5, HN001 – L. rhamnosus HN001, DSM – L. reuteri DSM 17938, 1101A – B. breve 1101A, 1191A – B. pseudolongum 1191A and 1622A – B. bifidum 1622A. Experiments were conducted in triplicate. (*) Statistically significant difference when compared to respective positive controls using One-way ANOVA with post hoc Tukey’s multiple comparisons (p < 0.05).
Probiotic CFS altered P. gingivalis proportion in multi-species biofilms
There were no differences in P. gingivalis cells number in mono-species biofilms exposed to probiotics CFS (data not shown). Data on absolute number of each bacteria strain/well are shown in supplemental material (Figure S1). Probiotic CFS have altered the relative abundance (%) of P. gingivalis in multi-species biofilms in a probiotic/P. gingivalis strain-specific manner (). CFS of L. acidophilus LA5, L. rhamnosus HN001, L. reuteri DSM, B. breve 1101A, and B. pseudolongum 1191A, but not of B. bifidum 1622A decreased the abundance of P. gingivalis ATCC 33277. On the other hand, CFS of all tested bifidobacteria, B. breve 1101A, B. pseudolongum 1191A and B. bifidum 1622A, but not the lactobacilli, increased P. gingivalis W83 abundance in multi-species biofilms (, and Figure S2 supplemental material, p < 0.05). The relative abundances of S. oralis and S. gordonii were higher in most groups treated with probiotics CFS in relation to controls in ATCC 33277 multispecies biofilms (p < 00.5) and similar to control in W83 multispecies biofilms (Figure S2 in supplemental material).
Figure 2. Effect of probiotics cell-free supernatants (CFS of LA5 – L. acidophilus LA5, HN001 – L. rhamnosus HN001, DSM – L. reuteri DSM 17938, 1101A – B. breve 1101A, 1191A – B. pseudolongum 1191A and 1622A – B. bifidum 1622A) on the relative abundance of P. gingivalis W83 (a) or ATCC 33277 (b) and the initial colonizers S. oralis and S. gordonii in multi-species biofilms, represented as the mean percentage of each bacteria determined by qPCR. (*) Significant difference in P. gingivalis counts when compared to respective positive controls using One-way ANOVA with post hoc Tukey’s multiple comparisons (p < 0.05).
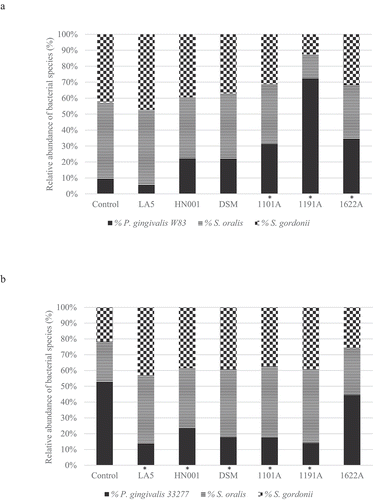
Figure 3. Effect of living Lactobacillus sp (L. acidophilus LA5, L. rhamnosus HN001, L. reuteri DSM 17938) and Bifidobacterium sp (B. breve 1101A, B. pseudolongum 1191A and B. bifidum 1622A) on the relative abundance of P. gingivalis W83 (a) or ATCC 33277 (b) and the initial colonizers S. oralis and S. gordonii after cell-to-cell interaction, represented as the mean percentage of each bacteria determined by qPCR (*). Significant difference in P. gingivalis abundance, when compared to respective positive controls using One-way ANOVA with post hoc Tukey’s multiple comparisons (p <0.05).
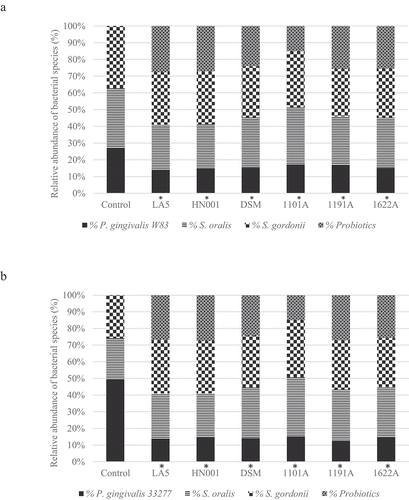
Living probiotic bacteria reduced P. gingivalis abundance in multi-species biofilms
Probiotic bacteria adhered to biofilm biomass and their abundance in the multi-species biofilms are shown in . Data on the number of each strain/well are shown in Figure S3 (supplemental material). Probiotics living cells induced a decrease in P. gingivalis abundance (, p < 0.05), which was more evident for ATCC 33277 than for W83 (Figure S4, supplemental material). Moreover, most of the living probiotics increased the relative abundance of commensal S. oralis and S. gordonii) in ATCC 33277 multi-species biofilms and promoted a slight, but significant, decrease in abundance of these commensals in W83 multi-species biofilms ( and Figure S4 of supplemental material).
Probiotic CFS alter the expression of P. gingivalis VAFs in mono- or multi-species biofilms
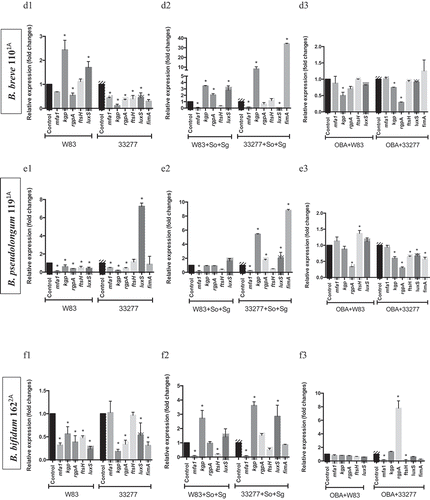
Since some probiotic CFS influenced biofilm biomass of P. gingivalis ATCC 33277 but not of W83, and altered their abundances in multi-species biofilms, we determined P. gingivalis transcription profiles of key VAFs under probiotics CFS. The effect of each CFS on P. gingivalis biofilms varied according to the environmental condition, mono- or multi-species biofilm, and VAFs transcription profiles were dependent on the environment. Changes in gene expression were not only related to the studied probiotic species but also dependent on the target P. gingivalis strain. Probiotics CFS altered the transcription of genes encoding fimbriae (mfa1 a fimA), proteases (fsH, kgp, and rgpA) and quorum sensing signaling molecules (luxS) (). Notably, L. acidophilus LA5 CFS down-regulated expression of: mfa1, in W83 and ATCC 33277 mono-species biofilms () and multi-species biofilms (); fimA, in ATCC 33277 (mono-and multi-species); fsH, in W83 and ATCC 33277 (mono- and multi-species); kgp, in W83 (mono-species) and ATCC 33277 (mono- and multi-species); rgp, in W83 and ATCC 33277 (mono-species); and luxS, in W83 (mono-species) and ATCC 33277 (mono- and multi-species) ().
Figure 4. Effect of probiotics on the relative transcription of P. gingivalis encoding virulence genes (mfa1 – minor fimbriae; fimA – major fimbriae; kgp – lysine gingipain; rgpA – arginine gingipain; ftsH – metalloproteinase, and luxS – quorum sensing components), determined by RT-qPCR. P. gingivalis strains W83 and ATCC 33277 biofilms formed in BHIHM broth added with the supernatant of cultures of probiotics (a1 and a2 - L. acidophilus LA5, b1 and b2 – L. rhamnosus HN001, c1 and c2 – L. reuteri DSM 17938, d1 and d2 – B. breve 1101A, e1 and e2 – B. pseudolongum 1191A and f1 and f2 – B. bifidum 1622A) diluted to 1:2.5, in mono-species (a1, b1, c1, d1, 1 and f1) and multi-species (a2, b2, c2, d2, e2 and f2), and infecting OBA-9 GECs with probiotic co-infection at a MOI of 1:1,000 (a3 - L. acidophilus LA5, b3 – L. rhamnosus HN001, c3 – L. reuteri DSM 17938, d3 – B. breve 1101A, e3 – B. pseudolongum 1191A and f3 – B. bifidum 1622A). Data are expressed as fold changes in relation to positive control conditions (biofilms without probiotic cell-free supernatants or GECs without probiotic bacteria), after normalization to the endogenous control gene 16SrRNA. (*) Significant difference when compared to respective positive controls using One-way ANOVA with post hoc Tukey’s multiple comparisons (p < 0.05).
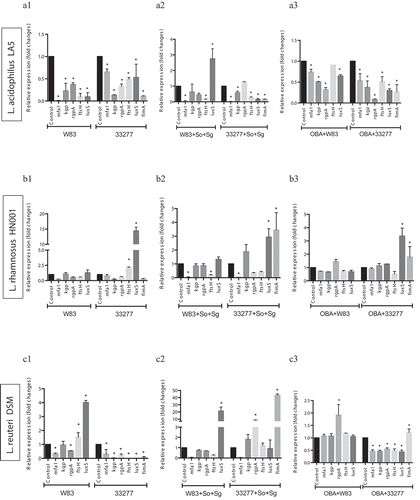
Living probiotics alter the expression of P. gingivalis VAFs under gingival epithelial cell co-infection
Our previous study [Citation9] reported that living Lactobacillus and Bifidobacterium reduced P. gingivalis ATCC 33277 and W83 adhesion and invasion of GECs, except for Lactobacillus which did not reduce invasion by strain W83. Moreover, these probiotics decreased pro-inflammatory cytokines synthesis induced by P. gingivalis. Data on the expression analysis of P. gingivalis VAF genes in co-infection with probiotic bacteria in GECs revealed that most studied genes were regulated by at least one probiotic, and the transcription profiles were probiotic-strain and P. gingivalis-strain specific (). L acidophilus LA5 showed prominent activity on VAF transcription, including the downregulation of the fimbriae encoding genes mfa1, in both strains of P. gingivalis, and fimA, in ATCC 33277 (); gingipains encoding genes kgp, rgpA, and the quorum sensing luxS in ATCC 33277 or W83 interacting with GECs ().
Discussion
An ideal probiotic to control periodontitis should keep the balance between the oral biofilm and the host by controlling pathogens colonization and modulating the inflammatory response. On this basis, we have recently shown that the studied probiotics were able to reduce P. gingivalis ATCC 33277 and W83 adherence to and invasion of GECs as well as to modulate the epithelial cell immune response against this periodontopathogen [Citation9]. Altogether, our present and previous data indicated that soluble by-products and living probiotic strains reduced P. gingivalis abundance in multi-species biofilms as well as its interaction with GECs. We have also shown that the probiotics can alter the transcription profile of P. gingivalis virulence-associated factors, thus interfering in its ability to colonize the host and subvert immune response.
Probiotics CFS showed little or no effect on the biomass of P. gingivalis mono-species biofilms. However, when tested on multi-species biofilms, which simulates a more realistic scenario in the oral cavity, some probiotics living cells and/or their soluble released products in the culture medium (CFS) reduced the abundance of P. gingivalis without little effect on the relative abundance of early colonizers, such as S. oralis and S. gordonii. Dysbiosis rate is referred to as the ratio between the relative abundances of disease-associated species to that of health-associated species [Citation29]. Hence, the selective effect of probiotics on the pathogen suggests that probiotics would reduce the dysbiosis in the oral biofilms. Furthermore, our results raise the hypothesis that P. gingivalis co-aggregation mechanisms should be disturbed by certain probiotics, since streptococci comprise 70% of the initial colonizers that interact with proteins/receptors on the acquired pellicle [Citation30], and the ability of P. gingivalis to form biofilms is dependent on co-aggregation with such bacteria [Citation31].
Nevertheless, P. gingivalis presents a high diversity as indicated by their different virulent surface components [Citation32]. P. gingivalis W83 is a capsulated/afimbriated highly virulent strain, whereas ATCC 33277 is a no capsulated/fimbriated less virulent strain [Citation32,Citation33]. These P. gingivalis strains also differ in relation to biofilm formation [Citation34] and chromosomal transposable elements, which may alter their gene transcription [Citation35], and physiology, such as their capacity to survive under oxidative stress [Citation36], which altogether can contribute to explain differences on biofilm composition and transcription of virulence factors induced by probiotics.
Interestingly, gene regulation of the two P. gingivalis strains promoted by the probiotics differed under different environmental conditions such as in mono-species and in multi-species biofilms. Recent data reported that P. gingivalis W83 causes a more severe dysbiosis than P. gingivalis ATCC 33277 in a multispecies biofilm model [Citation23]. On the other hand, transcriptome analysis reported that each strain differed in their ability to modulate the expression S. mitis genes [Citation23]. Our data contribute to this discussion, showing that products released by the commensal B. pseudolongum 1191A may reduce the dysbiosis promoted by strain ATCC 33277 but not by W83. Furthermore, not only P. gingivalis alters the transcription profile of commensals as shown previously [Citation23] but also the ability of commensal lactobacilli and bifidobacteria to alter the transcription profile of P. gingivalis was also dependent on the pathogen strain.
Our data have also indicated that lactobacilli and bifidobacteria may participate in in vitro multi-species biofilms with P. gingivalis and streptococci, and most inhibitory effects were obtained with living cells-to-cell interaction. Previous study, also adopting a multi-species biofilms model, reported that bifidobacteria induced a reduction in P. gingivalis levels [Citation37]. Lactobacillus spp. and Bifidobacterium spp. are part of the normal resident microbiota, and some strains of these genera such as L. acidophilus, L. rhamnosus and B. longum produce exopolysaccharides (EPS) that contribute to biofilm formation [Citation38]. Thus, our data suggest that interactions between probiotics surface components such as extracellular polysaccharides and surface-layer proteins with commensal bacteria [Citation39,Citation40] may compete with P. gingivalis W83 adhesion/aggregation mechanisms.
Virulence of P. gingivalis is mediated by an array of factors involved in several functions, such as attachment to host surfaces and other oral microorganisms, acquisition of nutrients, induction of a destructive inflammatory response and evasion of host response. Probiotics were not only able to decrease the abundance of P. gingivalis in multispecies biofilms but were also able to inhibit adhesion and invasion of P. gingivalis to GECs [Citation9]. Once again, these activities were dependent on the P. gingivalis strain, since the tested lactobacilli were able to inhibit P. gingivalis W83 adhesion to but not the invasion of GECs, whereas both adhesion and invasion of ATCC 33277 were inhibited.
In order to shed some light on the inhibitory mechanisms of probiotics, we have shown that these beneficial bacteria were able to alter the transcription of genes associated with several virulence-associated factors. Lactobacilli and bifidobacteria overall down-regulated the expression of fimA, which encodes the main fimbriae FIMA in P. gingivalis ATCC 33277. These fimbriae provide pathogen binding to saliva and serum components, and to extra-cellular matrix proteins and epithelial cells [Citation41,Citation42]. It also promotes auto-aggregation and co-aggregation to other bacterial species, including S. gordonii [Citation43] and reasonably its down-regulation can explain the reduction in P. gingivalis abundance in multi-species biofilm. Moreover, FIMA is associated with pathogen adhesion/invasion to epithelial cells [Citation44] and it is recognition by host cells through TLR2 and TLR4 receptors with downstream activation of pro-inflammatory cytokines [Citation45]. Thus, fimA down-regulation may explain our previous study that found a modulation of the immune response triggered in gingival epithelial cells followed by a reduction in the adhesion of P. gingivalis ATCC 33277 [Citation9]. Curiously, all tested probiotics supernatants were able to down-regulate the expression of mfa1 in multi-species biofilm assays. The minor fimbriae MFA1 is also involved in P. gingivalis auto-aggregation [Citation46] and co-aggregation with S. gordonii [Citation47]. Thus, a reduction in mfa1 transcription promoted by probiotic secretome would impair P. gingivalis interaction with streptococci.
Probiotics and their by-products were also able to regulate transcription of proteases encoding genes. FstH is an integral membrane zinc metallopeptidase that participates in a network that restrains biofilm accumulation in heterotypic P. gingivalis ATCC 33277-S. gordonii biofilms [Citation48]. Thus, alteration in this regulatory mechanism promoted by probiotics would alter P. gingivalis interaction with S. gordonii in multi-species biofilms.
Our data have also revealed that some probiotics down-regulated the transcription of kgp and rgp that encode lysine- and arginine-gingipains, respectively, in W83 and ATCC 33277 strains. Lysine-gingipain (Kgp) degrades tissue matrix and proteins that contain iron and hemin [Citation21,Citation49], and arginine-gingipain (Rgp, cysteine protease) regulates exopolysaccharide accumulation, and promotes hemagglutination and maturation of several P. gingivalis surface proteins such as fimbrilin of FIMA [Citation14,Citation20]. Negative regulation of kgp and rgpA transcription in W83 was achieved in most conditions by L. acidophilus LA5, whereas other probiotics even up-regulated transcription of these proteases encoding-genes. In fact, a proteomics of L. acidophilus revealed that an increased cysteine synthase activity may accumulate a cysteine pool relevant for protein stability and enzyme catalysis in this probiotic, which suggests its advantage in terms of protease regulation [Citation50]. Still, the decreased expression of kgp and rgp induced by some probiotics, especially L. acidophilus LA5, may not only impair uptake of iron and decrease tissue destruction by the periodontopathogen strains but should also play an important role adherence of P. gingivalis to epithelial cells [Citation20,Citation51], in consonance with our previous data [Citation9]. Furthermore, several surface components of bacteria are proteases-sensitive, including those of probiotics [Citation39,Citation40], and they could be degraded by P. gingivalis proteolytic activity [Citation52]. Hence, regulation of gingipains encoding genes kgp and rgpA may have influenced the effect of probiotics on biofilm formation by different P. gingivalis strains.
On the other hand, the effects of probiotics on the expression of luxS were contradictory. Such findings indicate differences in the regulatory mechanisms of transcription between P. gingivalis W83 and ATCC 33277. LuxS controls quorum sensing, a system in response to bacterial biofilm density through the release of auto-inducers (AI-2) [Citation21]. Different microorganisms use this system to communicate with each other, and AI-2 works like a ‘universal language’ for intra-species and inter-species communication. LuxS/AI-2 of P. gingivalis regulates proteinase and hemagglutinin activities [Citation21,Citation53,Citation54] and may affect the expression of biofilm-associated genes, such as fimbriae [Citation14]. Thus, a beneficial effect of L. acidophilus LA-5 could be associated with the down-regulation of transcription of P. gingivalis luxS in mono-species biofilms. However, LuxS is also expressed by probiotics, influencing their ability to adhere, produce exopolysaccharides and form biofilm [Citation55,Citation56]. In addition, LuxS/AI-2 from Bifidobacterium spp. plays an essential role in their metabolism by regulating iron acquisition [Citation57] but data on probiotic lactobacilli are sparse [Citation58]. Interestingly, quorum-sensing signaling promoted by commensal microorganisms may disturb pathogen communication, since P. gingivalis LuxS/AI-2 network is reduced by the AI-2 synthetized by S. gordonii [Citation59], which consequently points to a beneficial effect of increasing LuxS expression in multi-species biofilms as observed under L. acidophilus LA-5 secretome stimulus. Therefore, since luxS involves a system common to pathogens, probiotics and other commensals, distinct results would be expected, depending on the environmental conditions and evaluated strains.
In summary, probiotics may affect biofilm formation and adherence of P. gingivalis to host cells by regulating the transcription of its virulence-associated factors. However, their effects differ not only regarding the probiotic species but also according to the colonizing pathogen strain. Taken all together, L. acidophilus LA 5 exerted the most promising effects by its secretome or by direct probiotic-pathogen contact that reduced P. gingivalis proportion in multi-species biofilms and promoted overall down-regulation transcription of key virulence genes responsible for encoding fimbriae, proteases, and quorum sensing components. Although bacterial interaction plays a key role in establishing the oral microbial communities, most of the interaction between P. gingivalis and probiotics are poorly understood. None of the studied probiotic strains was well characterized and further evaluation on the probiotics composition and released products should be performed, in order to elucidate their distinct mechanisms on the interaction with P. gingivalis.
Further in vivo experimental models and clinical studies are needed to provide evidence on the potential of these probiotics, especially L. acidophilus LA5, to control periodontopathogens levels and tissue destruction in the context of the periodontitis.
Supplemental Material
Download Zip (626.4 KB)Disclosure statement
The authors report no conflicts of interest related to this study.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Hajishengallis G, Lambris JD. Complement and dysbiosis in periodontal disease. Immunobiology. 2012;217(11):1111–12.
- Ebersole JL, Dawson D 3rd, Emecen-Huja P, et al. The periodontal war: microbes and immunity. Periodontol 2000. 2017;75(1):52–115.
- Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10(10):717–725.
- Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366(9499):1809–1820.
- Chapple ILC, Mealey BL, Van Dyke TE, et al. Periodontal health and gingival diseases and conditions on an intact and a reduced periodontium: consensus report of workgroup 1 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J Periodontol. 2018;89(Suppl 1):S74–S84.
- Teughels W, Newman MG, Coucke W, et al. Guiding periodontal pocket recolonization: a proof of concept. J Dent Res. 2007;86(11):1078–1082.
- Meurman JH. Functional foods/ingredients and oral mucosal diseases. Eur J Nutr. 2012;51(Suppl 2):S31–38.
- Dongarra ML, Rizzello V, Muccio L, et al. Mucosal immunology and probiotics. Curr Allergy Asthma Rep. 2013;13(1):19–26.
- Albuquerque-Souza E, Balzarini D, Ando-Suguimoto ES, et al. Probiotics alter the immune response of gingival epithelial cells challenged by Porphyromonas gingivalis. J Periodontal Res. 2019;54(2):115-127.
- Alemka A, Clyne M, Shanahan F, et al. Probiotic colonization of the adherent mucus layer of HT29MTXE12 cells attenuates Campylobacter jejuni virulence properties. Infect Immun. 2010;78(6):2812–2822.
- Zhou M, Zhu J, Yu H, et al. Investigation into in vitro and in vivo models using intestinal epithelial IPEC-J2 cells and Caenorhabditis elegans for selecting probiotic candidates to control porcine enterotoxigenic Escherichia coli. J Appl Microbiol. 2014;117(1):217–226.
- Matsubara VH, Ishikawa KH, Ando-Suguimoto ES, et al. Probiotic bacteria alter pattern-recognition receptor expression and cytokine profile in a human macrophage model challenged with Candida albicans and Lipopolysaccharide. Front Microbiol. 2017;8:2280.
- Nissen L, Sgorbati B, Biavati B, et al. Lactobacillus salivarius and L. gasseri down-regulate Aggregatibacter actinomycetemcomitans exotoxins expression. Ann Microbiol. 2014;64:611–617.
- Kuboniwa M, Amano A, Hashino E, et al. Distinct roles of long/short fimbriae and gingipains in homotypic biofilm development by Porphyromonas gingivalis. BMC Microbiol. 2009;9:105.
- Lamont RJ, Bevan CA, Gil S, et al. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol Immunol. 1993;8(5):272–276.
- Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62(4):1244–1263.
- Weinberg A, Belton CM, Park Y, et al. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1997;65(1):313–316.
- Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15(1):30–44.
- Takahashi Y, Davey M, Yumoto H, et al. Fimbria-dependent activation of pro-inflammatory molecules in Porphyromonas gingivalis infected human aortic endothelial cells. Cell Microbiol. 2006;8(5):738–757.
- Shi Y, Ratnayake DB, Okamoto K, et al. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem. 1999;274(25):17955–17960.
- James CE, Hasegawa Y, Park Y, et al. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect Immun. 2006;74(7):3834–3844.
- Capestany CA, Kuboniwa M, Jung IY, et al. Role of the Porphyromonas gingivalis InlJ protein in homotypic and heterotypic biofilm development. Infect Immun. 2006;74(5):3002–3005.
- Zhang Y, Shi W, Song Y, et al. Metatranscriptomic analysis of an in vitro biofilm model reveals strain-specific interactions among multiple bacterial species. J Oral Microbiol. 2019;11(1):1599670.
- Souza TC, Silva AM, Drews JR, et al. In vitro evaluation of Bifidobacterium strains of human origin for potential use in probiotic functional foods. Benef Microbes. 2013;4(2):179–186.
- Hsu SD, Cisar JO, Sandberg AL, et al. Adhesive properties of viridans streptococcal species. Microbial Ecol Health Dis. 1994;7(3):125–137.
- Lee KH, Kim BS, Keum KS, et al. Essential oil of Curcuma longa inhibits Streptococcus mutans biofilm formation. J Food Sci. 2011;76(9):H226–230.
- Maeda H, Fujimoto C, Haruki Y, et al. Quantitative real-time PCR using TaqMan and SYBR green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol Med Microbiol. 2003;39(1):81–86.
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acid Res. 2001;29(9):e45.
- Baraniya D, Naginyte M, Chen T, et al. Modeling normal and dysbiotic subgingival microbiomes: effect of nutrients. J Dent Res. 2020; 99(6):695–702
- Sharon N, Lis H. Lectins as cell recognition molecules. Science. 1989;246(4927):227–234.
- Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21(3):172–183.
- Teixeira SR, D’Epiro TT, Pinheiro ET, et al. Lineage variability in surface components expression within Porphyromonas gingivalis. Microbial Pathogen. 2014;77:100–104.
- Polak D, Ferdman O, Houri-Haddad Y. Porphyromonas gingivalis capsule-mediated coaggregation as a virulence factor in mixed infection with Fusobacterium nucleatum. J Periodontol. 2017;88(5):502–510.
- Barbosa GM, Colombo AV, Rodrigues PH, et al. Intraspecies variability affects heterotypic biofilms of porphyromonas gingivalis and Prevotella intermedia: evidences of strain-dependence biofilm modulation by physical contact and by released soluble factors. PLoS One. 2015;10(9):e0138687.
- Nelson KE, Fleischmann RD, DeBoy RT, et al. Complete genome sequence of the oral pathogenic Bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185(18):5591–5601.
- Sonoi N, Maeda H, Murauchi T, et al. IS1598 (IsPg4) distributed to abscess-forming strains of Porphyromonas gingivalis may enhance virulence through upregulation of nrdD-like gene expression. New Microbiol. 2018;41(1):52–60.
- Jäsberg H, Söderling E, Endo A, et al. Bifidobacteria inhibit the growth of Porphyromonas gingivalis but not of Streptococcus mutans in an in vitro biofilm model. Eur J Oral Sci. 2016;124(3):251–258.
- Castro-Bravo N, Wells JM, Margolles A, et al. Interactions of surface exopolysaccharides from Bifidobacterium and Lactobacillus within the intestinal environment. Front Microbiol. 2018;9:2426.
- Yadav AK, Tyagi A, Kumar A, et al. Adhesion of Lactobacilli and their anti-infectivity potential. Crit Rev Food Sci Nutr. 2017;57(10):2042–2056.
- Keller MK, Hasslöf P, Stecksén-Blicks C, et al. Co-aggregation and growth inhibition of probiotic lactobacilli and clinical isolates of mutans streptococci: an in vitro study. Acta Odontol Scand. 2011;69(5):263–268.
- Nakagawa I, Amano A, Kuboniwa M, et al. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect Immun. 2002;70(1):277–285.
- Enersen M, Nakano K, Amano A. Porphyromonas gingivalis fimbriae. J Oral Microbiol. 2013;5:1–10. doi:10.3402/jom.v5i0.20265.
- Maeda K, Nagata H, Kuboniwa M, et al. Characterization of binding of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase to Porphyromonas gingivalis major fimbriae. Infect Immun. 2004;72(9):5475–5477.
- Yilmaz O, Watanabe K, Lamont RJ. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell Microbiol. 2002;4(5):305–314.
- Davey M, Liu X, Ukai T, et al. Bacterial fimbriae stimulate proinflammatory activation in the endothelium through distinct TLRs. J Immunol. 2008;180(4):2187–2195.
- Lin X, Wu J, Xie H. Porphyromonas gingivalis minor fimbriae are required for cell-cell interactions. Infect Immun. 2006;74(10):6011–6015.
- Park Y, Simionato MR, Sekiya K, et al. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun. 2005;73(7):3983–3989.
- Simionato MR, Tucker CM, Kuboniwa M, et al. Porphyromonas gingivalis genes involved in community development with Streptococcus gordonii. Infect Immun. 2006;74(11):6419–6428.
- Olczak T, Simpson W, Liu X, et al. Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol Rev. 2005;29(1):119–144.
- Calderini E, Celebioglu HU, Villarroel J, et al. Comparative proteomics of oxidative stress response of Lactobacillus acidophilus NCFM reveals effects on DNA repair and cysteine de novo synthesis. Proteomics. 2017;17:5.
- Kadowaki T, Nakayama K, Yoshimura F, et al. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J Biol Chem. 1998;273(44):29072–29076.
- Frece J, Kos B, Svetec IK, et al. Importance of S-layer proteins in probiotic activity of Lactobacillus acidophilus M92. J Appl Microbiol. 2005;98(2):285–292.
- Burgess NA, Kirke DF, Williams P, et al. LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiology. 2002;148(Pt 3):763–772.
- Chung WO, Park Y, Lamont RJ, et al. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J Bacteriol. 2001;183(13):3903–3909.
- Lebeer S, Claes IJ, Verhoeven TL, et al. Impact of luxS and suppressor mutations on the gastrointestinal transit of Lactobacillus rhamnosus GG. Appl Environ Microbiol. 2008;74(15):4711–4718.
- Wilson CM, Aggio RB, O’Toole PW, et al. Transcriptional and metabolomic consequences of LuxS inactivation reveal a metabolic rather than quorum-sensing role for LuxS in Lactobacillus reuteri 100-23. J Bacteriol. 2012;194(7):1743–1746.
- Christiaen SE, O’Connell Motherway M, Bottacini F, et al. Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PloS One. 2014;9(5):e98111.
- Yeo S, Park H, Ji Y, et al. Influence of gastrointestinal stress on autoinducer-2 activity of two Lactobacillus species. FEMS Microbiol Ecol. 2015;91:7.
- McNab R, Ford SK, El-Sabaeny A, et al. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol. 2003;185(1):274–284.
- Amano A, Nakagawa I, Kataoka K, et al. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J Clin Microbiol. 1999;37(5):1426–1430.
- Rinttilä T, Kassinen A, Malinen E, et al. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97(6):1166–1177.
- Zhou Y, Yang J, Zhang L, et al. Differential utilization of basic proline-rich glycoproteins during growth of oral bacteria in saliva. Appl Environ Microbiol. 2016;82(17):5249–5258.
- Capestany CA, Tribble GD, Maeda K, et al. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J Bacteriol. 2008;190(4):1436–1446.