 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Dermatophytosis are one of the most common fungal infections in the world. They compromise keratinized tissues and the main etiological agent is Trichophyton rubrum. Macrophages are key cells in innate immunity and prominent sources of IL-1β, a potent inflammatory cytokine whose main production pathway is by the activation of inflammasomes and caspase-1. However, the role of inflammasomes and IL-1 signaling against T.rubrum has not been reported. In this work, we observed that bone marrow-derived macrophages produce IL-1β in response to T.rubrum conidia in a NLRP3-, ASC- and caspase-1-dependent fashion. Curiously, lack of IL-1 signaling promoted hyphae development, uncovering a protective role for IL-1β in macrophages. In addition, mice lacking IL-1R showed reduced IL-17 production, a key cytokine in the antifungal defense, in response to T.rubrum. Our findings point to a prominent role of IL-1 signaling in the immune response to T.rubrum, opening the venue for the study of this pathway in other fungal infections.
Abbreviations
| ASC | = | Apoptosis-associated Speck-like protein containing a CARD |
| BMDM | = | bone marrow derived macrophages |
| CFU | = | colony-forming units |
| IL | = | interleukin |
| NLR | = | NOD-like receptor |
| NLRP3 | = | NOD-like receptor family, pyrin domain containing 3 |
| WT | = | wild-type |
Introduction
Interleukin-1β (IL-1β) is a potent inflammatory cytokine that is produced as an inactive precursor and whose main activation route is the inflammasomes.Citation1 These structures are associated to the surveillance of the intracellular environment and their main function is the activation of pro-caspase-1 into an enzyme (caspase-1) able to cleave (and activate) IL-1β and IL-18.Citation2-4
IL-1 family members signal through IL-1RI (IL-1R for short), a receptor composed of an extracellular immunoglobulin domain and a TLR/IL-1 receptor cytoplasmic domain.Citation1 Among the effects on innate immune cells, these cytokines promote microbicidal activities in myeloid cells, as induction of phagocytosis and production of cytokines by macrophages, recruitment of neutrophils, up regulation of their oxidative burst and induction of enzyme release.Citation1 Due to the strong inflammatory potential of IL-1 cytokines, they are under tight control in production (through inflammasomes) and action steps, e.g. IL-1RII, an alternative receptor that does not trigger any signaling pathway and, therefore, it helps to neutralize IL-1 cytokines.Citation5
The core of the majority of the inflammasomes is the NOD-like Receptor (NLR) and the most studied of them is the NOD-like receptor family, pyrin domain containing 3 (NLRP3). Once activated, NLRP3 recruits the adaptor molecule Apoptosis-associated Speck-like protein containing a CARD (ASC), that helps the subsequent recruitment of (pro-)caspase-1.Citation6 NLRP3 inflammasome is activated by a diverse set of stimuli, but, classically, it is associated to the sensing of noxious agents as monosodium urate crystals (associated to gout) and extracellular ATP, particles as silica and asbestos, and also pathogens.Citation7 Among infectious agents, bacteria, as Streptococcus agalactiaeCitation8; viruses, as Rift Valley fever virusCitation9; protozoans, as Plasmodium bergheiCitation10 and fungi, as Candida albicansCitation11 were shown to activate this system. This multitude of potential activators places NLRP3 as a prominent actor in many diseases associated to inflammatory conditions.
Mycoses as dermatophytosis are characterized by chronic inflammation. These cutaneous infections affect keratinized tissues (skin, nails and hair) and, in humans, the main etiological agent is Trichophyton rubrum.Citation12 Previously, our group has shown that macrophages phagocytose T.rubrum conidia but, instead of eliminating the fungal cells, conidia turn into hyphae inside the phagocytes and disrupt them. Associated to this process, we detect production of inflammatory cytokines, e.g., TNF-α, that would feed the inflammation observed in this disease.Citation13,14
However, up to date, the current role of inflammasomes and IL-1 signaling in response to T.rubrum has not been characterized. In this work, we investigated the role of the NLRP3 inflammasome in macrophages against T.rubrum conidia and how IL-1 signaling influences the outcome of this interaction. We observed that T.rubrum is able to induce IL-1β production by macrophages in an NLRP3-dependent manner. ASC absence impaired not only cytokine production but also conidia phagocytosis itself. Remarkably, impaired IL-1 signaling led to increase susceptibility of macrophages to hyphae growth and reduced IL-17 production in mice, putting IL-1 as an important element in the immune response against T.rubrum.
Material and Methods
Animals
Female C57BL/6 mice, 8-12 weeks old, wild-type (WT) phenotype, were obtained from the animal facilities at the Faculty of Pharmaceutical Sciences / Institute of Chemistry of the University of São Paulo. Male C57BL/6 mice, 8-12 weeks old, knockout for NLRP3, ASC, Caspase-1/-11 and IL-1R were obtained from the animal facilities at the School of Medicine of Ribeirão Preto (University of São Paulo). The experimental protocols involving animals were previously approved by the Institutional Ethics Committee for Animal Care and Research at the Faculty of Pharmaceutical Sciences (CEUA/FCF Protocol 293).
Fungal strain and conidia preparation
An ATCC T.rubrum strain (ATCC 28188) was used throughout this study. Conidia were obtained as previously described.Citation13 Briefly, a 14 days-old culture growing in Sabouraud Agar (Difco) at 30°C was transferred to Potato-broth medium (Difco) and kept at 30°C and constant rotation (200 rpm) for 3 days. The suspension was filtered in cell strainer 40 μm (BD) and the suspension was washed 2 times with 0.1% Tween 80 (Synth) in PBS 1× (Na2HPO4 18 mM; NaH2PO4.H2O 3 mM and NaCl 140 mM in miliQ water, salts from Synth). The conidia were, then, ressuspended in PBS 1× and stored at 4°C.
Bone marrow derived macrophages (BMDM)
BMDM were obtained as described.Citation15 Briefly, bone marrow from the femurs were collected and cultured in RPMI-1640 medium supplemented with 20% FBS, 30% L929-cell conditioned medium and gentamicin (40 mg/L) – R20/30 media – at 37°C in a 5% CO2 atmosphere. After four days, the cells were supplemented with the same medium and incubated for additional 3 days. Macrophages were collected and incubated in RPMI-1640 medium supplemented with 10% FBS, 5% L929-cell conditioned medium and gentamicin (40 mg/L) – R10/5 media – at 37°C in a 5% CO2 atmosphere.
Phagocytosis assay and CFU assay
BMDMs were plated over glass slides in 24-wells plates (2.0 × 105 cell/mL.well) and kept overnight at 37°C and 5% CO2 for cell adhesion. The cells were primed with LPS (E.coli O55:B5, 1 μg/mL.well, Sigma-Aldrich) in fresh R10/5 media for 30 minutes and washed with PBS 1×. T.rubrum conidia produced as described above were diluted in fresh R10/5 media and incubated with the BMDMs at MOI 1:1 (1 mL/well). The systems were maintained at 37°C and 5% CO2 for 4, 6, 8 and 10 hours. After each time, samples were collected, centrifuged at 3000 rpm for 10 minutes and supernatants were stored at 20°C for cytokine measurements. Glass slides were stained with the hematological staining kit InstantProv (NewProv).
For Colony-Forming Units (CFU) assay, the above phagocytosis assay was performed without glass slides. At each time point, wells were washed with PBS 1× to remove free fungi and BMDMs were lysed with 0.1% Triton X-100 solution. A ten-fold dilution of the samples were plated on Sabouraud Agar (Difco) and kept at 30°C up to 7 days. Recovered colonies were counted, multiplied by dilution factor and expressed as mean log.
Phagocytosis index
The phagocytosis index (PI) was calculated according to the following formula:
In vivo infection
Groups of 3 mice were infected intraperitoneally with 5.0 × 106 T.rubrum conidia diluted in PBS and euthanized after 7 or 14 days. Liver and spleen from each animal were homogenized in PBS and fungal burden was determined by CFU assay. Briefly, 10-fold dilutions of each sample were plated on Sabouraud agar (Difco) and incubated at 30°C for 5 days. Recovered colonies were counted, multiplied by dilution factor and expressed as mean log (CFU/gram of organ). Organ homogenates were then centrifuged at 3000 rpm for 10 minutes and the supernatants were collected for cytokines measurements.
Cytokines measurements
Cytokines were measured by sandwich ELISA assay kits (R&D Systems) according to the manufacturer's instructions.
Statistical analysis
Data were expressed as mean±SEM. and analyzed in the software GraphPad Prism (version 5.00 for Windows, GraphPad Software, San Diego California USA, www.graphpad.com) by Two-way ANOVA and Bonferroni post test or unpaired t-test. Differences were considered statistically significant at P < 0.05.
Results
T.rubrum conidia induces IL-1β production in BMDMs
Our group had previously shown that peritoneal macrophages are able to phagocytose T.rubrum conidia but they succumb to the hyphae development.Citation13 In order to assess the role of inflammasomes in response to T.rubrum, we first analyzed the interaction between T.rubrum conidia and BMDMs (). As expected, BMDMs were able to phagocytose conidia but remain susceptible to hyphae development, whose growth led to BMDMs destruction at latter time points. This BMDMs susceptibility was also verified by LDH release (Supporting information, Fig. S1A).
Figure 1. BMDMs produce IL-1β in response to T.rubrum conidia but succumb to hyphae growth. LPS-primed BMDMs were incubated with T.rubrum conidia for 4, 6, 8 and 10 h (no fungi in control wells). (A) Interaction outcome was analyzed by optical microscopy (1000×) and arrows indicate fungal structures. (B) IL-1β levels quantified in the culture supernatants expressed as mean ± SEM. Two-way ANOVA and Bonferroni posttest: ***P < 0.001. Cytokine data are pooled from 3 independent experiments and microscopy photos taken from one experiment (representative of 3 independent experiments) are shown.
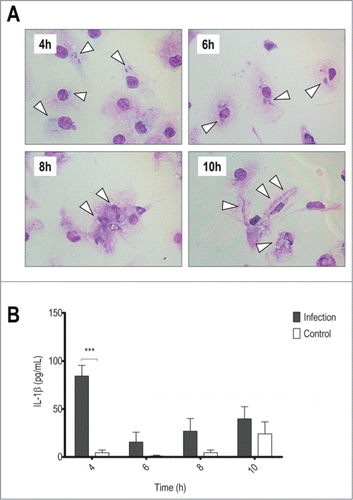
Associated to this process we detected IL-1β in culture supernatants at the beginning of the interaction (), suggesting that inflammasomes could be activated in response to T.rubrum.
IL-1β production relies on NLRP3, ASC and Caspase-1
Since the most studied inflammasome associated to fungal infections is the NLRP3,Citation11,16-19 we sought to investigate whether this molecule could be necessary in our system. By analyzing the interaction between NLRP3−/− BMDMs and T.rubrum conidia, we observed that the lack of this receptor did not change the interaction outcome (). However, NLRP3−/− BMDMs showed reduced IL-1β secretion (), while no differences on TNF-α production were observed (Fig. S2). Our findings add T.rubrum as another NLRP3 inflammasome activator, supporting this receptor as an important response element in fungal / dermatophyte infections.
Figure 2. NLRP3 and ASC deficiency reduces IL-1β secretion, but only lack of ASC interferes in phagocytosis. LPS-primed NLRP3−/− or ASC−/− BMDMs were incubated with T.rubrum conidia for 4, 6, 8 and 10 h (no fungi in control wells). (A) NLRP3−/− Interaction outcome and (B) ASC−/− Interaction outcome analyzed by optical microscopy (1000×). Arrows indicate fungal structures. (C) IL-1β production by NLRP3−/− BMDMs and (D) IL-1β levels produced by ASC−/− BMDMs. (E) ASC−/− BMDMs were incubated with T.rubrum conidia for 4 h and conidia internalization was counted by optical microscopy. Results expressed as mean ± SEM. Two-way ANOVA and Bonferroni post test: *P < 0.05; **P < 0.01; ***P < 0.001. Cytokine data pooled from 2 independent experiments performed, in triplicate wells each, are shown. Microscopy photos taken from one experiment (representative of 2 independent experiments triplicate wells each) are shown.
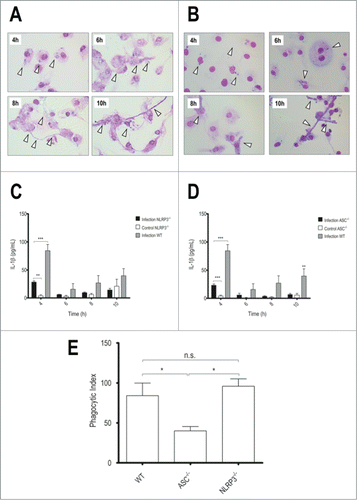
In order to recruit caspase-1, the NLRP3 inflammasome requires the adaptor ASCCitation6 and the role of this molecule was also investigated in our system. As expected, ASC deficiency reduced IL-1β () but not TNF-α levels (Fig. S2), supporting our hypothesis that a classical NLRP3-ASC axis is involved.
It should be noticed that, like NLRP3 absence, no changes were observed in the outcome of phagocytosed conidia in ASC−/− BMDMs (). However, these macrophages showed reduced fungal internalization, as measured by the phagocytosis index (). This finding indicates that, besides impaired inflammasome activation, ASC−/− BMDMs cannot internalize T.rubrum conidia efficiently. However, once inside the BMDMs, conidia have the same fate as in WT macrophages.
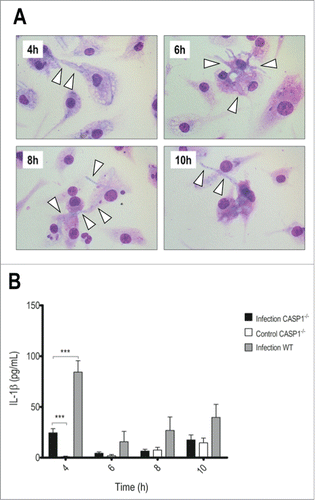
Finally, the last step of inflammasome activation is the recruitment of caspase-1, the protease responsible for IL-1β activation,Citation6 and, therefore, we investigated the outcome of the interaction in caspase-1/-11−/− BMDMs (). Strikingly, they showed increased susceptibility to fungal growth (), measured also by LDH release (Fig. S1B). In the first time point, we could already observe hyphae development inside macrophages, leading to their destruction. In addition, they showed reduced IL-1β production () while TNF-α secretion was even higher than in WT cells (Fig. S2). These results point to caspases as important to both T.rubrum conidia restriction and cytokine production.
Figure 3. Caspase-1/-11 absence accelerates hyphae development and reduces IL-1β secretion. LPS-primed caspase-1/-11−/− BMDMs were incubated with T.rubrum conidia for 4, 6, 8 and 10 h (no fungi in control wells). (A) Interaction outcome was analyzed by optical microscopy (1000×) and arrows indicate fungal structures. (B) IL-1β levels quantified in the culture supernatants expressed as mean ± SEM. Two-way ANOVA and Bonferroni posttest: ***P < 0.001. Cytokine data are pooled from 2 independent experiments performed in triplicate wells each. Microscopy photos taken from one experiment (representative of 2 independent experiments, triplicate wells each) are shown.
IL-1β signaling through IL-1R is necessary for fungal restriction
In order to establish the importance of this cytokine in our system, we analyzed the impact of IL-1R absence over BMDMs response.
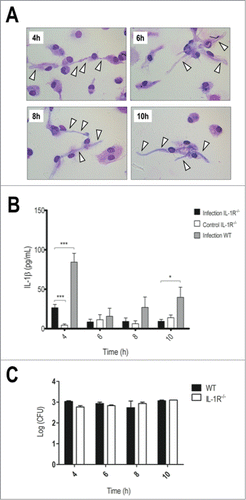
IL-1R−/− BMDMs seemed to recapitulate the susceptible phenotype. In 4 h of interaction, we could observe a profuse hyphae development in IL-1R−/− BMDMs (), also detected indirectly by LDH release (Fig. S1B). Further, they showed lower IL-1β levels () without impairments in TNF-α production (Fig. S2). Since the fungal load in IL-1R−/− BMDMs did not differ from WT BMDMs () we ruled out the possibility that these knockout cells display altered fungal internalization. Therefore, these findings indicate that IL-1 signaling promotes fungal restriction while deficiency IL-1R turns BMDMs more permissive to conidia development into hyphae.
Figure 4. Impaired IL-1 signaling leads to faster hyphae development and reduced IL-1β production. LPS-primed IL-1R−/− BMDMs were incubated with T.rubrum conidia for 4, 6, 8 and 10 h (no fungi in control wells). (A) Interaction outcome was analyzed by optical microscopy (1000×) and arrows indicate fungal structures. (B) IL-1β levels quantified in the culture supernatants. Two-way ANOVA and Bonferroni posttest: *P < 0.05; ***P < 0.001. (C) Fungal loads in WT and IL-1R−/− BMDMs determined by CFU assay. Data expressed as mean ± SEM. Cytokine and CFU data were pooled from 2 independent experiments performed in triplicate wells each. Microscopy photos taken from one experiment (representative of 2 independent experiments, triplicate wells each) are shown.
IL-1R signaling helps to shape the immune response in vivo
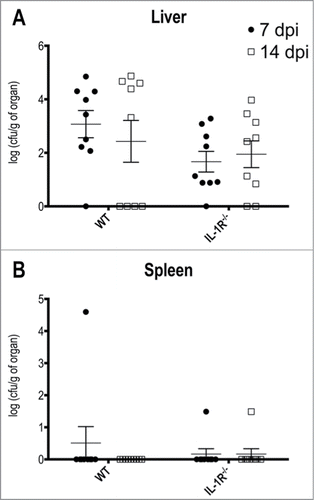
Our in vitro findings prompted us to investigate the in vivo importance of IL-1 signaling in the response to T.rubrum. To evaluate the reliability of our approach, we assessed the fungal burden in 2 main organs: liver and spleen (). Indeed, we were able to rescue fungi from liver homogenates after 7 days post infection and, although without statistical significance, we observed a slightly reduction in the fungal burden after 14 days in wild type animals, but not in IL-1R−/− counterparts (). Curiously, the spleen was resistant to T.rubrum colonization in both mice groups ().
Figure 5. T.rubrum affects preferentially the liver but not the spleen. Animals were infected i.P. with T.rubrum conidia (5 × 106/ animal) and fungal burden were determined 7 and 14 days post infection (dpi). (A) Fungal burden in Liver. (B) Fungal burden in Spleen. Unpaired t-test: no significance found. Data pooled from 3 to 4 independent experiments (groups of 3 animals in each experiment) expressed as mean ± SEM.
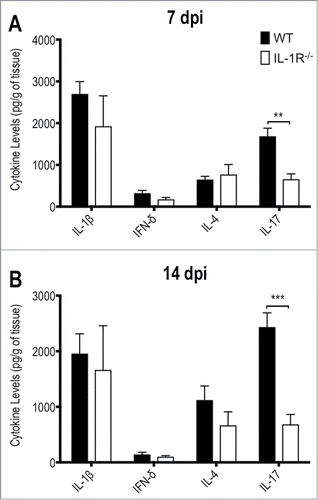
Besides the fungal burden, we also analyzed the cytokines profile in liver homogenates (). Even though they did not reach statistical significance, levels of IL-1β, IL-4 and IFN-γ were slightly reduced in knockout animals in response to T.rubrum infection mainly after 14 days. Strikingly, IL-17 levels were greatly impaired by IL-1R deficiency, uncovering an important role for IL-1 signaling in IL-17 response triggered in response to T.rubrum. The cytokine production seems to require fungal colonization since cytokines levels in the spleen did not show any statistical differences between both groups (data not shown).
Figure 6. IL-1R−/− mice show defective IL-17 production in response to T.rubrum. Animals were infected i.p. with T.rubrum conidia (5 × 106/ animal) and cytokines levels (IL-1β; IFN-γ; IL-4 and IL-17) were determined in liver homogenates (A) 7 and (B) 14 days post infection (dpi). Two-way ANOVA and Bonferroni posttest: *P < 0.05; ***P < 0.001. Data, expressed as mean ± SEM, were pooled from 2 to 4 independent experiments (groups of 3 animals in each experiment).
Discussion
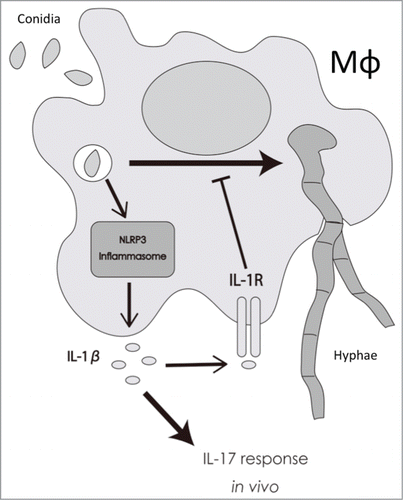
In this work we investigated the importance of IL-1β in the response to the dermatophyte T.rubrum in in vitro and in vivo systems. We observed that IL-1β is produced by BMDMs in response to T.rubrum conidia and that this process relies on the classical NLRP3 inflammasome. In addition, we showed that IL-1 signaling is crucial to delay the fungal development into hyphae, protecting macrophages at the beginning of the interaction, and uncovered an important role for this pathway in shaping IL-17 response in vivo ().
Figure 7. Proposed model for the role of IL-1β in response to T.rubrum. Macrophages phagocytose T.rubrum, leading to the activation of NLRP3 inflammasome and, consequently, to IL-1β production. By your turn, IL-1β, signaling through IL-1R, delays fungal development into hyphae inside the macrophages and also helps to shape the IL-17 response in vivo.
Our findings that IL-1β production in response to T.rubrum is impaired in the absence of NLRP3 () is in accordance to the role of this receptor in the response against other fungal pathogens, as Candida albicans,Citation11 Aspergillus fumigatus,Citation16 Paracoccidioides brasiliensisCitation19 and dermatophytes as Trichophyton schoenleiniiCitation18 and Microsporum canis.Citation17 However, the works on dermatophytes focused mainly on THP-1 monocytes, which show a constitutive caspase-1 activity and rely on pro-IL-1β transcription/expression as the limiting step for IL-1β production, while BMDMs obey to a 2-signal model to activate inflammasomes: signal 1, TLR activation for transcription; and signal 2, NLR activation for inflammasome assemblage.Citation20,21 Therefore, by working with LPS-primed cells to rule out signal 1 interference, our data provides a better understanding of the role of the inflammasome strictu sensu in response to a dermatophyte. Even though we could not determine yet which molecule(s)/fungal determinant(s) is (are) involved, it is known that NLRP3 activation relies on K+ efflux (induced by channels as ATP-dependent P2X receptor), reactive oxygen species (ROS) production and lysosomal damage (mainly, cathepsin B release).Citation22 Nevertheless, we observed that neither K+ efflux nor ROS production are important in our case (Fig. S3).
Interestingly, the molecule ASC was needed for IL-1β secretion and in phagocytosis process (). Ippagunta et.al. demonstrated that dendritic cells lacking ASC showed impaired actin polymerization, leading to defective internalization of zymosan (a particle that resembles the fungal cell wall).Citation23 A similar defect may happen in our system and ASC−/− BMDMs could not phagocytose T.rubrum as efficiently as WT cells. Nevertheless, even though phagocytosis might be impaired, some conidia are indeed internalized and, once inside, they share the same outcome, turning into hyphae inside the macrophages.
Next, we observed that Caspase-1/-11−/− BMDMs did show a reduced secretion of IL-1β in response to T.rubrum (). Since direct IL-1β processing by caspase-11 is negligible,Citation24,25 we conclude that the classical NLRP3/ASC/Caspase-1 inflammasome is important for IL-1β production in response to T.rubrum.
Remarkably, this cytokine showed a prominent role in fungal restriction since IL-1R−/− cells succumb to the accelerated hyphae growth (). We also observed a reduction in IL-1β levels in the absence of this receptor () that can be explained by the ability of IL-1β to induce its own transcription in a positive feedback mechanism.Citation5,26,27 Despite their reduced levels of IL-1β, NLRP3−/− and ASC−/− cells do not show enhanced susceptibility to T.rubrum because they have a functional IL-1R and their residual cytokine production should be enough to keep the fungus in check. This residual IL-1β may indicate the participation of others systems besides classical inflammasomes. An interesting candidate is the caspase-8 inflammasome triggered in response to dectin-1 signaling.Citation28 It would be interesting to evaluate the role of this system since dectin-1 is a classical receptor in immune response to fungi.
In other hand, caspase-1/-11−/− BMDMs were more susceptible to fungal growth despite a residual amount of IL-1β and a functional IL-1R. Akhter et.al. showed that caspase-11 absence impairs the fusion of phagosomes to lysosomes and this defect promoted the survival of pathogenic bacteria in the intracellular environment.Citation29 A similar problem could be happening in our case: phagosomes containing T.rubrum conidia cannot fusion with lysosomes and the fungus finds an appropriate environment to develop faster. Therefore, even though lack of caspase-1/-11 and IL-1R led to similar outcomes, their mechanism of action might be different. Further experiments will shed more light to the importance of caspase-11 in the response to T.rubrum.
Since IL-1 signaling showed an unexpected role in vitro, we investigated its importance in vivo ( and ). IL-1β levels showed a tendency toward reduction in IL-1R−/− mice that resembles the in vitro findings. However, since others cellular sources of IL-1β, as neutrophils, that produce this cytokine through caspase-1 independent pathways,Citation5 cannot be ruled out, IL-1β pool in vivo may be the result of multiple contributions. Strikingly, IL-1R deficiency led to an impaired IL-17 response (). Indeed, IL-1β is needed for the development of TH17 cells together with IL-6 and TGF-βCitation1,30 and this seems to be our cases: IL-1β produced in T.rubrum infection is fundamental to shape the IL-17 response.
At the time of this manuscript preparation, Baltazar et.al. proposed an alternative approach for experimental dermatophytosis that relied on IL-12 and IFN-γ to promote fungal restriction,Citation31 however, we did not observed significant contribution of IFN-γ in our system. Differences in the models, as administration route/site of infection and mice background or housing may account for these different outcomes. However, they did not analyze the importance of inflammasomes and IL-1 signaling as our model have uncovered.
It is interesting to note that IL-17 levels were impaired within 7 days of infection, while a full adaptive response should take longer time to establish until cytokine-secreting effector T cells differentiate. Even though TH17 cells are the classical source of IL-17, it should not be ignored that this cytokine can also be secreted by other kinds of cells. One example is the innate-like lymphoid cells (ILCs), a recently identified lymphoid lineage population whose functions show innate immunity-like features, as a fast-response timing.Citation32,33 Kim et.al. showed that obesity-associated airway hyperactivity is driven by ILCs-derived IL-17 and the recruitment of these cells relied on NLRP3-derived IL-1β.Citation34 IL-17-producing ILCs were also reported to determine protection against C.albicans in a murine model of oropharyngeal candidiasis.Citation33 Even neutrophils can be a source of IL-17,Citation35 and their importance in a model of keratitis by Aspergillus was also documented.Citation36
In conclusion, our findings add T.rubrum as another NLRP3 inflammasome activator, supporting this system as another component of the inflammatory process in dermatophytosis. Also, we showed a remarkable role for IL-1 signaling in the shaping of the response of macrophages to this pathogen and in IL-17 response in vivo. The importance of these pathways could be extrapolated to other pathogenic fungi and point them as prominent players in the immunity to fungal infections.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
FSYY and LGF performed the experiments; FSYY and SRA designed the study; FSYY wrote and SRA edited the manuscript.
2014VIRULENCE0173R_Supplemental_files.pdf
Download PDF (231.1 KB)Acknowledgments
We thank Maira Cristina Nakamura and Dario Simões Zamboni from School of Medicine of Ribeirão Preto (University of São Paulo) for providing knockout animals.
Funding
This work was supported by São Paulo Research Foundation (FAPESP): Grant FAPESP 2012/14684-6.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Sims JE, Smith DE. The IL-1 family: regulators of immunity. Nat Rev Immunol 2010; 10:117; http://dx.doi.org/10.1038/nri2691
- Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature 2012; 481:278-86; PMID:22258606; http://dx.doi.org/10.1038/nature10759
- Franchi L, Muñoz-Planillo R, Núñez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol 2012; 13:325-32; PMID:22430785; http://dx.doi.org/10.1038/ni.2231
- Horvath GL, Schrum JE, De Nardo CM, Latz E. Intracellular sensing of microbes and danger signals by the inflammasomes. Immunol Rev 2011; 243:119-35; PMID:21884172; http://dx.doi.org/10.1111/j.1600-065X.2011.01050.x
- Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol 2009; 27:519-50; PMID:19302047; http://dx.doi.org/10.1146/annurev.immunol.021908.132612
- Davis BK, Wen H, Ting JP-Y. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol 2011; 29:707-35; PMID:21219188; http://dx.doi.org/10.1146/annurev-immunol-031210-101405
- Henao-Mejia J, Elinav E, Strowig T, Flavell RA. Inflammasomes: far beyond inflammation. Nat Immunol 2012; 13:321-4; PMID:22430784; http://dx.doi.org/10.1038/ni.2257
- Costa A, Gupta R, Signorino G, Malara A, Cardile F, Biondo C, Midiri A, Galbo R, Trieu-Cuot P, Papasergi S, et al. Activation of the NLRP3 inflammasome by group B streptococci. J Immunol 2012; 188:1953-60; PMID:22250086; http://dx.doi.org/10.4049/jimmunol.1102543
- Ermler ME, Traylor Z, Patel K, Schattgen SA, Vanaja SK, Fitzgerald KA, Hise AG. Rift valley fever virus infection induces activation of the NLRP3 inflammasome. Virology 2014; 449:174-80; PMID:24418550; http://dx.doi.org/10.1016/j.virol.2013.11.015
- Kalantari P, DeOliveira RB, Chan J, Corbett Y, Rathinam V, Stutz A, Latz E, Gazzinelli RT, Golenbock DT, Fitzgerald KA. Dual engagement of the NLRP3 and AIM2 inflammasomes by plasmodium-derived hemozoin and DNA during malaria. Cell Rep [Internet] 2014 [cited 2014 Jan 25]; Available from: http://linkinghub.elsevier.com/retrieve/pii/S2211124713007596
- Hise AG, Tomalka J, Ganesan S, Patel K, Hall BA, Brown GD, Fitzgerald KA. An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans. Cell Host Microbe 2009; 5:487-97; PMID:19454352; http://dx.doi.org/10.1016/j.chom.2009.05.002
- Mendez-Tovar LJ. Pathogenesis of dermatophytosis and tinea versicolor. Clin Dermatol 2010; 28:185-9; PMID:20347661; http://dx.doi.org/10.1016/j.clindermatol.2009.12.015
- Campos MRM, Russo M, Gomes E, Almeida SR. Stimulation, inhibition and death of macrophages infected with Trichophyton rubrum. Microbes Infect Inst Pasteur 2006; 8:372-9; http://dx.doi.org/10.1016/j.micinf.2005.07.028
- Almeida SR. Immunology of dermatophytosis. Mycopathologia 2008; 166:277-83; PMID:18478362; http://dx.doi.org/10.1007/s11046-008-9103-6
- Marim FM, Silveira TN, Lima DS, Zamboni DS. A method for generation of bone marrow-derived macrophages from cryopreserved mouse bone marrow cells. PLoS One 2010; 5:e15263; PMID:21179419; http://dx.doi.org/10.1371/journal.pone.0015263
- Saïd-Sadier N, Padilla E, Langsley G, Ojcius DM. Aspergillus fumigatus stimulates the NLRP3 inflammasome through a pathway requiring ROS production and the syk tyrosine kinase. PLoS One 2010; 5:e10008; http://dx.doi.org/10.1371/journal.pone.0010008
- Mao L, Zhang L, Li H, Chen W, Wang H, Wu S, Guo C, Lu A, Yang G, An L, et al. Pathogenic fungus microsporum canis activates the NLRP3 inflammasome. Infect Immun 2013; 82:882-92; PMID:24478101; http://dx.doi.org/10.1128/IAI.01097-13
- Li H, Wu S, Mao L, Lei G, Zhang L, Lu A, An L, Yang G, Abliz P, Meng G. Human pathogenic fungus Trichophyton schoenleinii activates the NLRP3 inflammasome. Protein Cell 2013; 4:529-38; PMID:23686720; http://dx.doi.org/10.1007/s13238-013-2127-9
- Tavares AH, Magalhães KG, Almeida RDN, Correa R, Burgel PH, Bocca AL. NLRP3 inflammasome activation by paracoccidioides brasiliensis. PLoS Negl Trop Dis 2013; 7:e2595; PMID:24340123; http://dx.doi.org/10.1371/journal.pntd.0002595
- Netea MG, Nold-Petry CA, Nold MF, Joosten LA, Opitz B, van der Meer JH, van de Veerdonk FL, Ferwerda G, Heinhuis B, Devesa I, et al. Differential requirement for the activation of the inflammasome for processing and release of IL-1β in monocytes and macrophages. Blood 2009; 113:2324-35; PMID:19104081; http://dx.doi.org/10.1182/blood-2008-03-146720
- Netea MG, Simon A, van de Veerdonk F, Kullberg B-J, Van der Meer JWM, Joosten LAB. IL-1beta processing in host defense: beyond the inflammasomes. PLoS Pathog 2010; 6:e1000661; PMID:20195505; http://dx.doi.org/10.1371/journal.ppat.1000661
- Von Moltke J, Ayres JS, Kofoed EM, Chavarría-Smith J, Vance RE. Recognition of bacteria by inflammasomes. Annu Rev Immunol 2013; 31:73-106; PMID:23215645; http://dx.doi.org/10.1146/annurev-immunol-032712-095944
- Ippagunta SK, Malireddi RKS, Shaw PJ, Neale GA, Walle LV, Green DR, Fukui Y, Lamkanfi M, Kanneganti T-D. The inflammasome adaptor ASC regulates the function of adaptive immune cells by controlling Dock2-mediated Rac activation and actin polymerization. Nat Immunol 2011; 12:1010-6; PMID:21892172; http://dx.doi.org/10.1038/ni.2095
- Wang S, Miura M, Jung Y, Zhu H, Gagliardini V, Shi L, Greenberg AH, Yuan J. Identification and characterization of Ich-3, a member of the interleukin-1β converting enzyme (ICE)/Ced-3 family and an upstream regulator of ICE. J Biol Chem 1996; 271:20580-7; PMID:8702803; http://dx.doi.org/10.1074/jbc.271.34.20580
- Viganò E, Mortellaro A. Caspase-11: The driving factor for noncanonical inflammasomes. Eur J Immunol 2013; 43:2240-5; http://dx.doi.org/10.1002/eji.201343800
- Dinarello CA, Ikejima T, Warner SJ, Orencole SF, Lonnemann G, Cannon JG, Libby P. Interleukin 1 induces interleukin 1. I. Induction of circulating interleukin 1 in rabbits in vivo and in human mononuclear cells in vitro. J Immunol Baltim Md 1950 1987; 139:1902-10
- Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev 2008; 223:20-38; PMID:18613828; http://dx.doi.org/10.1111/j.1600-065X.2008.00624.x
- Gringhuis SI, Kaptein TM, Wevers BA, Theelen B, van der Vlist M, Boekhout T, Geijtenbeek TBH. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1β via a noncanonical caspase-8 inflammasome. Nat Immunol 2012; 13:246-54; PMID:22267217; http://dx.doi.org/10.1038/ni.2222
- Akhter A, Caution K, Abu Khweek A, Tazi M, Abdulrahman BA, Abdelaziz DHA, Voss OH, Doseff AI, Hassan H, Azad AK, et al. Caspase-11 promotes the fusion of phagosomes harboring pathogenic bacteria with lysosomes by modulating actin polymerization. Immunity 2012; 37:35-47; PMID:22658523; http://dx.doi.org/10.1016/j.immuni.2012.05.001
- Kryczek I, Wei S, Vatan L, Escara-Wilke J, Szeliga W, Keller ET, Zou W. Cutting edge: opposite effects of IL-1 and IL-2 on the regulation of IL-17+ T cell pool IL-1 subverts IL-2-mediated suppression. J Immunol 2007; 179:1423-6; PMID:17641006; http://dx.doi.org/10.4049/jimmunol.179.3.1423
- Baltazar Lde M, Santos PC, de Paula TP, Rachid MA, Cisalpino PS, Souza DG, Santos DA. IFN-γ impairs Trichophyton rubrum proliferation in a murine model of dermatophytosis through the production of IL-1β and reactive oxygen species. Med Mycol 2014; 52:293-302; PMID:24577006; http://dx.doi.org/10.1093/mmy/myt011
- Walker JA, Barlow JL, McKenzie ANJ. Innate lymphoid cells – how did we miss them? Nat Rev Immunol 2013; 13:75-87; PMID:23292121; http://dx.doi.org/10.1038/nri3349
- Gladiator A, Wangler N, Trautwein-Weidner K, LeibundGut-Landmann S. Cutting edge: IL-17-secreting innate lymphoid cells are essential for host defense against fungal infection. J Immunol 2012; 190:521-5; PMID:23255360; http://dx.doi.org/10.4049/jimmunol.1202924
- Kim HY, Lee HJ, Chang Y-J, Pichavant M, Shore SA, Fitzgerald KA, Iwakura Y, Israel E, Bolger K, Faul J, et al. Interleukin-17-producing innate lymphoid cells and the NLRP3 inflammasome facilitate obesity-associated airway hyperreactivity. Nat Med [Internet] 2013 [cited 2014 Jan 8]; Available from: http://www.nature.com/doifinder/10.1038/nm.3423
- Katayama M, Ohmura K, Yukawa N, Terao C, Hashimoto M, Yoshifuji H, Kawabata D, Fujii T, Iwakura Y, Mimori T. Neutrophils are essential as a source of Il-17 in the effector phase of arthritis. PLoS One 2013; 8:e62231; PMID:23671588; http://dx.doi.org/10.1371/journal.pone.0062231
- Taylor PR, Roy S, Leal SM Jr, Sun Y, Howell SJ, Cobb BA, Li X, Pearlman E. Activation of neutrophils by autocrine IL-17A-IL-17RC interactions during fungal infection is regulated by IL-6, IL-23, RORγt and dectin-2. Nat Immunol 2014; 15:143-51; PMID:24362892; http://dx.doi.org/10.1038/ni.2797
