Abstract
Candida parapsilosis is a fungal pathogen that is associated with hematogenously disseminated disease in premature neonates, acutely ill or immunocompromised patients. In cell culture, C. parapsilosis cells are actively and avidly endocytosed by endothelial cells via actin polymerization mediated by N-WASP. Here we present evidence that C. parapsilosis that were internalized by endothelial cells remained alive, and avoided being acidified or otherwise damaged via the host cell. Internalized fungal cells reproduced intracellularly and eventually burst out of the host endothelial cell. When neutrophils were added to endothelium and C. parapsilosis, they patrolled the endothelial surface and efficiently killed most adherent fungal cells prior to endocytosis. But after endocytosis by endothelial cells, internalized fungal cells evaded neutrophil killing. Silencing endothelial N-WASP blocked endocytosis of C. parapsilosis and left fungal cells stranded on the cell surface, where they were susceptible to neutrophil killing. These observations suggest that for C. parapsilosis to escape from the bloodstream, fungi may adhere to and be internalized by endothelial cells before being confronted and phagocytosed by a patrolling leukocyte. Once internalized by endothelial cells, C. parapsilosis may safely replicate to cause further rounds of infection. Immunosurveillance of the intravascular lumen by leukocytes crawling on the endothelial surface and rapid killing of adherent yeast may play a major role in controlling C. parapsilosis dissemination and infected endothelial cells may be a significant reservoir for fungal persistence.
Abbreviations
| N-WASP | = | Neuronal Wiskott-Aldrich Syndrome protein |
| NETs | = | Neutrophil Extracellular Traps |
| HUVEC | = | Human Umbilical Vein Endothelial Cells |
| PMN | = | Polymorphonuclear leukocytes |
| YPD | = | Yeast Peptone Dextrose medium |
Introduction
Candida spp. fungi are widely distributed in the environment and are also present on skin and mucosal surfaces as a part of the apparently healthy commensal microbiome.Citation1 In immunocompromised or acutely ill patients, including premature neonates, Candida may become blood borne and cause disseminated disease, with high morbidity and mortality. Systemic candidiasis has been estimated to kill more than 15,000 people annually in the US.Citation2 Most disseminated Candida infections are due to C. albicans, however C. parapsilosis is emerging as a significant cause of sepsis in some populations such as low birthweight infants.Citation3-6
For circulating fungi to leave the bloodstream and invade tissues, they must cross the endothelial barrier. C. albicans can convert between a yeast form and a hyphal form. The filamentous hyphae adhere to endothelial cells, and penetrate and damage the endothelial monolayer.Citation7,8 However, C. albicans that has been trapped in the yeast form can still leave the blood stream to invade organs, albeit with reduced virulence.Citation9-12 C. parapsilosis is related to C. albicans, but does not form true hyphae; although it can form pseudohyphae in the presence of specific amino acids.Citation13
Circulating Candida is rapidly cleared from the bloodstream.Citation14 In mice, >90% of circulating C. albicans is cleared from the blood in a time scale of an hour or less.Citation15 This clearance may represent a combination of adhesion or entrapment of fungi along blood vessel surfaces, as well as uptake by host phagocytic mechanisms.Citation16 Several families of adhesion molecules have been identified in C. albicans, including the ALS, HWP and IFF/HYR families.Citation17 Expression of these proteins is typically induced upon hyphal formation. Als3p is highly expressed on C. albicans hyphae, and binds to E- or N-cadherin on host cells where it leads to endocytosis of fungi.Citation18,19 Ssa1 is another adhesion molecule that binds to host cadherins and leads to endocytosis.Citation20 Putative homologs to C. albicans adhesins have been identified in the C. parapsilosis genome, however their actual function has not been tested.Citation21 In C. glabrata, the Epa and Pwp families of adhesins have been identified, which bind to glycosyl ligands on epithelial and endothelial cells.Citation17
Phagocytic leukocytes in the blood are ideally positioned to provide the first line of defense against circulating pathogens.Citation22 Neutrophils are well known to play a major role against Candida.Citation2 Neutropenia or otherwise impaired neutrophil function are known risk factors for candidiasis, although perhaps less so for C. parapsilosis than other Candida spp.Citation23 In phagocytosis assays in vitro, neutrophils ingest C. parapsilosis more avidly than C. albicans yeast forms.Citation24 Phagocytic killing mechanisms may include reactive oxidative and non-oxidative pathways.Citation25,26 Hyphae of C. albicans are also killed by Neutrophil Extracellular Traps or NETs,Citation27-30 which consist of extruded chromatin with antimicrobial granule proteins bound to the DNA. Monocytes are another leukocyte lineage long known to phagocytose yeast.Citation31 After phagocytosis by monocyte-derived macrophages, live C. glabrata avoid being acidified.Citation32 C. glabrata have other adaptations to increase intracellular survival, and phagocytes may be an important reservoir for fungal persistence.Citation33,34
We previously observed that C. parapsilosis yeast cells were endocytosed over several hours by primary human umbilical vein endothelial cells (HUVEC). Heat-killed yeast cells were trafficked to an acidic compartment, and for both heat-killed and live yeast, endocytosis was dependent on endothelial N-WASP and the actin cytoskeleton.Citation35 Endothelial N-WASP was also involved in internalization of hyphae of C. albicans. At early time points, yeast adhered to the endothelial surface before becoming enshrouded in endothelial membranous sheet-like or lamellipodial projections, and eventually became completely internalized in the cytoplasm.
In vivo, endothelial cells are exposed to blood borne pathogens, however circulating phagocytes are abundant and undoubtedly mitigate interactions between the endothelium and microbes. In the current study we investigated C. parapsilosis yeast adherent to the endothelial cell surface as a target for neutrophil killing. We also examined the fate of live yeast that had been internalized by endothelial cells.
Materials and Methods
Yeast strain and media
C. parapsilosis invasive isolate 14–72931–101 (referred to here as JMB81) was used in this study.Citation36 This isolate was obtained from a premature infant blood culture between March 2004 and July 2007. Fungi were maintained on YPD agar (1% yeast extract, 2% peptone, 2% dextrose, 2% agar). Overnight (O/N) cultures were grown in YPD broth for 16 hours with vigorous agitation at 37°C. For heat killing, fungi were incubated at 65°C for 30 min with occasional mixing to reduce clumping. Heat-killed C. parapsilosis fungi were routinely cultured on YPD agar and incubated at 37°C overnight to confirm killing. For assays involving fluorescent detection, endothelial monolayers that had been co-incubated with fungi were labeled with 5 µM calcofluor white (Sigma, #F3543) in cell culture medium. For detection of Neutrophil Extracellular Traps (NETs), SYTOX Green at 5 uM was usedCitation28 to stain extracellular nucleic acids (Life Technologies, #S7020).
Endothelial cells
Primary human umbilical vein endothelial cells (HUVEC) (Lonza, #C2519A) were subcultured in EGM2 medium (without antibiotics or antimycotics) on gelatin coated tissue culture plastic dishes. All data were generated using passage 4 or 5 cells. In imaging experiments, HUVEC were grown on fibronectin coated coverslip slides or dishes (Nunc Labtek II #154534 and Mat-Tek #P35G-1.5–14-C).
Microscopy and imaging
Images were obtained with a Nikon inverted microscope (TE2000E) equipped with a charge-coupled-device (CCD) camera (Roper Coolsnap HQ) and excitation and emission filter wheels (Prior Proscan II). A stage incubator (20–20 Technologies Inc..) at 37°C and 5% CO2 was used for live cell imaging. Microscope control and image acquisition was via MetaVue 6.42r6 software (Molecular Devices). Images were overlaid and further analyzed in ImageJ 1.45 s (National Institutes of Health). DIC images were flatfielded to achieve contrast homogeneity and uniform backgrounds. The only adjustments to fluorescence images were linear changes in overall contrast and global changes in brightness. Figures were assembled and edited in Photoshop Elements 6.0 (Adobe Systems Inc.).
Neutrophil isolation
Neutrophils were isolated from whole blood according to the method of BoyumCitation37 with modifications. Whole blood was obtained from healthy human volunteers by venipuncture after approval by the Institutional Review Board. Heparinized blood was subjected to Ficoll-Paque density gradient centrifugation. Polymorphonuclear leukocytes (PMN) were further purified by dextran sedimentation of the Ficoll pellet followed by hypotonic lysis of contaminating erythrocytes. The resulting population was typically >90% pure by modified Wright Giemsa stain. PMN were re-suspended in HBSS +/+ and kept on ice for less than 1 hour before being used in yeast killing assays and/or live imaging experiments.
Killing assays
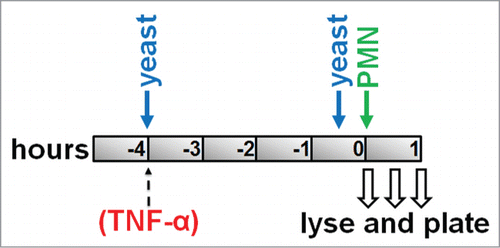
Killing assays were performed on HUVEC monolayers grown to postconfluence in 6-well tissue culture plates. To prevent experimental variation due to loss of neutrophil activity over time, killing experiments were designed so that fungi were added at different time points but neutrophils were only added once (). A ratio of 1 HUVEC:3 yeast cells:9 PMN was used. C. parapsilosis (from overnight culture in YPD) was washed 3x in DPBS −/−, and then added to appropriate wells in EGM2 to mark the start of the experimental time course at t = −4 hours (). Over the next 4 hours, some of these fungi were internalized by endothelial cells as previously described.Citation35 Freshly prepared C. parapsilosis cells were also added at t = −0.5 hours to a different set of wells. At this time point, fungi adhere firmly to the endothelial surface, but almost no internalization occurs.Citation35 At t = 0, all wells were washed 3 times with DPBS +/+ to remove excess non-internalized/non-adherent fungi, then freshly isolated PMN were added (). Surviving yeast were then counted for adherent or internalized yeast in an identical method. Immediately after PMN addition, 30 and 60 minutes later, media was removed by aspiration, and endothelial cells and PMN were lysed by scraping in one milliliter of cold water. Some wells did not receive any PMN to measure total number of adherent or internalized fungi (no PMN controls). Following harvesting of wells, lysates were diluted and plated on YPD agar. Plates were incubated for 16–18 hours at 37°C and then C. parapsilosis colonies were imaged under white light epi-illumination (Chemidoc XRS, Biorad). Image stacks were imported into ImageJ 1.45 s where colonies were thresholded and counted automatically for each plate using the particle analyzer plugin. Colony counts were normalized to no PMN controls and shown as percentages. The endothelial monolayer appeared damaged at time points beyond one hour, possibly due to non-specific damage by neutrophils. For killing assays with TNFα, TNFα was diluted in EGM2 and added to appropriate wells at 25 ng/ml at time point t = −4 hours. Fungi were co-incubated with HUVEC in the presence of TNFα.
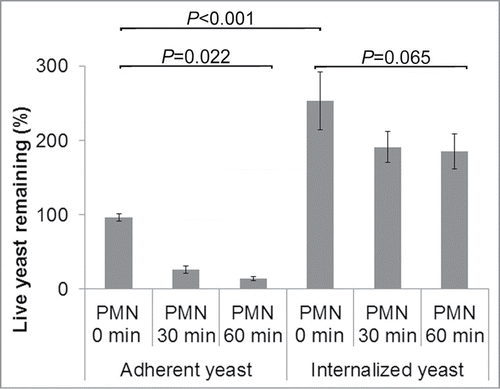
Figure 1. Killing assay timeline: Confluent monolayers of HUVEC were pre-incubated with C. parapsilosis for either 4 hours (to allow internalization) or 30 minutes (to only permit adhesion). Freshly isolated PMN were added at t = 0. Monolayers were lysed at specific time points and plated for surviving yeast. In some experiments, TNFα (25 ng ml−1) was added to HUVEC at the same time as yeast cells.
siN-WASP treatment
HUVEC were transiently transfected with equimolar pooled siRNA oligonucleotides, with maximum knockdown of N-WASP protein occurring at 72 hours post transfection as described previously.Citation35
Lysotracker
Live or heat-killed C. parapsilosis fungi were incubated with HUVEC overnight in EGM2 at a ratio of 3 yeast cells:1 HUVEC. Post-incubation, wells were washed in HBSS+/+ and stained with Lysotracker red to highlight acidified endosomes, and calcofluor white to highlight external yeast. Wells were then imaged with a 40x Plan-Apo lens. Extracellular calcofluor+ fungi were excluded from analysis. Internal fungi (calcofluor−) were counted manually for each condition and scored as “acidified” (Lyso+) or “non-acidified” (Lyso−). The sum of (Lyso+) and (Lyso−) is equal to the total number of internalized C. parapsilosis cells.
Live imaging of internalized C. parapsilosis budding and PMN Phagocytosis
HUVEC were grown to confluence in a 35mm coverglass bottom dish (MatTek). Fungi were added at 3 fungal cells per HUVEC (as in the killing assays) and co-incubated for 4 hours at 37°C and 5% CO2. Following this incubation, wells were aspirated, washed with DPBS −/− to remove excess non-adherent fungi, and then stained with calcofluor white in EGM2 to highlight external yeast. Time lapse fluorescent and DIC imaging was then performed for varying periods of time as previously described,Citation35 Since HUVEC actively exclude calcofluor from their cytoplasm, external and adherent fungi take up the stain while internalized fungi are protected from staining and are negative. For imaging of fungal budding, calcofluor was used to distinguish external from internalized C. parapsilosis. For imaging of phagocytosis, calcofluor pre-labeled fungi were tracked during their phagocytosis by PMN. Imaging was with a Nikon 20× Plan Apo objective lens.
Statistics
Comparisons between groups were made by t-test for experiments involving 2 groups and one-way analysis of variance (ANOVA) for more than 2 groups. Between-group comparisons following ANOVA were made using the student Newman–Keuls test with P values <0.05 considered significant.
Results
Live and dead C. parapsilosis undergo differential trafficking
The fluorescent dyes Lysotracker Red DND-99 and calcofluor white were used to examine the fates of live vs. dead C. parapsilosis. Lysotracker is a fluorescent marker that stains acidic membranous compartments such as lysosomes. At the concentrations used, Lysotracker did not appreciably stain fungi, but did stain endothelial cells (for example, see calcofluor+ extracellular C. parapsilosis cells in , which were completely negative for Lysotracker). Calcofluor fluorescently stains fungal cell walls. Calcofluor is excluded from live endothelial cells, and so will not stain endocytosed yeast when added after endocytosis has occurred. Thus extracellular yeast cells may be eliminated from analysis as calcofluor+ (blue clusters in ).
Figure 2. (A) Heat-killed and live C. parapsilosis were differentially trafficked after internalization. Overlay of brightfield DIC (gray), red (Lysotracker) and blue (calcofluor) channel fluorescence. Internalized yeast cells were negative for calcofluor. Internalized heat-killed yeast cells typically were present in a Lysotracker+ compartment (arrows). Internalized live yeast cells on the other hand were rarely stained red but were in a Lysotracker− compartment (arrowheads). Extracellular yeast cells (adherent or free floating) were brightly stained with calcofluor, and were disregarded for analysis. (B) Quantification of trafficking. The number of internalized (calcofluor−) Lysotracker+ and Lysotracker− yeast cells were manually counted, and the number of LysoT+ yeast was expressed as a percentage of the total (left panel). The right panel shows the number of internalized yeast per endothelial cell for heat-killed vs. live. The data represents the mean and SEM of 10 fields of view from each of 3 independent experiments.
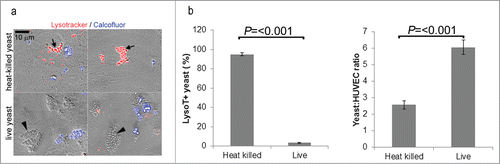
Endothelial monolayers were incubated with heat-killed or live C. parapsilosis overnight in EGM2 (containing serum). The next day monolayers were washed gently to remove non-adherent fungi, and stained with Lysotracker and calcofluor. shows an overlay of calcofluor (blue channel) and Lysotracker (red channel) fluorescence with DIC (Differential Interference Contrast, gray scale). With heat-killed C. parapsilosis, intracellular yeast cells (calcofluor-) were observed in a lysosomal (Lysotracker+) compartment (arrows, upper panels). In contrast, intracellular live yeast cells (calcofluor−) were present in a Lysotracker− compartment (arrowheads, lower panels). In quantification of multiple fields of view (FOV), 95% of internalized heat-killed C. parapsilosis cells were in an acidic endothelial subcompartment, but live fungi were rarely in an acidic compartment (3%; ). We also observed that live fungi were present in greater numbers than heat-killed yeast after overnight incubation (). This may be due to more efficient uptake or proliferation of live yeast.
Live C. parapsilosis cells replicate within endothelial cells
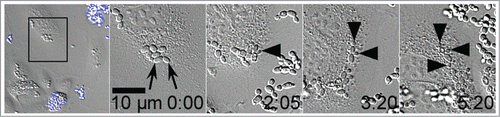
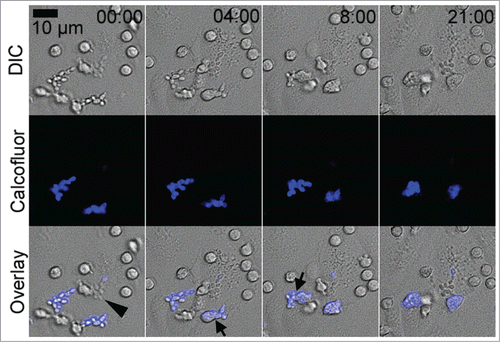
We had previously shown that live and heat-killed C. parapsilosis was taken up by endothelial cells between 4 and 24 hours.Citation35 Although smaller clusters of yeast cells were visible at 4 hours, and larger clusters after 24 hours, we attributed this increase in size to ongoing endothelial endocytosis, since both live and heat-killed yeast showed similar enlargement of clusters. C. glabrata is known to replicate within host cells.Citation32,38-40 To more critically examine if internalized live C. parapsilosis cells became dormant or remained active, we used live cell imaging, focusing on the period after active endocytosis had occurred. Endothelial monolayers and live yeast cells were incubated together in EGM2 medium with serum for 4 hours to allow fungal uptake to occur. Free-floating fungi were removed by gentle washing and culture dishes were then imaged in a live-cell imaging chamber in EGM2 medium containing Calcofluor (). Yeast cells were confirmed to be completely intracellular by negative staining (calcofluor−). Panel 1 shows a wide field of view, with clusters of calcofluor+ extracellular yeast cells at the top and bottom of the frame (). A cluster of calcofluor− yeast cells in the black rectangle is followed over the next 4 panels. Individual yeast cells within the cluster were observed frequently forming buds (), indicating that the fungi were actively replicating within their intracellular compartment, while protected from contact with calcofluor. Note that in an unusually well spread endothelial cell is shown where yeast budding is clearly visible. Despite its spread morphology, the host endothelial cell migrated toward the bottom right corner over this time period indicating that it was still alive and actively motile. In most endothelial cells, internalized C. parapsilosis cells were present in a cluster or ball making difficult the quantification of yeast cells or unambiguous evidence for budding (for example see , live yeast cells).
Figure 3. Live yeast replicated within endothelial cells. HUVEC were preincubated with C. parapsilosis for 4 hours to allow internalization to occur prior to imaging. Panel 1 shows a wide field of view at t = 0. Extracellular yeast are brightly stained blue with calcofluor and disregarded. Intracellular yeast are visible as gray ovals. Panel 2 shows a higher magnification of the area marked by a frame in the previous panel. The two arrows indicate the pair of yeast cells that will bud over the next frames. The time stamp is in hours and minutes, and does not include the 4 hour preincubation time. Arrowheads track 3 budding events in frames 3, 4 and 5. Image sequences are representative of 3 independent experiments. Data are also shown as Movie S1, where the budding process may be observed.
Internalized yeast cells may escape by bursting out of host endothelial cells
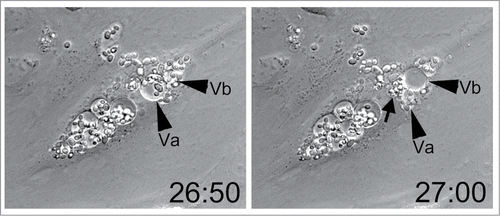
After prolonged co-incubation with endothelial cells C. parapsilosis cells were sometimes observed within fluid vesicles (). These vesicles were occasionally observed to undergo rapid changes in volume and shape, appearing to burst or deflate and release the cluster of yeast contained within them. Vesicle bursting was observed 24–48 hours after endocytosis, and occurred apparently at random in a few infected endothelial cells. Similar vesicles were never observed in uninfected endothelial cells. Vesicle bursting may represent an escape mechanism for endocytosed and proliferating C. parapsilosis. Endothelial cells appear to remain motile during this process, implying that release of yeast cells is not immediately lethal (Movie S2). A major technical limitation in these imaging studies is the long time periods required for yeast intracellular replication and escape and the concomitant proliferation of extracellular yeast in the culture medium. This overgrowth obscures the underlying endothelium and makes imaging difficult.
Figure 4. Live C. parapsilosis escaped from endothelial vesicles. After prolonged internalization, yeast cells were occasionally observed in fluid filled vesicles (arrowheads, Va and Vb). Ten minutes later, Va has collapsed, while Vb has enlarged. A cluster of newly expelled yeast cells is visible at the arrow. The time stamp is in hours and minutes and was initiated at the time of addition of live yeast cells to HUVEC. Data are also shown as Movie S2.
Neutrophils efficiently kill adherent but not endocytosed yeast cells
Neutrophils isolated from blood tend to spontaneously activate over time. To avoid this loss of function, a killing assay was designed to use PMN directly after isolation (). Yeast cells were incubated with confluent endothelial monolayers for 4 hours (in EGM2 medium containing serum allowing significant internalization by endothelial cells to occur) or for 30 minutes (allowing endothelial adhesion but minimal internalization). Monolayers were washed in DPBS+/+ to remove any free-floating fungi, and freshly isolated PMN were added (in DPBS+/+). Immediately, or after 30 or 60 minutes, monolayers were washed again and endothelial cells and neutrophils were hypotonically lysed in water, and surviving yeast were quantified by plating and colony count (). Colony counts were normalized to the number of colonies in the absence of any neutrophils (100%). For adherent yeast cells, addition of PMN for 30 or 60 minutes showed rapid killing of fungi, with only 14% surviving after 1 hour coincubation (). In contrast, after yeast cells had been allowed to become internalized over 4 hours, they became largely resistant to killing (73% survival at 1 hour). Internalized yeast cells also showed 2.5-fold increase due to proliferation over 4 hours.
Figure 5. Killing of adherent C. parapsilosis. PMN efficiently killed adherent yeast cells (14% remaining at 1 h, P = 0 .022), but killing of internalized yeast was much less efficient (73% remaining, P = 0 .065). The number of live yeast cells remaining from each condition was measured by plating and colony count, with 100% representing the colony count in the absence of PMN for adherent yeast cells. For adherent C. parapsilosis most of the killing occurred in the first 30 minutes of neutrophil treatment. For internalized C. parapsilosis, relatively few yeast cells were killed even after 1 hour PMN treatment. During the 4 hours of the internalization process, C. parapsilosis replicated within endothelial cells, as can be seen by the increased live yeast remaining (2.5-fold increase). Data represent means and SEMs from 7 independent experiments.
Internalization of C. parapsilosis by HUVEC did not result in endothelial expression of E-selectin, ICAM-1, VCAM-1
Resting HUVEC express extremely low levels of adhesion molecules such as E-selectin, ICAM-1, or VCAM. Their expression is upregulated in response to inflammatory factors such as TNFα. Infection with C. parapsilosis did not result in their significant upregulation at the protein level by Western blot (). Similar lack of strong upregulation was obtained by indirect immunofluorescence for E-selectin, ICAM and VCAM, as well as for N-cadherin (). Occasional increases in E-selectin were observed in C. parapsilosis colonized HUVEC, however this was not consistent, and much smaller in magnitude than compared to TNFα ().
Figure 6. (A) Infection with C. parapsilosis did not induce endothelial expression of ICAM-1, VCAM-1 or E-selectin at the protein level. Western blot of lysates from control endothelial cells (ctrl), cells treated with low doses of TNFα (1.25 ng/mL for 4 or 24 hours), or live C. parapsilosis for 4 or 24 hours. Standard TNFα (25 ng/ml) for 24 hours resulted in such high expression of ICAM, VCAM and E-selectin that small changes in their baseline expression could not be accurately measured. Note that at this concentration, ICAM-1 and VCAM-1 still showed strong upregulation with TNFα, but E-selectin showed only modest increases. The ICAM blot was subsequently reprobed for actin to confirm equal loading. (B) Infection with C. parapsilosis did not induce endothelial expression of ICAM-1, VCAM-1 or E-selectin by immunofluorescence. HUVEC were incubated overnight with C. parapsilosis, or for 4 hours with TNFα (25 ng/ml). Monolayers were fixed in neutral buffered formalin and stained by indirect immunofluorescence. (C) Effect of TNFα on killing. Endothelial cells were pretreated with TNFα (25 ng/mL) for 4 hours (as described in ), and the efficiency of killing measured as before. TNFα had no effect on killing efficiency for adherent yeast, but significantly reduced yeast internalization (P = 0 .005). Data represent means and SEM from 3 independent experiments.
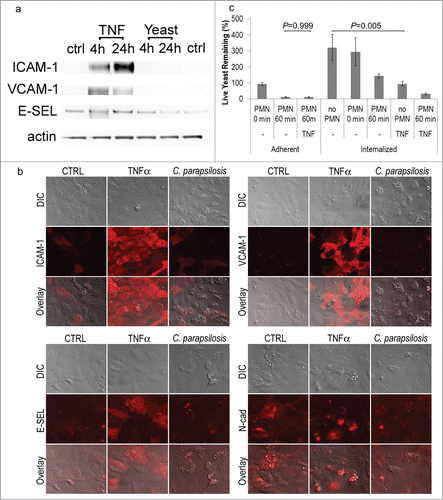
TNFα activation may reduce yeast internalization by HUVEC
To test the effect of inflammation on PMN adhesion, migration and killing, we treated endothelial monolayers with TNFα at the time of addition of yeast. For adherent yeast, the presence of TNFα made no significant difference in yeast survival (12%, ). For internalized yeast, although the presence of TNFα appeared to reduce survival at 1 hour post PMN contact from 45% to 11%, this was not statistically significant (P = 0.5). Interestingly, treatment with TNFα was observed to have a profound effect on yeast internalization by HUVEC (compare Internalized no PMN to Int/TNF no PMN). However, this decrease in colony count may be also be due to decreased proliferation of internalized yeast in the presence of TNFα.
Blocking endothelial N-WASP makes C. parapsilosis susceptible to neutrophil killing
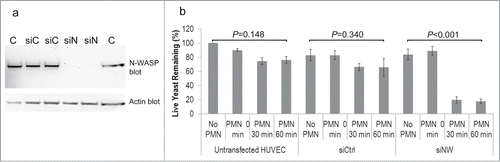
We have previously shown that endothelial N-WASP is required for endocytosis of C. parapsilosis yeast.Citation33 To determine whether blocking endocytosis would increase susceptibility of C. parapsilosis to neutrophils, N-WASP was silenced in HUVEC by transient transfection with siRNA (). Yeast were co-incubated with HUVEC for 4 hours, and neutrophils were added as before. Knockdown of N-WASP protein resulted in significantly increased vulnerability of yeast to killing by PMN at 60 minutes as compared to untransfected HUVEC or siRNA control ().
Figure 7. (A) Western blot showing that N-WASP was knocked down following transient transfection of HUVEC with siRNA to N-WASP (siN) but not by control siRNA (siC). (B) Silencing endothelial expression of N-WASP left yeast cells vulnerable to PMN killing. Untransfected HUVEC or control siRNA allowed yeast cells to escape neutrophil killing by endothelial internalization. Graph represents the means and SEM from 3 independent experiments.
Imaging neutrophil phagocytosis
In imaging experiments, HUVEC were incubated with yeast for 4 hours to allow internalization. Calcofluor was used to distinguish between extracellular yeast (calcofluor+) and endocytosed yeast (calcofluor−). Freshly isolated PMN were then added and their behavior was monitored using live cell imaging (). Approximately 25% of PMN were observed to flatten and crawl on the endothelium, while the rest remained rounded and did not move. Actively crawling PMN engulfed clusters of extracellular yeast (, calcofluor+, arrows), which could subsequently be observed packed tightly within the neutrophils. In contrast, yeast that were previously endocytosed by endothelial cells (, calcofluor−, arrowhead) did not alter neutrophil behavior. Instead, neutrophils migrated apically on the endothelial surface overlying the area of internalization with no apparent activation. SYTOX green is a fluorescent DNA stain that has been used to detect neutrophil extracellular traps (NETs).Citation28 In similar experiments, we observed no evidence of NET formation under these conditions (not shown).
Figure 8. PMN migrated on the endothelial monolayer to phagocytose adherent C. parapsilosis. Top row shows DIC, middle row shows calcofluor+ extracellular yeast cells, and the bottom row shows the overlay. Time is shown in minutes and seconds. Arrows show PMN in the process of engulfing a cluster of adherent yeast cells. Internalized yeast cells (calcofluor−, arrowhead) appeared to be ignored by apically migrating PMN. Data are also shown as movie 3 in the supplemental material.
Discussion
We had previously shown that heat-killed C. parapsilosis cells were endocytosed by endothelial cells, where they were transported to an acidic subcompartment, using the pH sensitive fluorescent dye, pHrodo.Citation35 Live yeast cells were taken up similarly by a mechanism that required endothelial actin polymerization and N-WASP activity. Other Candida spp have been shown to be endocytosed by host cells including immune phagocytes such as neutrophils and the monocyte/macrophage lineage,Citation2 as well as by epithelial and endothelial cells.Citation41 C. glabrata multiply within macrophages and interfere with phagolysosome acidification.Citation32,33 We show here that live C. parapsilosis cells likewise avoid acidification, whereas heat-killed C. parapsilosis cells become acidified. Our data also suggest that live C. parapsilosis may replicate within endothelial cells, and that after prolonged internalization, yeast may be observed bursting out of the host cell. In keeping with our results, it has recently been shown that C. parapsilosis can also replicate within macrophages after phagocytosis.Citation42
Endothelial monolayers have been previously shown to be damaged by germinating hyphae of C. albicans, and protected from damage by neutrophils.Citation43 Using videomicroscopy, neutrophils were observed to wrap around hyphae, and were described to be removing or even pulling them out of endothelial cells. To our knowledge, the equivalent experiment has never been conducted with a non-albicans species that lacks a true hyphal form. In the current study we show that PMN can patrol an endothelial surface and efficiently clear it of adherent C. parapsilosis cells () by phagocytosis (). However, if neutrophil clearance is delayed, some yeast cells will be endocytosed by endothelial cells (), where they remain alive and replicate, potentially allowing further rounds of infection (). Thus in vivo, leukocyte patrolling of the endothelial surface may be an important aspect of innate immunity toward yeast, and patrolling efficiency may be a significant factor in the dynamics of infection. Because blood vessels present a large surface area and perfuse most tissues, patrolling may represent significant effort for the innate immune system. Patrolling deficiencies such as leukopenia, problems in locomotion or killing may have severe consequences for the host if infected endothelial cells present a major reservoir for Candida persistence.
Figure 9. Model of intravascular surveillance and immune evasion. C. parapsilosis in the blood adheres to endothelial cells (1 and 2). A patrolling neutrophil can engulf and kill yeast cells on the surface (3). However, if yeast cells become internalized by endothelial cells, they are protected from PMN killing (4). Internalized C. parapsilosis may replicate and burst out of endothelial cells to cause further rounds of infection or be killed by patrolling neutrophils (5).
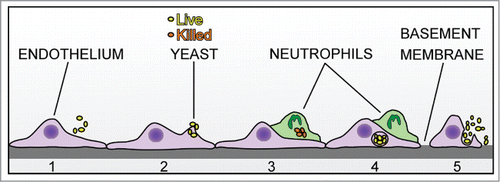
Our results suggest that neutrophils appear not to be able to detect intracellular C. parapsilosis antigens on the endothelial surface 4 hours after internalization, however it is possible that cells of the adaptive immune system may be capable of this. Detection and engulfment of apoptotic cells, or efferocytosis, is important in the immune response to other microbial pathogens.Citation44 It may be that neutrophils or monocytes may be able to detect endothelial damage at a much later time point, if damage occurs when endothelial cells are about to burst. Endothelial damage was not addressed in the current study, however, non-lytic release of Cryptococcus from host macrophages has been described by othersCitation45 and might possibly be occurring here.
Although PMN were used in the current study, it is possible that neutrophils are not the primary lineage responsible for intravascular immunity toward C. parapsilosis, or that different subpopulations are responsible for specific organs or anatomic locations. These may include CXCR3+ monocytes in the skin and kidney,Citation46,47 and invariant NK cells in the liver.Citation48
Our data provide some insight into the role of inflammatory mediators and adhesion molecules in this process. Interestingly TNFα treatment may decrease endocytosis of C. parapsilosis by endothelial cells, or decrease fungal proliferation. Similarly, uptake of C. albicans hyphae by endothelial cells is also inhibited by interferon-γ.Citation49 Knockdown of endothelial N-WASP also reduces efficiency of internalization of C. parapsilosis,Citation35 and increased PMN killing. These findings suggest that other strategies that might reduce endothelial internalization may have potential therapeutic value. We observed that in the absence of exogenous TNFα, HUVEC with internalized yeast did not strongly upregulate classic inflammatory adhesion molecules such as E-selectin, ICAM-1 or VCAM-1. A small upregulation of E-selectin was occasionally observed, however the significance of this is not currently clear. The low levels of inflammatory adhesion molecules suggests that C. parapsilosis may avoid activation of the endothelial NFκB pathway as a part of its immune evasion strategy. Despite the presence of low levels of E-selectin, ICAM and VCAM, neutrophils were still able to crawl on the endothelial surface and capture adherent yeast. Crawling leukocytes may instead be binding to constitutively expressed endothelial adhesion molecules such as ICAM-2. Citation50-52
The current experimental model does not examine the role of fluid shear on leukocyte recruitment. Although PMN crawling on the HUVEC monolayer was observed here under static conditions, it is not known if neutrophil crawling will occur under blood flow conditions in normal healthy humans. Mouse neutrophils have been observed to exhibit a basal level of intravascular crawling in certain models, but this may be due to surgical trauma due to tissue exteriorization for in vivo imaging.Citation53-55 Others have described low levels of basal leukocyte rolling in apparently unstimulated cultured endothelium.Citation56 It may be that leukocytes rolling in flow may make transient contact with adherent yeast during rolling and then switch to crawling mode. Another important question not addressed here is if PMN find adherent yeast by random migration, or is it directed by immobilized chemokine gradients or fungal pathogen-associated molecular patterns (PAMPs). Macrophages have been recently reported to exhibit directed migration toward C. albicans, C. glabrata and C. parapsilosis.Citation42
In the current study, efficient neutrophil killing was observed in the absence of serum (although serum was required for uptake by endothelial cells). However, in vivo clearance will occur in whole blood that contains antibodies or other opsonizing factors that may significantly modulate the neutrophil response. Other factors in whole blood that may play a role in yeast clearance may include interactions with other leukocyte subtypes. The presence of erythrocytes may also enhance neutrophil interactions with the vessel wall due to a crowding effect.Citation57
Extrusion of neutrophil chromatin in NETS has been described with C. albicans.Citation27,28,30 We observed frequent phagocytosis of adherent yeast cells, but no evidence of NETs for C. parapsilosis. Recent studies indicate that NETs may be preferentially used when the target is too large for ingestion.Citation58 It is possible that hyphal forms may elicit production of NETs, while the smaller yeast forms such as C. parapsilosis may be efficiently cleared by phagocytosis. Neutrophil degranulation is another mechanism whereby anti-microbial defense may be mounted, however its importance is unclear in the bloodstream, where soluble factors may be rapidly carried downstream.
In conclusion, we show that live C. parapsilosis yeast are efficiently killed on the endothelial surface by patrolling neutrophils. However, after endocytosis by endothelial cells, yeast avoid PMN killing and may safely replicate to cause further rounds of infection. Immunosurveillance of the intravascular lumen may play an important role in controlling dissemination of C. parapsilosis, and infected endothelial cells may be a significant reservoir for fungal persistence.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1042643_supplemental_movies__3_.zip
Download Zip (5.6 MB)Acknowledgments
We are grateful to the Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network for providing clinical isolates.
Funding
This work was supported by grants from the National Institute of General Medical Sciences (P20GM103537) and the National Heart, Lung, and Blood Institute (R21HL093561) of the National Institutes of Health.
References
- Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 2010; 6:e1000713; PMID:20072605; http://dx.doi.org/10.1371/journal.ppat.1000713.
- Lionakis MS. New insights into innate immune control of systemic candidiasis. Med Mycol 2014; 52:555-64; PMID:25023483; http://dx.doi.org/10.1093/mmy/myu029.
- Trofa D, Gacser A, Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev 2008; 21:606-25; PMID:18854483; http://dx.doi.org/10.1128/CMR.00013-08.
- Neu N, Malik M, Lunding A, Whittier S, Alba L, Kubin C, Saiman L. Epidemiology of candidemia at a Children's hospital, 2002 to 2006. Pediatr Infect Dis J 2009; 28:806-9; PMID:19636286; http://dx.doi.org/10.1097/INF.0b013e3181a0d78d.
- Chow BD, Linden JR, Bliss JM. Candida parapsilosis and the neonate: epidemiology, virulence and host defense in a unique patient setting. Expert Rev Anti Infect Ther 2012; 10:935-46; PMID:23030332; http://dx.doi.org/10.1586/eri.12.74.
- Pammi M, Holland L, Butler G, Gacser A, Bliss JM. Candida parapsilosis is a significant neonatal pathogen: a systematic review and meta-analysis. Pediatr Infect Dis J 2013; 32:e206-16; PMID:23340551; http://dx.doi.org/10.1097/INF.0b013e3182863a1c.
- Ibrahim AS, Filler SG, Ghannoum MA, Edwards JE, Jr. Interferon-gamma protects endothelial cells from damage by Candida albicans. J Infect Dis 1993; 167:1467-70; PMID:8501342; http://dx.doi.org/10.1093/infdis/167.6.1467.
- Filler SG, Swerdloff JN, Hobbs C, Luckett PM. Penetration and damage of endothelial cells by Candida albicans. Infect Immun 1995; 63:976-83; PMID:7868270.
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell 1997; 90:939-49; PMID:9298905; http://dx.doi.org/10.1016/S0092-8674(00)80358-X.
- Bendel CM, Hess DJ, Garni RM, Henry-Stanley M, Wells CL. Comparative virulence of Candida albicans yeast and filamentous forms in orally and intravenously inoculated mice. Crit Care Med 2003; 31:501-7; PMID:12576958; http://dx.doi.org/10.1097/01.CCM.0000049954.48239.A1.
- Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell 2003; 2:1053-60; PMID:14555488; http://dx.doi.org/10.1128/EC.2.5.1053-1060.2003.
- Spellberg B, Johnston D, Phan QT, Edwards JE, Jr., French SW, Ibrahim AS, Filler SG. Parenchymal organ, and not splenic, immunity correlates with host survival during disseminated candidiasis. Infect Immun 2003; 71:5756-64; PMID:14500497; http://dx.doi.org/10.1128/IAI.71.10.5756-5764.2003.
- Kim SK, El Bissati K, Ben Mamoun C. Amino acids mediate colony and cell differentiation in the fungal pathogen Candida parapsilosis. Microbiology 2006; 152:2885-94; PMID:17005970; http://dx.doi.org/10.1099/mic.0.29180-0.
- Sawyer RT, Moon RJ, Beneke ES. Hepatic clearance of Candida albicans in rats. Infect Immun 1976; 14:1348-55; PMID:793993.
- MacCallum DM, Odds FC. Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses 2005; 48:151-61; PMID:15842329; http://dx.doi.org/10.1111/j.1439-0507.2005.01121.x.
- Mavor AL, Thewes S, Hube B. Systemic fungal infections caused by Candida species: epidemiology, infection process and virulence attributes. Curr Drug Targets 2005; 6:863-74; PMID:16375670; http://dx.doi.org/10.2174/138945005774912735.
- de Groot PW, Bader O, de Boer AD, Weig M, Chauhan N. Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot Cell 2013; 12:470-81; PMID:23397570; http://dx.doi.org/10.1128/EC.00364-12.
- Liu Y, Filler SG. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell 2011; 10:168-73; PMID:21115738; http://dx.doi.org/10.1128/EC.00279-10.
- Yang W, Yan L, Wu C, Zhao X, Tang J. Fungal invasion of epithelial cells. Microbiol Res 2014; 169:803-10; PMID:24670964; http://dx.doi.org/10.1016/j.micres.2014.02.013.
- Sun JN, Solis NV, Phan QT, Bajwa JS, Kashleva H, Thompson A, Liu Y, Dongari-Bagtzoglou A, Edgerton M, Filler SG. Host cell invasion and virulence mediated by Candida albicans Ssa1. PLoS Pathog 2010; 6:e1001181; PMID:21085601; http://dx.doi.org/10.1371/journal.ppat.1001181.
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009; 459:657-62; PMID:19465905; http://dx.doi.org/10.1038/nature08064.
- Hickey MJ, Kubes P. Intravascular immunity: the host-pathogen encounter in blood vessels. Nat Rev Immunol 2009; 9:364-75; PMID:19390567; http://dx.doi.org/10.1038/nri2532.
- Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, Marr KA, Pfaller MA, Chang CH, Webster KM. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis 2009; 48:1695-703; PMID:19441981; http://dx.doi.org/10.1086/599039.
- Linden JR, Maccani MA, Laforce-Nesbitt SS, Bliss JM. High efficiency opsonin-independent phagocytosis of Candida parapsilosis by human neutrophils. Med Mycol 2010; 48:355-64; PMID:19672781; http://dx.doi.org/10.3109/13693780903164566.
- Gazendam RP, van Hamme JL, Tool AT, van Houdt M, Verkuijlen PJ, Herbst M, Liese JG, van de Veerdonk FL, Roos D, van den Berg TK, et al. Two independent killing mechanisms of Candida albicans by human neutrophils: evidence from innate immunity defects. Blood 2014; 124:590-7; PMID:24948657; http://dx.doi.org/10.1182/blood-2014-01-551473.
- Kaloriti D, Jacobsen M, Yin Z, Patterson M, Tillmann A, Smith DA, Cook E, You T, Grimm MJ, Bohovych I, et al. Mechanisms underlying the exquisite sensitivity of Candida albicans to combinatorial cationic and oxidative stress that enhances the potent fungicidal activity of phagocytes. MBio 2014; 5:e01334-14; PMID:25028425; http://dx.doi.org/10.1128/mBio.01334-14.
- Urban CF, Reichard U, Brinkmann V, Zychlinsky A. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol 2006; 8:668-76; PMID:16548892; http://dx.doi.org/10.1111/j.1462-5822.2005.00659.x.
- Byrd AS, O'Brien XM, Johnson CM, Lavigne LM, Reichner JS. An extracellular matrix-based mechanism of rapid neutrophil extracellular trap formation in response to Candida albicans. J Immunol 2013; 190:4136-48; PMID:23509360; http://dx.doi.org/10.4049/jimmunol.1202671.
- Bianchi M, Hakkim A, Brinkmann V, Siler U, Seger RA, Zychlinsky A, Reichenbach J. Restoration of NET formation by gene therapy in CGD controls aspergillosis. Blood 2009; 114:2619-22; PMID:19541821; http://dx.doi.org/10.1182/blood-2009-05-221606.
- Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, Brinkmann V, Jungblut PR, Zychlinsky A. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog 2009; 5:e1000639; PMID:19876394; http://dx.doi.org/10.1371/journal.ppat.1000639.
- Cline MJ, Lehrer RI. Phagocytosis by human monocytes. Blood 1968; 32:423-35; PMID:4877597.
- Seider K, Brunke S, Schild L, Jablonowski N, Wilson D, Majer O, Barz D, Haas A, Kuchler K, Schaller M, et al. The facultative intracellular pathogen Candida glabrata subverts macrophage cytokine production and phagolysosome maturation. J Immunol 2011; 187:3072-86; PMID:21849684; http://dx.doi.org/10.4049/jimmunol.1003730.
- Seider K, Gerwien F, Kasper L, Allert S, Brunke S, Jablonowski N, Schwarzmuller T, Barz D, Rupp S, Kuchler K, et al. Immune evasion, stress resistance, and efficient nutrient acquisition are crucial for intracellular survival of Candida glabrata within macrophages. Eukaryot Cell 2014; 13:170-83; PMID:24363366; http://dx.doi.org/10.1128/EC.00262-13.
- Kasper L, Seider K, Gerwien F, Allert S, Brunke S, Schwarzmuller T, Ames L, Zubiria-Barrera C, Mansour MK, Becken U, et al. Identification of Candida glabrata genes involved in pH modulation and modification of the phagosomal environment in macrophages. PLoS One 2014; 9:e96015; PMID:24789333; http://dx.doi.org/10.1371/journal.pone.0096015.
- Shintaku T, Glass KA, Hirakawa MP, Longley SJ, Bennett RJ, Bliss JM, Shaw SK. Human endothelial cells internalize Candida parapsilosis via N-WASP-mediated endocytosis. Infect Immun 2013; 81:2777-87; PMID:23690407; http://dx.doi.org/10.1128/IAI.00535-13.
- Benjamin DK, Jr., Stoll BJ, Gantz MG, Walsh MC, Sanchez PJ, Das A, Shankaran S, Higgins RD, Auten KJ, Miller NA, et al. Neonatal candidiasis: epidemiology, risk factors, and clinical judgment. Pediatrics 2010; 126:e865-73; PMID:20876174; http://dx.doi.org/10.1542/peds.2009-3412.
- Boyum A. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand J Clin Lab Invest Suppl 1968; 97:77-89; PMID:4179068.
- Kaur R, Ma B, Cormack BP. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc Natl Acad Sci U S A 2007; 104:7628-33; PMID:17456602; http://dx.doi.org/10.1073/pnas.0611195104.
- Otto V, Howard DH. Further studies on the intracellular behavior of Torulopsis glabrata. Infect Immun 1976; 14:433-8; PMID:987021.
- Roetzer A, Gratz N, Kovarik P, Schuller C. Autophagy supports Candida glabrata survival during phagocytosis. Cell Microbiol 2010; 12:199-216; PMID:19811500; http://dx.doi.org/10.1111/j.1462-5822.2009.01391.x.
- Sheppard DC, Filler SG. Host Cell Invasion by Medically Important Fungi. Cold Spring Harb Perspect Med 2014; 5:a019687; PMID:25367974.
- Toth R, Toth A, Papp C, Jankovics F, Vagvolgyi C, Alonso MF, Bain JM, Erwig LP, Gacser A. Kinetic studies of Candida parapsilosis phagocytosis by macrophages and detection of intracellular survival mechanisms. Front Microbiol 2014; 5:633; PMID:25477874.
- Edwards JE, Jr., Rotrosen D, Fontaine JW, Haudenschild CC, Diamond RD. Neutrophil-mediated protection of cultured human vascular endothelial cells from damage by growing Candida albicans hyphae. Blood 1987; 69:1450-7; PMID:3552077.
- Martin CJ, Peters KN, Behar SM. Macrophages clean up: efferocytosis and microbial control. Curr Opin Microbiol 2014; 17:17-23; PMID:24581688; http://dx.doi.org/10.1016/j.mib.2013.10.007.
- Stukes SA, Cohen HW, Casadevall A. Temporal kinetics and quantitative analysis of Cryptococcus neoformans nonlytic exocytosis. Infect Immun 2014; 82:2059-67; PMID:24595144; http://dx.doi.org/10.1128/IAI.01503-14.
- Auffray C, Fogg D, Garfa M, Elain G, Join-Lambert O, Kayal S, Sarnacki S, Cumano A, Lauvau G, Geissmann F. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science 2007; 317:666-70; PMID:17673663; http://dx.doi.org/10.1126/science.1142883.
- Lionakis MS, Swamydas M, Fischer BG, Plantinga TS, Johnson MD, Jaeger M, Green NM, Masedunskas A, Weigert R, Mikelis C, et al. CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. J Clin Invest 2013; 123:5035-51; PMID:24177428; http://dx.doi.org/10.1172/JCI71307.
- Lee WY, Moriarty TJ, Wong CH, Zhou H, Strieter RM, van Rooijen N, Chaconas G, Kubes P. An intravascular immune response to Borrelia burgdorferi involves Kupffer cells and iNKT cells. Nat Immunol 2010; 11:295-302; PMID:20228796; http://dx.doi.org/10.1038/ni.1855.
- Fratti RA, Ghannoum MA, Edwards JE, Jr., Filler SG. Gamma interferon protects endothelial cells from damage by Candida albicans by inhibiting endothelial cell phagocytosis. Infect Immun 1996; 64:4714-8; PMID:8890230.
- de Fougerolles AR, Stacker SA, Schwarting R, Springer TA. Characterization of ICAM-2 and evidence for a third counter-receptor for LFA-1. J Exp Med 1991; 174:253-67; PMID:1676048; http://dx.doi.org/10.1084/jem.174.1.253.
- Schenkel AR, Mamdouh Z, Muller WA. Locomotion of monocytes on endothelium is a critical step during extravasation. Nat Immunol 2004; 5:393-400; PMID:15021878; http://dx.doi.org/10.1038/ni1051.
- Halai K, Whiteford J, Ma B, Nourshargh S, Woodfin A. ICAM-2 facilitates luminal interactions between neutrophils and endothelial cells in vivo. J Cell Sci 2014; 127:620-9; PMID:24317296; http://dx.doi.org/10.1242/jcs.137463.
- Wojciechowski JC, Sarelius IH. Preferential binding of leukocytes to the endothelial junction region in venules in situ. Microcirculation 2005; 12:349-59; PMID:16020081; http://dx.doi.org/10.1080/10739680590934763.
- Phillipson M, Heit B, Colarusso P, Liu L, Ballantyne CM, Kubes P. Intraluminal crawling of neutrophils to emigration sites: a molecularly distinct process from adhesion in the recruitment cascade. J Exp Med 2006; 203:2569-75; PMID:17116736; http://dx.doi.org/10.1084/jem.20060925.
- Sumagin R, Prizant H, Lomakina E, Waugh RE, Sarelius IH. LFA-1 and Mac-1 define characteristically different intralumenal crawling and emigration patterns for monocytes and neutrophils in situ. J Immunol 2010; 185:7057-66; PMID:21037096; http://dx.doi.org/10.4049/jimmunol.1001638.
- Patel KD. Mechanisms of selective leukocyte recruitment from whole blood on cytokine-activated endothelial cells under flow conditions. J Immunol 1999; 162:6209-16; PMID:10229866.
- Migliorini C, Qian Y, Chen H, Brown EB, Jain RK, Munn LL. Red blood cells augment leukocyte rolling in a virtual blood vessel. Biophys J 2002; 83:1834-41; PMID:12324405; http://dx.doi.org/10.1016/S0006-3495(02)73948-9.
- Branzk N, Lubojemska A, Hardison SE, Wang Q, Gutierrez MG, Brown GD, Papayannopoulos V. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat Immunol 2014; 15:1017-25; PMID:25217981; http://dx.doi.org/10.1038/ni.2987.
