Abstract
K-12 Escherichia coli cells grown in static media containing a critical phosphate (Pi) concentration ≥25 mM maintained a high polyphosphate (polyP) level in stationary phase, impairing biofilm formation, a phenomenon that is triggered by polyP degradation. Pi concentration in human urine fluctuates according to health state. Here, the influence of environmental Pi concentration on the occurrence of virulence traits in uropathogenic E. coli (UPEC) isolated from acute prostatitis patients was evaluated. After a first screening, 3 isolates were selected according to differential biofilm formation profiles depending on media Pi concentration. For each isolate, biofilm positive and negative conditions were established. Regardless of the isolate, biofilm formation capacity was accompanied with curli and cellulose production and expression of some key virulence factors associated with adhesion. When the selected isolates were grown in their non-biofilm-forming condition, low concentrations of nalidixic acid and ciprofloxacin induced biofilm formation. Interestingly, similar to laboratory strains, polyP degradation induced biofilm formation in the selected isolates. Data demonstrated the complexity of UPEC responses to environmental Pi and the importance of polyP metabolism in the virulence of clinical isolates.
INTRODUCTION
Pathogenic Escherichia coli comprise a diverse group of strains associated with both intestinal and extraintestinal infections.Citation1 Uropathogenic E. coli (UPEC) is the predominant causative agent of urinary tract infections (UTIs), representing up to 85% of community acquired UTIs.Citation2 In comparison to commensal strains, UPEC has several virulence factors that allow it to colonize host mucosal surfaces, being important for establishing infection; these include adhesins, toxins, iron acquisition systems, and capsular antigens.Citation3 Biofilm formation may be considered another pathogenic determinant, which allows these strains to persist a long time in the genito-urinary tract and interfere with bacterial eradication by host defense mechanisms and antibiotics.Citation4,5
Curli fimbriae and/or cellulose are produced in E. coli, Salmonella spp. and other Enterobacteriaceae influencing the adherence properties of several biofilm-forming microorganisms.Citation6-10 As a virulence factor associated to biofilm formation, flagella, allows fixation of bacteria to epithelial cells.Citation11 In pathogenic bacteria such as Salmonella spp, E. coli, Bordetella bronchiseptica y Bordetella pertussis, it has been demonstrated that motility plays an essential role, mainly in initial phase of infection.Citation12,13
Inorganic polyphosphate (polyP) was shown to be critical for attributes such as motility, quorum sensing, biofilm formation, resistance to oxidative, osmotic, heat, nutritional and alkaline stress, and stationary-phase survival in several microorganisms.Citation14-23 PolyP is a linear chain of tens to many hundreds of phosphate residues linked by high-energy phosphoanhydride bonds.Citation14, 24 PolyP is usually accumulated during exponential phase of growth, and degraded at the beginning of stationary phase.Citation15 Previous reports from our laboratory shown that E. coli cells grown in static media containing a critical phosphate (Pi) concentration ≥25 mM maintained a high polyP level in stationary phase.Citation25 This characteristic was related to the impairment in biofilm formation, since polyP degradation is a cellular mechanism necessary to trigger this phenomenon.Citation25
Human urine Pi concentration ranges are between 0.02–2 mM, although values higher than 2 mM could be found depending on eating habits, age, muscle mass, renal function, and other factors.Citation26 Hyperphosphaturia, an excess of phosphate in urine, was described under several health disorders, such as cases of multiple myeloma, kidney diseases or excess of vitamin D in the body, among others.Citation27-33 Depending on the clinical case, Pi concentrations in urine from 20 to 50 mM were found.
In the present study, the capacity to form biofilm of clinical UPEC isolates from acute prostatitis patients was analyzed in response to the media Pi concentration. Also, the relationship between in vitro biofilm formation and expression/production of some virulence traits was determined.
RESULTS
Biofilm formation of uropathogenic E. coli isolates
A total of 55 UPEC strains causing acute prostatitis were isolated from 44 patients and analyzed for biofilm formation in media containing different Pi concentrations (2 to 60 mM). A total of 38 isolates were positive for in vitro biofilm formation. Out of these isolates, 16 isolates formed biofilm independently of media Pi concentration. However, 11 isolates increased biofilm formation with increasing Pi concentrations, 7 presented maximum biofilm formation in media containing 20 mM Pi, and 4 decreased biofilm formation with increasing Pi concentrations.
Considering previously reported data about Pi influence on biofilm formation capacity in laboratory strains, the present study was focused in those isolates in which biofilm formation capacity varied with Pi concentration. Thus, 3 isolates, named MGP16, MGP12 and MGP45, were selected according to differential biofilm formation profiles depending on media Pi concentration (). For MGP16, a decrease in biofilm formation was observed when Pi concentration increased, conversely to the behavior of MGP45 isolate. For MGP12, biofilm formation reached a maximum at 20 mM Pi decreasing at higher concentrations. For further studies, two Pi concentrations corresponding to biofilm positive (BF+) and biofilm negative (BF-) conditions were established for each of the 3 selected isolates (see ), according to results in .
Figure 1. Biofilm formation in different Pi concentration media. Selected isolates (MGP16, MGP12, and MGP45) were grown in static conditions for 48 h in M63 medium modified with the indicated Pi concentrations. Biofilm formation was quantified by cristal violet technique. Data represent the mean ± SD of at least 3 independent experiments. Different letters indicate significant differences according to Tukey's test with a p-value ≥ 0.05.
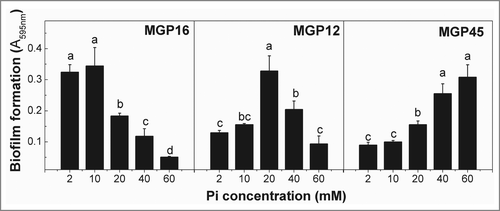
Table 1. Pi concentrations established to each biofilm formation conditions
Biofilm structure and related phenotypes
To evaluate biofilm structures formed by each isolate, confocal laser scanning microscopy (CLSM) and scanning electronic microscopy (SEM) microscopies were carried out. CLSM analysis allowed the reconstruction of 3D-images of biofilms from MGP45 and MGP12 strains (). In BF+ condition, MGP45 produced a biofilm thicker than the one from MGP12. For both strains, projections could be distinguished on the biofilm surfaces, giving them the appearance of a “Christmas tree forest.” (left panels) shows SEM images of the 3 isolates in BF+ conditions. In general, cells had sizes ranging between 1.2 and 2 μm in length. MGP16 and MGP12 showed similar biofilm phenotypes with abundant extracellular material, presenting the latter higher number of cells attached to the glass surface. At higher magnification, cells exhibited smooth or slightly rough surfaces, a great amount of extracellular vesicular material, and potential conjugating structures (arrows in inserts i and ii of ). In MGP45 biofilm, formation of protrusions that rise to the surface was observed (arrows). Using higher magnification, the presence of extracellular matrix was observed (arrow in insert iii). In addition, a peculiarity identified in this isolate was the presence of chains of bacilli as larger as 7 to 8 μm forming a nest in which most of the cells presented no septum (insert iv of ). Using CLSM and SEM, no structure was visualized in BF- conditions (right panels in ).
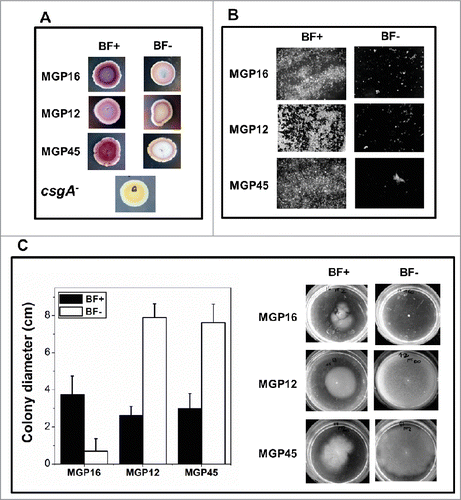
For further characterization, curli, cellulose and motility were analyzed as aspect related to the biofilm formation in the selected E. coli isolates. On Congo red plates (), the 3 isolates presented violet colonies in BF+ conditions, indicating rdar morpho-type which is characterized by curli fiber and cellulose production. In BF- conditions, isolates grew as pink colonies, indicative of pdar morphotype, characteristic of cells expressing only cellulose. Indeed, production of cellulose, cellulose-like, or extracellular material was observed in both BF+ and BF- conditions, given by a strong Calcofluor White fluorescence in biofilms and unattached single cells, respectively (). All isolates presented swimming motility in BF+ conditions (). It is noteworthy that MGP12 and MGP45 presented an exacerbated motility in BF- conditions and, conversely, no motility was observed for MGP16 in BF- ().
Figure 2. Microscopy analysis of E. coli isolates. Biofilm formation of the indicated E. coli isolates grown in static conditions for 48 h in M63 medium in BF+ and BF– conditions was analyzed (A) by CLSM and represented by 3-dimensional reconstruction of biofilm image or (B) by SEM microscopy, magnification: 5,000x. Inserts i and ii, 30,000x; insert iii, 80,000x; insert iv, 20,000x. Data are representative from 2 independent experiments.
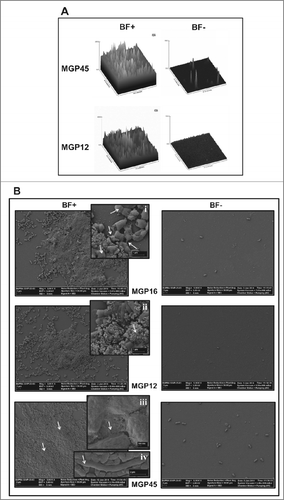
Figure 3. Biofilm associated phenotypes of E. coli isolates. (A) Aliquots of bacteria in an OD560nm = 0 .1 were spotted on solid M63 plates supplemented with different Pi concentrations (BF+ or BF- conditions). Plates containing Congo red dye and brilliant blue were incubated at 30°C for 48 h. The presence of curli fiber was observed as purple or red colonies. The colorless colony by csgA mutant, defective in an essential curli component, was used as negative control. (B) Bacteria were grown statically on glass covers in M63 liquid medium supplemented with the indicated Pi concentrations. After 48 h, attached cells were treated with Calcofluor White. The presence of cellulose was observed in an optical fluorescence microscope with 100x magnification. (C) Bacteria were grown for 48 h on semisolid 0.3% agar M63 plates with the indicated Pi concentrations. Motility was expressed as colony diameter. In all cases, results represent at least 4 independent experiments performed in duplicate.
Another isolate of each group was selected to determine curli and motility phenotypes (supplementary Fig. S1 A and B, top panels). The newly analyzed 3 isolates were MGP42 forming biofilm similarly to MGP16, MGP18 to MGP12 and MGP04 to MGP45. MGP42, MGP18 and MGP04 presents similar curli phenotypes than isolates from the corresponding group described above.
Virulence factor expression
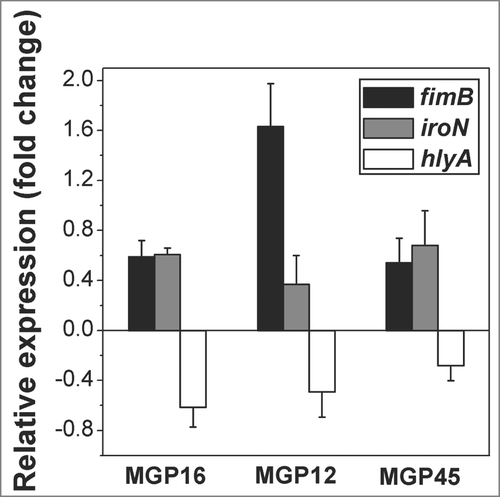
To assess whether selected known virulence genes (including adhesin-related encoding genes (fimB, fimA, papA, agn-43), siderophore-related genes (iroN, iutA), and toxin-encoding genes (cnf-1, hlyA) were present in the 3 isolates, PCR was used. fimB, agn-43, iroN, iutA, and hlyA, were detected in MGP16, MGP45 and MGP12, while fimA, papA, and cnf-1 were not found in any of the selected strains. Differential expression of genes related to each virulence trait, e.g. fimB, iroN, and hlyA, were determined by q-PCR in BF+ conditions relative to BF- conditions. In all isolates, fimB and iroN were induced, while hlyA was repressed ().
Figure 4. Relative expression of virulence factors related to biofilm formation. Expression of fimB, iroN and hlyA genes of the selected isolates was determined by q-PCR. Values are depicted as x-fold of up- or down-regulation of BF+ conditions compared to BF- conditions. Data represent the mean ± SD of at least 2 independent experiments performed in triplicate.
Antibiotic susceptibility
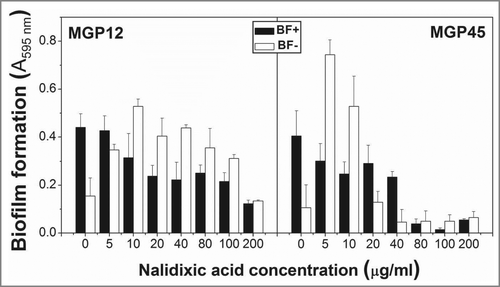
Nalidixic acid and ciprofloxacin susceptibility of the selected isolates was determined as MIC values. MGP12 and MGP16 isolates were found to be resistant up to 200 µg ml−1. For MGP45, MICs for ciprofloxacin and nalidixic acid were 1 and 30 μg ml−1, respectively. Additionally, the effect on biofilm formation of nalidixic acid added at zero time was tested in MGP12 and MGP45 isolates (). Biofilm formation was inhibited in BF+ conditions by high antibiotic concentration for both isolates, mainly in MGP45. In BF- conditions, low concentrations of antibiotic strongly induced biofilm formation.
Figure 5. Antibiotic resistance in static conditions. ON cultures of the selected isolates were diluted to an OD560nm=0 .1 and incubated in multiwell plates in M63 medium containing different Pi concentrations and the indicated nalidixic acid concentrations. Plates were incubated at 30°C for 48 h and biofilm formation was assessed by cristal violet staining. Results represent the mean ± SD of 3 different experiments.
Modulation of polyP levels related to biofilm formation
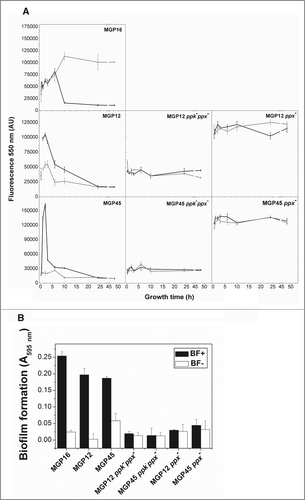
To analyze whether intracellular polyP was implicated in the biofilm formation phenotype of the selected isolates, polyP levels were measured in cells grown in BF+ and BF- conditions at 30°C during 48 h. Regarding the isolates grown in the BF+ conditions, polyP was synthesized and subsequently degraded in all cases (). Peaks of polyP levels were achieved between 6–10 h of growth for MGP16 isolate and in about 2 h for MGP12 and MGP45. It should be noted that in BF- conditions, isolates MGP12 and MGP45 were deficient in polyP synthesis during the entire growth curve and MGP16 was unable to degrade the polymer in stationary phase (). To further analyze the involvement of polyP metabolism in biofilm formation, polyP levels () and biofilm formation () were assayed in double mutants MGP12ppkppx and MGP45ppkppx and in the corresponding complemented strains (ppk+). Regarding the polymer levels, both ppkppx mutants were unable to reach the peak of polyP characteristic of parental strains in BF+ conditions (, middle panels). However, complemented strains were unable to degrade the polymer, showing elevated polyP levels in the entire growth curve in all tested conditions (, right panels). Note that all mutant strains did not show biofilm formation as the parental isolates (). As expected, polyP degradation was observed in MGP42, MGP18 and MGP04 isolates in biofilm forming conditions (Fig. S1C).
Figure 6. Intracellular polyP levels and biofilm formation in parental and mutants strains. (A) Cells of the indicated strains were grown for 48 h in static M63 medium modified with the Pi concentrations corresponding to BF+ (black line) and BF- (gray line) conditions. At the indicated time of growth, polyP was quantified using DAPI fluorescence, as described in Methods. DAPI emission was undetectable in cell free controls. (B) Indicated strains were grown in static conditions for 48 h in M63 medium modified with the indicated Pi concentrations corresponding to BF+ conditions (black columns) and to BF- conditions (white columns). Biofilm formation was quantified by cristal violet technique. Data are expressed as average ± SD of 4 independent experiments.
DISCUSSION
Here, a high variability among the capacities to form biofilms in vitro of UPEC isolated from prostatitis patients was observed, in agreement with previous studies with other pathogenic E. coli strains.Citation34–37 Remarkably, the ability of UPEC isolates to form biofilms was shown to be different according to medium Pi concentration.
Considering differential biofilm formation phenotype related to media Pi concentration, characterization of virulence aspects were carried out. Selected isolates were found to form prominent biofilms, presenting extracellular matrix or vesicular material. High quantities of extracellular material such as outer membrane vesicles are common among clinical isolates due to the necessity to secrete virulence effectors to attack the host.Citation38 In BF+ conditions, selected isolates presented rdar (red, dry and rough) morphotype, as reported by Römling et al.Citation39 This phenotype includes adhesion to abiotic surfaces (biofilm formation) and expression of curli fimbriae and cellulose as extracellular matrix components.Citation35 Thus, the capacity of these strains to form biofilms at solid-liquid interfaces would be a consequence of curli and cellulose production. Similarly, it was previously reported that expression of curli fimbriae and cellulose (rdar morphotypes) determine the medium or high biofilm-forming capacity of UTI isolates under at least one of the growth conditions tested.Citation35 On the other hand, the cellulose production found in BF- conditions is in agreement with the fact that cellulose is associated with both single and biofilm forming cells.Citation40, 41 Additionally, motility seems to be required for biofilm formation in the selected isolates. It is worth to note that MGP12 and MGP45 showed an exacerbated motility in BF- condition, which may lead to a reversible attachment of bacteria to the surface, as previously described by Dunne.Citation42 However, variability in respect to motility results within the isolates and their mutants among conditions (Fig. S1 B) suggest that this feature is not directly related to biofilm phenotype.
In E. coli, initiation, attachment and subsequent maturation of biofilm requires the expression of a set of genes, encoding a variety of virulence factors such as haemolysin, fimbriae, secreted proteins, capsules, and iron-acquisition systems, among others.Citation43,44 Consistently, iutA (not shown), fimB and iroN genes were induced in BF+ conditions in the selected isolates. Haemolysin was also expressed in the selected isolates but it was repressed in BF+ conditions. These results may be explained by the fact that, although associated to higher potential to form biofilm,Citation4,45 haemolysin is a trait involved in invasiveness and could be secreted in the planktonic lifestyle in order to injure host tissue and then colonize it by forming the biofilm. Unexpectedly, even when fimB was found in the genetic pool of the 3 isolates, fimA gene was not detected as the other virulence factors; this may be due to the loss of genes or pathogenicity islands in UPEC associated with different environmental conditions, according to Middendorf et al.Citation46 Induction of siderophore related genes in BF+ conditions indicate that UPEC may use a broad repertoire of systems to acquire iron in order to survive within the iron-limited urinary tract.Citation47 Interestingly, UPEC strains causative of asymptomatic bacteriuria, while lacking classical virulence factors such as fimbriae, express the full complement of iron acquisition systems, providing further evidence for the requirement of iron uptake during successful urinary tract colonization.Citation48
Fluoroquinolones have been the antimicrobial treatment of choice in febrile and acute UTIs.Citation49-52 Thus, susceptibility to nalidixic acid and ciprofloxacin was tested. From the 55 isolates tested, 83% and 36% were sensitive to ciprofloxacin and nalidixic acid, respectively. Biofilm producing strains were significantly more resistant than those unable to form biofilm (not shown). However, Soto et al.Citation49 described that strains able to form biofilms were significantly less resistant to nalidixic acid than non-biofilm producers. This discrepancy could be due to genome plasticity in pathogen isolates, which is responsible for the phenotypic diversity.Citation53 Interestingly, when antibiotics were added at low concentration in a BF- conditions, biofilm formation induction was achieved. Similar result were previously observed in Staphylococcus epidermidis isolates using macrolidesCitation54 and in Salmonella enteric serovar Typhimurium in which biofilm and EPS production was improved after treatment with sub-inhibitory concentrations of cefotaxime.Citation55 It is worth to note that, when antibiotic was added at 24 h, the above mentioned induction of biofilm formation was not observed in the BF- conditions (not shown).
Pathogenic strains associated with human diseases are remarkably diverse, reacting differently to environmental conditions.Citation56 E. coli has a collection of more than 30 genes, which respond to variation in media Pi concentrations, known as Pho regulon. This regulon is involved in transport and assimilation of phosphorylated compounds and its transcription is activated by Pi deficiency in the external environment.Citation57,58 E. coli posses two Pi transport systems, a high-affinity phosphate transport and low affinity Pi transporter called Pst and Pit system, respectively.Citation59 Inactivation of the Pst system attenuate virulence of both extraintestinal pathogenic E. coli (ExPEC) and enteropathogenic E. coli (EPEC) strainsCitation60–64 (7, 15, 19, 24, 42). In pst operon UPEC mutants, Pho regulon is activated even under phosphate-replete conditions, reducing colonization of the murine urinary tract.Citation65, 66
A very important aspect reported here is that, regardless of the media Pi concentration, biofilm formation in all of the isolates requires the presence and the degradation of polyP. These data were corroborated with mutant strains and are in agreement with previous results from our laboratory.Citation25 Further studies would be necessary to elucidate the regulatory mechanism involved in this process. Additionally, it would be interesting to consider the development of alternative therapies with ppk and/or ppx as target genes to modify synthesis or degradation of the polymer. Although the complexity of isolates genetic backgrounds, our study highlights the implication of Pi as a possible physiological signal to regulate biofilm phenotype in E. coli species.
METHODS
Bacterial strains and growth media
E. coli strains collected from acute prostatitis patients at the Hospital Clinic of Barcelona were analyzed. Patients with prostatitis had a mean age of 59.6 +/− 16 years-old and remained hospitalized for at least 24 h. All urinary tract infection episodes were community acquired and uncomplicated, which means that no patient had an underlying comorbidity, apparent urological abnormality or an urethral catheter in place. MGP12 ppk−ppx− and MGP45 ppk−ppx− were constructed by P1 phage transduction.Citation67 Complemented strains MGP12 ppk−ppx− (ppk+) and MGP45 ppk−ppx− (ppk+) were obtained by transformation with pSPK1 plasmid,Citation68 containing ppk gene. Cells were grown using M63 minimal medium supplemented with 0.4% glucose. M63 medium contained: 100 mM KH2PO4, 15 mM (NH4)2SO4, 1.7 μM FeSO4·7H2O and 1 mM MgSO4.Citation69 When indicated, medium was prepared with Pi concentrations other than 100 mM.
Quantification of biofilm formation
Biofilm formation was assayed on the basis of the ability of cells to adhere and grow on polystyrene microtiter plates and stained by crystal violet.Citation70 Overnight (ON) cultures in Luria Broth (LB) (L3022, Sigma) were diluted to an OD560 = 0.1 (corresponding to around 6 × 10Citation7 CFU ml−1) with fresh M63 glucose medium with different Pi concentrations. Cells were grown in microtiter plates under static conditions at 30°C for 48 h. Then, unattached cells were removed by washing the plates with deionized water. Two hundred microliters of 0.1% crystal violet solution was added to each well and plates were incubated at room temperature for 5 min. Then, wells were rinsed 3 times with water. Finally, the absorbed crystal violet was extracted with 200 μl of 95% ethanol and absorbance at 595nm was measured (Spectra MaxPlus384 Absorbance Microplate Reader, US). Six replicates were performed for each experimental condition.
PolyP level measurement
Intracellular polyP was measured by a fluorescence approach using 4′,6-diamidino-2-phenylindole (DAPI) in cell suspensions growing in static conditions.Citation71 Briefly, cells were washed and resuspended in T buffer (100 mM Tris–HCl, pH 8). Seventeen µM DAPI (D9542, Sigma) was added to cuvettes containing cell suspensions (OD560 = 0.02) in T buffer, with 0.00075% SDS and chloroform for cell permeabilization.Citation25 After incubation for 5 min at 37°C with agitation, the DAPI fluorescence spectra (excitation, 415 nm; emission, 445–650 nm) were recorded using an ISS PCI spectrofluorometer (ISS Inc., Champaign, IL). Fluorescence of the DAPI-polyP complex at 550 nm was used as a measurement of intracellular polyP, since emissions from free DAPI and DAPI-DNA are minimal at this wavelength.Citation71
Microscopic analysis
Microscopic images of biofilms were obtained by CLSM or by SEM. E. coli strains were grown under the same conditions used in the biofilm formation assay in a 6-well polystyrene plate containing M63 medium with different Pi concentration with a glass coverslip inside. After incubation for 48 h, unattached cells were removed by pipetting and cells on coverslips were then rinsed with water and dried at 37°C for 10 min. For CLSM the adhered cells were stained with 15 μl Calcofluor White (18909 Sigma) for 10 min in the dark. The coverslips were then rinsed with sterile water and removed from the wells. Images were captured using a Leica Confocal Microscope TCS–SP5 (Unitat de Microscopìa confocal, UB, Barcelona, Spain). For SEM, cells on coverslips were fixed with 2.5% glutaraldehyde and 2.5% paraformaldehyde, acetone and ethanol dehydrated, and gold coated with anion sputter JFC-1100 (JEOL). Coverslips containing the biofilms were then attached to aluminum holders and analyzed using a Carl Zeiss SUPRA-55 scanning electron microscope from CIME (UNT-CONICET) with a resolution of 1.0 nm at 15 kV and 1.7 nm at 1 kV in high-vacuum (HV) mode and 2 nm at 30 kV in variable-pressure mode (VP).
Curli and cellulose production assays
Curli production was examined by the Congo red-binding assay according to Da Re and Ghigo.Citation72 Briefly, 5 microliters of each ON culture grown at 37°C in LB medium were spotted onto M63 agar plates, supplemented with 40 μg ml−1 Congo red and 20 μg ml−1 brilliant blue containing different Pi concentrations and incubated for 48 h. Colony morphologies on Congo red plates were scored according to the basic morphotypes previously detected in S. Typhimurium: rdar (violet colony, expresses curli fimbriae and cellulose), pdar (pink colony, expresses cellulose), bdar (brown colony, expresses curli fimbriae) and saw (colorless colony, no expression of curli fimbriae nor cellulose).Citation39
Cellulose production was followed using the Calcofluor White qualitative assay with modification.Citation73 Strains were grown onto a coverslip immersed in M63 medium containing different Pi concentrations at 30°C during 48 h. After incubation attached cells were washed twice with distilled water and stained with 15 μl of Calcofluor White for 10 min. Celullose were visualized under a fluorescence microscope Olympus BX51TF equipped with an Olympus QColor5 digital camera (Q-imaging, Surrey, BC, Canada) at excitation and emission wavelengths of 395 and 440 nm, respectively.
Motility assays
Motility was evaluated according to Ulett et al. with modifications.Citation74 Briefly, ON LB cultures were washed and resuspended with fresh M63 medium to an OD560nm = 0.1. Cells were spotted onto the center of 0.3 % agar M63 plates supplemented with different Pi concentrations, using a sterile toothpick for inoculation. Plates were incubated at 30°C for 48 h and swimming motility was determined by measuring diameters of growth. For irregular halos, colony diameters were calculated as an average of several diameters around the colony. Data are expressed as the mean diameter (cm) of movement for 3 independent experiments.
Antibiotic susceptibility assays
Susceptibility to nalidixic acid or ciprofloxacin was determined by a disc-plate diffusion method, according to the Clinical and Laboratory Standards Institute (CSLI). Antibiotic minimal inhibitory concentrations (MICs) were determined by E-test strips (AB Biodisk, Solna, Sweden), as the intersection of the ellipse of inhibition with the strip. To test the antibiotic concentration able to inhibit biofilm formation or to disassemble a preformed biofilm, increasing antibiotic concentrations (0–200 μg ml−1) were added from the beginning or after 24 h of growth, respectively. Plates were incubated at 30°C and biofilm was quantified at 48 h.
Detection and expression of virulence factors genes
The presence of the virulence factor genes, haemolysin (hlyA gene), cytotoxic necrotizing factor (cnf1), type I fimbriae regulator (fimB gene), type I fimbriae (fimA gene), P-fimbriae (papA gene), siderophores (iroN and iutA genes), was analyzed by PCR with specific primers (), using as template genomic DNA extracted from each strain with Wizard® Genomic DNA Purification Kit (A1120, Promega).
Table 2 Primers used for detection and expression of virulence factor genes
RNA was extracted from each strain culture grown in M63 medium supplemented with different Pi concentrations at 30°C for 48 h, using the SV Total RNA Isolation System (Z3100, Promega), according to the manufacturer's recommendations. Reverse transcription-PCR (RT-PCR) was carried out using M-MLV Reverse Transcriptase (M1701, Promega) with random nonamer primers (Sigma) and Quantitative PCR (q-PCR) was performed using IQ Sybr Green Super Mix (1708880, Bio-Rad) with gene-specific primers in an Applied Biosystems 7500 Real-Time PCR System. Specific primers used for q-PCR analysis are listed in . Each q-PCR reaction was done in triplicate and the calculated threshold cycle (CT) was normalized to the CT of the 16S gene (used as a reference internal gene) amplified from the corresponding sample. The fold change was calculated using the 2ΔCT method. Genes with a fold change above or below the defined threshold of 2 were considered differentially expressed.
Statistical analysis
Data were subjected to analysis of variance (ANOVA) followed by Tukey's test with Statitix 9.0 Analytical Software 2008 for Windows (USA). Differences at p-value ≥ 0.05 were considered significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
1059561_Supplementary.pdf
Download PDF (220.1 KB)Funding
This research was supported by Argentinean grants of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), the Agencia Nacional de Promoción Científica y Técnica (ANPCyT) and the Consejo de Investigaciones de la Universidad Nacional de Tucumán (CIUNT).
References
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol 2004; 2:123-140; PMID:15040260; http://www.nature.com/nrmicro/journal/v2/n2/full/nrmicro818.html.
- Griebling TL. Urinary tract infection in women. In: Litwin MS, Saigal CS, ed. Urologic diseases in America: National Institutes of Health publication no. 07-5512.U.S. GPO, Washington, DC, US Government Printing Office. pp. 587-620., 2007.
- Nielubowicz GR, Mobley HL. Host-pathogen interactions in urinary tract infection. Nat Rev Urol 2010; 7:430-441; PMID:20647992; http://www.nature.com/nrurol/journal/v7/n8/full/nrurol.2010.101.html.
- Soto SM, Smithson A, Martinez JA, Horcajada JP, Mensa J, Vila J. Biofilm formation in uropathogenic Escherichia coli strains: relationship with prostatitis, urovirulence factors and antimicrobial resistance. J Urol 2007; 177:365-368; PMID:17162092; http://www.jurology.com/article/S0022-5347(06)02198-7.
- Johnson JR, Kuskowski MA, O'Bryan TT, Maslow JN. Epidemiological correlates of virulence genotype and phylogenetic background among Escherichia coli blood isolates from adults with diverse-source bacteremia. J Infect Dis 2002; 15:1439-1447; PMID:11992279; http://jid.oxfordjournals.org/content/185/10/1439.full.
- Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol 1991; 173:4773-4781; PMID:1677357; http://jb.asm.org/content/173/15/4773.long.
- Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol 1995; 18:661-670; PMID:8817489; http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2958.1995.mmi_18040661.x/pdf.
- Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 1989; 338:652-655; PMID:2649795; http://www.nature.com/nature/journal/v338/n6217/abs/338652a0.html.
- Römling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol 1998; 180:722-731; PMID:9457880; http://jb.asm.org/content/180/3/722.long.
- Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 2001; 39:1452-1463; PMID:11260463; http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2958.2001.02337.x/abstract.
- Lindsay D, von Holy A. Bacterial biofilms within the clinical setting: what healthcare professionals should know. J Hosp Infect 2006; 64:313-325; PMID:17046102; http://www.journalofhospitalinfection.com/article/S0195-6701(06)00335-5/abstract.
- Josenhans C, Suerbaum S. The role of motility as a virulence factor in bacteria. Int J Med Microbiol 2002; 291:605-614; PMID:12008914; http://www.sciencedirect.com/science/article/pii/S1438422104700788.
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 1998; 30:285-293; PMID:9791174; http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2958.1998.01061.x/abstract.
- Kulaev IS. The biochemistry of inorganic polyphosphates. New York: Wiley, Pp: 255; 1979. http://onlinelibrary.wiley.com/doi/10.1016/0307-4412%2881%2990065-0.
- Rao NN, Kornberg A. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J Bacteriol 1996; 178:1394-1400; PMID:8631717; http://jb.asm.org/content/178/5/1394.long.
- Rashid MH, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci 2000; 97:4885-4890; PMID:10758151; http://www.pnas.org/content/97/9/4885.full.
- Rashid MH, Rao NN, Kornberg, A. Inorganic polyphosphate is required for motility of bacterial pathogens. J Bacteriol 2000a; 182:225-227; PMID: 10613886; http://jb.asm.org/content/182/1/225.long.
- Rashid MH, Rumbaugh K, Passador L, Davies DG, Hamood AN, Iglewski BH, Kornberg A. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc Natl Acad Sci 2000b; 97:9636-9641; PMID:10931957; http://www.pnas.org/content/97/17/9636.long.
- Jahid IK, Silva AJ, Benitez JA. Polyphosphate stores enhance the ability of Vibrio cholerae to overcome environmental stresses in a low-phosphate environment. Appl Environ Microbiol 2006; 72:7043-7049; PMID:16950899; http://aem.asm.org/content/72/11/7043.long.
- Kim KS, Rao NN, Fraley CD, Kornberg A. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc Natl Acad Sci USA 2002; 99:7675-7680; PMID:12032342; http://www.pnas.org/content/99/11/7675.long.
- Ogawa N, Tzeng CM, Fraley CD, Kornberg A. Inorganic polyphosphate in Vibrio cholerae: genetic, biochemical, and physiologic features. J Bacteriol 2000; 182:6687-6693; PMID:11073913; http://www.ncbi.nlm.nih.gov/pmc/articles/PMC111411/.
- Price-Carter M, Fazzio TG, Vallbona EI, Roth JR. Polyphosphate kinase protects Salmonella enterica from weak organic acid stress. J Bacteriol 2005; 187:3088-3099; PMID:15838036; http://jb.asm.org/content/187/9/3088.long.
- Tan S, Fraley CD, Zhang M, Dailidiene D, Kornberg A, Berg DE. Diverse phenotypes resulting from polyphosphate kinase gene (ppk1) inactivation in different strains of Helicobacter pylori. J Bacteriol 2005; 187:7687-7695; PMID:16267293; http://jb.asm.org/content/187/22/7687.short.
- Kornberg A, Rao NN, Ault-Riche D. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem 1999; 68:89-125; PMID:10872445; http://link.springer.com/chapter/10.1007/978-3-642-58444-2_1.
- Grillo-Puertas M, Villegas JM, Rintoul MR, Rapisarda VA. Polyphosphate degradation in stationary phase triggers biofilm formation via LuxS quorum sensing system in Escherichia coli. PloS one 2012; 7:e50368; PMID:23226268; http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0050368.
- Gethke K, Herbst H, Montag D, Bruszies D, Pinnekamp J. Phosphorus recovery from human urine. Water Practice & Technology IWA Publishing 2006; 1(4); http://dx.doi.org/10.2166/WPT.20060701
- Ollayos RW, Winkler AW. Urinary excretion and serum concentration of inorganic phosphate in man. J Clin Invest. 1943; 22(2):147-54; http://dx.doi.org/10.1172/JCI101377.
- Aubia J, Bosch J, Lloveras J, Chine M, Hojman L, Masramon J. Relative hyperphosphaturia in diabetic chronic renal failure: a protective factor of hyperparathyroidism. Miner Electrolyte Metab 1987; 13:311-5; PMID:3670226 http://europepmc.org/abstract/med/3670226.
- Dash T, Parker MG, Lafayette RA. Profound Hypophosphatemia and Isolated Hyperphosphaturia in Two Cases of Multiple Myeloma. Am J Kidney Dis. 1997, 29:445-8; PMID:9041222; doi:10.1016/S0272-6386(97)90207-9.
- Narvaez J, Domingo-Domenech E, Narvaez JA, Nolla JM, and Valverde J. Acquired hypophosphatemic osteomalacia associated with multiple myeloma. Joint Bone Spine 2005; 72:424-6; PMID:16112595; doi:10.1016/j.jbspin.2004.10.012.
- Slatopolsky E, Robson AM, Elkan I, Bricker NS. Control of phosphate excretion in uremic man. Clin Invest 1968; 47:1865-1874; PMID:5666116; http://dx.doi.org/10.1172/JCI105877.
- Green J, Debby H, Lederer E, Levi M, Zajicek HK, Bick T. Evidence for a PTH-independent humoral mechanism in post-transplant hypophosphatemia and phosphaturia. Kidney Int. 2001; 60:1182-96; PMID:11532115; doi:10.1046/j.1523-1755.2001.0600031182.x.
- Rosenbaum RW, Hruska KA, Korkor A, Anderson C, Slatopolsky E. Decreased phosphate reabsorption after renal transplantation: Evidence for a mechanism independent of calcium and parathyroid hormone. Kidney International 1981; 19:568-578;PMID:6264200; http://dx.doi.org/10.1038/ki.1981.54.
- Reisner A, Krogfelt KA, Klein BM, Zechner EL, Molin S. In vivo biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J Bacteriol 2006; 188:3572-81; PMID:16672611; http://jb.asm.org/content/188/10/3572.long.
- Bokranz W, Wang X, Tschäpe H, Römling U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J Med Microbiol 2005; 54:1171-82; PMID:16278431; http://jmm.sgmjournals.org/content/54/12/1171.long.
- Naves P, del Prado G, Huelves L, Gracia M, Ruiz V, Blanco J, Dahbi G, Blanco M, Ponte M, Soriano F. Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microb Pathogenesis 2008; 45: 86-91; PMID: 18486439; http://www.sciencedirect.com/science/article/pii/S0882401008000363.
- Mendez-Arancibia E, Vargas M, Soto S, Ruiz J, Kahigwa E, Schellenberg D, Urassa H, Gascón J, Vila J. Prevalence of different virulence factors and biofilm production in enteroaggregative Escherichia coli isolates causing diarrhea in children in Ifakara (Tanzania). Am J Trop Med Hyg 2008; 78:985-9; PMID:18541781; http://www.ajtmh.org/content/78/6/985.long.
- Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev 2005; 19:2645-55; PMID:16291643; http://www.iwaponline.com/wpt/001/0070/0010070.pdf.
- Römling U, Rohde M, Olsen A, Normark S, Reinköster, J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol Microbiol 2000; 36:10-23; PMID:10760159; http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2958.2000.01822.x/abstract.
- Römling U. Molecular biology of cellulose production in bacteria. Res Microbiol 2002; 153:205-12; PMID:12066891; http://www.sciencedirect.com/science/article/pii/S0923250802013165.
- Ude S, Arnold DL, Moon CD, Timms-Wilson T, Spiers AJ. Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environm Microbiol 2011; 8:1997-2011; PMID:17014498; http://onlinelibrary.wiley.com/doi/10.1111/j.1462-2920.2006.01080.x/pdf.
- Dunne WM Jr. Bacterial Adhesion: Seen Any Good. Biofilms Lately? Clin Microbiol Rev 2002; 15:155-66; PMID:11932228; http://cmr.asm.org/content/15/2/155.full.
- Bower JM, Eto DS, Mulvey MA. Covert operations of uropathogenic Escherichia coli within the urinary tract. Traffic 2005; 6:18-31; PMID:15569242; http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2523259.
- Wright KJ, Hultgren SJ. Sticky fibers and uropathogenesis: bacterial adhesins in the urinary tract. Future Microbiol 2006; 1:75-87; PMID:17661687; http://www.futuremedicine.com/doi/pdf/10.2217/17460913.1.1.75.
- Kanamaru S, Kurazono H, Terai A, Monden K, Kumon H, Mizunoe Y, Ogawa O, Yamamoto S. Increased biofilm formation in Escherichia coli isolated from acute prostatitis. Int J Antimicrob Agents 2006; 28S:21-25; PMID:16828264; http://www.sciencedirect.com/science/article/pii/S0924857906001622.
- Middendorf B, Hochhut B, Leipold K, Dobrindt U, Blum-Oehler G, Hacker J. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J Bacteriol 2004; 186:3086-96; PMID:15126470; http://www.ncbi.nlm.nih.gov/pmc/articles/PMC400636/.
- Torres AG, Redford P, Welch RA, Payne SM. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun 2001; 69:6179-85;PMID:11553558; doi:10.1128/IAI.69.10.6179-6185.2001.
- Roos VGC, Ulett M, Schembri A, Klemm P. The asymptomatic bacteriuria Escherichia coli strain 83972 outcompetes uropathogenic E. coli strains in human urine. Infect Immun 2006; 74:615-24; PMID:16369018; http://dx.doi.org/10.1128/IAI.74.1.615-624.
- Soto SM, Zúñiga S, Ulleryd P, Vila J. Acquisition of a pathogenicity island in an Escherichia coli clinical isolate causing febrile urinary tract infection. Eur J Clin Microbiol Infect Dis 2011; 30:1543-1550; PMID:21499969; http://link.springer.com/article/10.1007%2Fs10096-011-1258-2
- Talan DA, Stamm WE, Hooton TM, Moran GJ, Burke T, Iravani A, Reuning-Scherer J, Church DA. Comparison of ciprofloxacin (7 days) and trimethoprim-sulfamethoxazole (14 days) for acute uncomplicated pyelonephritis in women: a randomized trial. JAMA Int Med 2000; 283:1583-90; PMID:10735395; http://jama.jamanetwork.com/article.aspx?articleid=192526.
- Andriole VT. The Quinolones: Past, Present, and Future. CID 2005; 41:113-19; PMID:15942877; http://cid.oxfordjournals.org/content/41/Supplement_2/S113.long.
- Kallen AJ, Welch GH, Sirovich BE. Current Antibiotic Therapy for Isolated Urinary Tract Infections in Women. JAMA Int Med 2006; 166:635-39; PMID:16567602; http://archinte.jamanetwork.com/article.aspx?articleid=410047.
- Brzuszkiewicz E, Brüggemann H, Liesegang H, Emmerth M, Ölschläger T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emődy L et al. How to become a uropathogen: Comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci USA 2006; 103:12879-84; PMID:16912116; http://www.pnas.org/cgi/pmidlookup?view=long&pmid=16912116.
- Wang Q, Sun FJ, Liu Y, Xiong LR, Xie LL,Xia, PY. Enhancement of biofilm formation by subinhibitory concentrations of macrolides in icaADBC-positive and -negative clinical isolates of Staphylococcus epidermidis. Antimicrob Agents Chemother 2010; 54:2707-2711; PMID:20231401; http://aac.asm.org/content/54/6/2707.long.
- Majtán J, Majtánová L, Xu M, Majtán, V. In vitro effect of subinhibitory concentrations of antibiotics on biofilm formation by clinical strains of Salmonella enteric serovar typhimurium isolated in Slovakia. J Appl Microbiol 2008; 104:1294-1301; PMID:18028358; http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2672.2007.03653.x/abstract.
- Dobrindt U, Agerer F, Michaelis K, Janka A, Buchrieser C, Samuelson M, Svanborg C, Gottschalk G, Karch H, Hacker J. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J Bacteriol 2003; 185:1831-40; PMID:12618447; http://jb.asm.org/content/185/6/1831.long.
- Wanner BL. Phosphorus assimilation and control of the phosphate regulon. En: Escherichia coli and Salmonella: Celular and Molecular Biology, (1996) Neidhardt FC, Curtis RI, Ingraham J. L., Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M., Schaechter M., Umbarger HE. 51 (Eds.).Washington DC: Amer. Soc. Microbiology. 1357-81.
- Rao NN, Torriani A. Molecular aspects of phosphate transport in Escherichia coli. Mol Microbiol 1990; 4:1083-90; PMID:1700257; http://dx.doi.org/10.1111/j.1365-2958.1990.tb00682.x.
- Rosenberg H, Gerdes RG, Chegwidden K. Two systems for the uptake of phosphate in Escherichia coli. J Bacteriol; 1977; 131:505-11. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC235458.
- Bertrand N, Houle S, LeBihan G, Poirier É, Dozois CM, Harel J. Increased Pho regulon activation correlates with decreased virulence of an avian pathogenic Escherichia coli O78 strain. Infect. Immun; 2010; 78:5324-5331; http://dx.doi.org/10.1128/IAI.00452-10.
- Cheng C, Tennant SM, Azzopardi KI, Bennett-Wood V, Hartland EL, Robins-Browne RM, Tauschek M. Contribution of the pst-phoU operon to cell adherence by atypical enteropathogenic Escherichia coli and virulence of Citrobacter rodentium. Infect. Immun. 2009; 77:1936-44; PMID:19255191; http://dx.doi.org/10.1128/IAI.01246-08.
- Crépin S, Chekabab SM, Le Bihan G, Bertrand N, Dozois CM, Harel J. The Pho regulon and the pathogenesis of Escherichia coli. Vet. Microbiol 2011; 153:82-88;PMID:21700403; http://dx.doi.org/10.1016/j.vetmic.2011.05.043.
- Ferreira GM, Spira B. The pst operon of enteropathogenic Escherichia coli enhances bacterial adherence to epithelial cells. Microbiology 2008; 154:2025-36; PMID:18599831: http://dx.doi.org/10.1099/mic.0.2008/016634-0.
- Lamarche MG, Dozois CM, Daigle F, Caza M, Curtiss R 3rd, Dubreuil JD, Harel J. Inactivation of the pst system reduces the virulence of an avian pathogenic Escherichia coli O78 strain. Infect. Immun. 2005; 73:4138-45; PMID:15972503; http://dx.doi.org/10.1128/IAI.73.7.4138-4145.2005.
- Buckles EL, Wang X, Lockatell CV, Johnson DE, Donnenberg MS. PhoU enhances the ability of extraintestinal pathogenic Escherichia coli strain CFT073 to colonize the murine urinary tract. Microbiology 2006; 152:153-60. http://dx.doi.org/10.1099/mic.0.28281-0.
- Crépin S, Houle S, Charbonneau MÈ, Mourez M, Josée Harel, Dozois CM. Decreased Expression of Type 1 Fimbriae by a pst Mutant of Uropathogenic Escherichia coli Reduces Urinary Tract Infection. Infect. Immun. 2012. 80:2802-15; PMID:22665376; http://dx.doi.org/10.1128/IAI.00162-12.
- Miller J. Procedures for working with lac. A short course in bacterial genetics: A laboratory manuals and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY; 1992; 1:72-74.
- Van Dien SJ, Keyhani S, Yang C, Keasling JD. Manipulation of independent synthesis and degradation of polyphosphate in Escherichia coli for investigation of phosphate secretion from the cell. Appl. Environ. Microbiol. 1997; 63:1689-95. http://aem.asm.org/content/63/5/1689.short.
- Danese P, Pratt LA, Kolter R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol 2000; 182:3593-96; PMID:10852895; http://jb.asm.org/content/182/12/3593.long.
- O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol 1998; 42:449-61; PMID:9632250; http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2958.1998.00797.x/abstract.
- Aschar-Sobbi R, Abramov AY, Diao C, Kargacin ME, Kargacin GJ, French RJ, Pavlov E. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J fluoresc 2008; 18:859-66; PMID:18210191; http://link.springer.com/article/10.1007%2Fs10895-008-0315-4.
- Da Re S, Ghigo JMA. CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol 2006; 188:3073-87; PMID:16585767; http://jb.asm.org/content/188/8/3073.long.
- Treat J, James WD, Nachamkin I, Seykora JT. Growth Inhibition of Trichophyton Species by Pseudomonas aeruginosa. Arch Dermatol 2007; 143:61-64; PMID:17224543; http://archderm.jamanetwork.com/article.aspx?articleid=410766.
- Ulett GC, Webb RI, Schembri MA. Antigen-43-mediated autoaggregation impairs motility in Escherichia coli. Microbiology 2006; 152:2101-110; PMID:16804184; http://mic.sgmjournals.org/content/152/7/2101.long.
