Abstract
Neonatal meningitis Escherichia coli K1 (NMEC) are thought to be transmitted from mothers to newborns during delivery or by nosocomial infections. However, the source of E. coli K1 causing these infections is not clear. Avian pathogenic E. coli (APEC) have the potential to cause infection in humans while human E. coli have potential to cause colibacillosis in poultry, suggesting that these strains may lack host specificity. APEC strains are capable of causing meningitis in newborn rats; however, it is unclear whether these bacteria use similar mechanisms to that of NMEC to establish disease. Using four representative APEC and NMEC strains that belong to serotype O18, we demonstrate that these strains survive in human serum similar to that of the prototypic NMEC strain E44, a derivative of RS218. These bacteria also bind and enter both macrophages and human cerebral microvascular endothelial cells (HCMEC/D3) with similar frequency as that of E44. The amino acid sequences of the outer membrane protein A (OmpA), an important virulence factor in the pathogenesis of meningitis, are identical within these representative APEC and NMEC strains. Further, these strains also require FcγRI-α chain (CD64) and Ecgp96 as receptors for OmpA in macrophages and HCMEC/D3, respectively, to bind and enter these cells. APEC and NMEC strains induce meningitis in newborn mice with varying degree of pathology in the brains as assessed by neutrophil recruitment and neuronal apoptosis. Together, these results suggest that serotype O18 APEC strains utilize similar pathogenic mechanisms as those of NMEC strains in causing meningitis.
Abbreviations
| APEC | = | Avian pathogenic E. coli |
| NMEC | = | Neonatal meningitis E. coli |
| BBB | = | blood-brain barrier |
| HCMEC | = | Human cerebral microvascular endothelial cells |
| OmpA | = | Outer membrane protein A. |
Introduction
E. coli K1 is the second most common pathogen associated with neonatal meningitis particularly during the first month after birth. Even with the use of very effective third-generation antibiotics, the morbidity and mortality rates associated with E. coli meningitis (5-30%–) have remained unchanged for the last few decades.Citation1,2 It is known that infants acquire the bacterium from the mother during the delivery or by nosocomial infection in the intensive care units.Citation3 NMEC is a sub-group of the extraintestinal pathogenic E. coli (ExPEC) group, which also includes uropathogenic E. coli and avian pathogenic E. coli (APEC). Despite their isolation from various hosts, tissues, and disease syndromes, ExPEC could harbor similar virulence associated genes leading to the hypothesis that APEC strains may have zoonotic potential.Citation4,5
Previously, we demonstrated that E. coli K1 infection of 3-day-old mice causes meningitis for which outer membrane protein A (OmpA) expression is essential.Citation5 The disease pathogenesis in this mouse model of meningitis closely mimics the pathogenesis in humans. In addition, our studies have shown that OmpA is an important surface structure required to evade complement attack during the initial stages of infection.Citation6 Subsequently, E. coli K1 enters neutrophils and macrophages in an OmpA-dependent fashion to down-regulate the bactericidal mechanisms of these 2 immune cells. We further showed that specific amino acid mutations in the extracellular loops of OmpA reduced the capacity of E. coli K1 to evade complement, invade immune cells, and cross the blood-brain barrier. These mutations did not affect the assembly of OmpA, expression of other virulence factors (IbeA, IbeB, CNF1, etc) and content of the polysaccharides, thereby demonstrating that OmpA expression is critical for E. coli K1 virulence in experimental meningitis.Citation7 Of note, a heat shock protein, HSP90β, which we have previously designated as gp96, and CD64 (Fcγ-receptor I α chain) act as receptors for OmpA on neutrophils and macrophages, respectively.Citation8-10 Our studies have shown that OmpA directly interacts with 3 N-glycosylation sites in the extracellular domains of CD64 in macrophages, and E. coli K1 does not require IgG opsonization for internalization. Since the entry of E. coli K1 is driven by the OmpA interaction with CD64, we used the term “invasion” here afterward to depict this bacterial-mediated entry process in macrophages.Citation12 After entering macrophages via the OmpA-CD64 mediated interaction, E. coli K1 survives, multiplies, and is released into the blood, an event that is critical for reaching a threshold level of bacteremia.Citation11-13 The bacterium then crosses the blood-brain barrier (BBB), which contains a lining of human brain microvascular endothelial cells (HBMEC), to reach the cerebrospinal fluid. E. coli K1 interacts with endothelial cell gp96 (Ecgp96) on the BBB to invade the cells, in which process it disrupts the tight junctions between HBMEC leading to increased permeability.Citation14,15
APEC strains, responsible for colibacillosis in poultry, and human ExPEC strains share some common phenotypic and genotypic characteristics to cause disease in certain animal models. APEC strains may cause meningitis in a newborn rat model of meningitis, and some NMEC strains cause avian colisepticemia, providing additional support that these strains may have zoonotic potential.Citation16 However, it is unknown whether APEC strains follow the same modality of infection as NMEC strains in causing meningitis in animal models. Therefore, we sought to test whether APEC strains use strategies similar to that of NMEC in the pathogenesis of meningitis. In addition to such well-known characteristics, such as K1 capsule, IbeA, etc., NMEC frequently harbors large putative virulence plasmids.Citation17,18 These plasmids bear a strong resemblance to APEC plasmids that are characteristic of the APEC pathotype.Citation19 Therefore, some NMEC and APEC strains were cured of their plasmids following serial growth at elevated temperatures, and their cell associative and invasive capabilities and abilities to cause meningitis are compared in various models. We report that all the tested strains entered macrophages, and furthermore, 2 representative APEC and NMEC clinical strains avoid serum killing and invade human cerebral microvascular endothelial cells (HCMEC/D3), for which the expression of gp96 in HCMEC/D3 and CD64 in macrophages are critical. Importantly, APEC strains also caused meningitis in the newborn mouse model, similar to RS218 and other NMEC strains, albeit causing varying degrees of pathology in the brain.
Results
APEC and NMEC strains exhibit varying degrees of invasion in RAW 264.7 macrophages
The E. coli strains used in this study belong to serogroup O18 except APEC 59 and are shown in . All strains exhibited similar growth kinetics over 8 hours of logarithmic phase growth, except E91 that showed a minor lag between 2 and 6 hours (Fig. S1). Nevertheless, when performing invasion assays, we verified that all multiplicity of infection (MOIs) were approximately the same. The panel of APEC and NMEC strains showed distinct patterns of cell association and invasion of RAW 264.7 macrophages. E44 (a rifampicin resistant strain of RS218) was used as a positive control, and E91 (OmpA negative strain derived from E44), which was poorly taken up by RAW 264.7 macrophages, served as a negative control for the experiments. The graphs for cell association and invasion data represent the data as a relative percentage compared to E44 taken as 100%. The raw CFU per well values are provided to represent relevant counts of cell-associated and invading bacteria and are not calculated relative to E44. In these experiments, E44 entry was approximately 1.37 × 104 CFU per well, whereas its ompA mutant (E91) was phagocytosed at a frequency of 4.16 × 103 CFU per well. All other APEC and NMEC strains entered macrophages with invasion frequencies ranging from ∼1.16 × 103 to ∼1.8 × 104 CFU per well. While most of the APEC and NMEC strains bound and entered efficiently, APEC 353, APEC 380, and NMEC 18 entered with lower frequency than their respective plasmid-cured versions (the plasmid-cured versions are denoted with the suffix ‘C’) (). Previous studies have demonstrated that OmpA expression in E44 is important for the onset of meningitis in newborn rat and mouse models.Citation20,21 Therefore, to determine whether the sequences of the ompA genes of the APEC and NMEC strains are similar to that of E44, we performed PCR using primers flanking the entire open reading frame of ompA on genomic DNA. E91 was used as a negative control. Sequencing analysis and sequence alignment revealed the presence of 100% homology among E44 and all of the APEC and NMEC strains, except for APEC 59 and APEC 79, which were 99% and 94% homologous, respectively (Fig. S2). The complete ompA sequence from E44 has been submitted to GenBank, accession number KT159980. For the ompA sequences of APEC and NMEC strains, the GenBank accession numbers are from KT336761 to KT336776 (Fig. S3).
Figure 1. APEC and NMEC strains exhibit distinct cell association and invasion patterns in RAW 264.7 macrophages. Cell association (A) and invasion (B) patterns of APEC and NMEC strains in RAW 264.7 macrophages were performed as described in the Materials and Methods. The OmpA− mutant of E44 (E91) was used as a negative control. Cell association and invasion experiments were performed at least 3 times in triplicate, and the values are presented as percent means ± S.D considering E44 invasion as 100%. Increase or decrease in cell association/invasion was statistically significant for some strains compared to E44, *p < 0.05 by Student's t test.
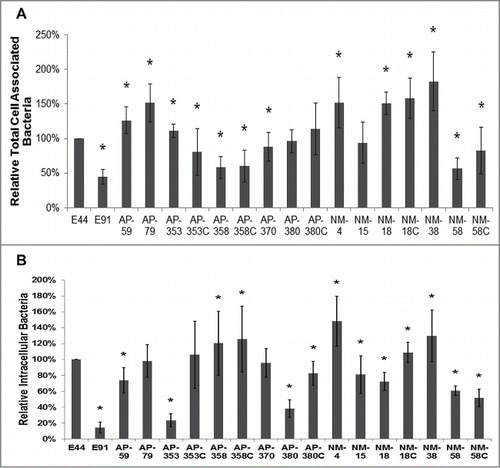
Table 1. The genotypes of all strains of E. coli used in this study
APEC and NMEC strains evade complement attack and invade macrophages using CD64
Two important “survival” strategies employed by E44 en route to the BBB are complement evasion and survival inside macrophages. We have previously demonstrated that E44 avoids complement attack by binding to C4bp, a classical complement pathway regulator, for which OmpA expression is essential, since a majority of its ompA mutant (E91) were killed within 15 min post-incubation with serum.Citation6 Therefore, APEC and NMEC strains were subjected to serum survival assays along with E44 and E91. All of the strains survived in 50% pooled human serum except E91, which was comparatively more susceptible to serum bactericidal activity by 15 min post-infection (). Heat-inactivated serum used as a control showed no effect on these strains, except for NMEC 4, indicating that the observed effect was complement mediated and that the presence of OmpA may enable E44, APEC, and NMEC to survive complement attack. Our speculation for lack of NMEC 4 survival in heat-inactivated human serum is that some essential components for the growth of the bacteria might be destroyed by the inactivation of serum as shown in the case of Aeromonas hydrophila.Citation22 However, lack of this effect during the invasion assays, which were performed in the presence of 5% heat-inactivated FBS, might be due to the presence of essential factors in the growth media for NMEC 4 survival.
Figure 2. APEC and NMEC strains evade serum complement killing and survive inside bone marrow derived macrophages (BMDMs). Serum survival assays were performed with the APEC (AP) or NMEC (NM) strains as described in the Materials and Methods in the presence of normal and heat-inactivated serum, and percentage survival after 15 minutes exposure was compared to the corresponding values at 0 minutes for each individual strain. The decrease in survival with normal serum or increase in survival with heat-inactivated serum is statistically significant, *p < 0.05 by Student's t test. (A). Cell association (B) and invasion (C) patterns of the strains in BMDMs were also analyzed. The serum survival and cell association/assays were performed at least 3 times in triplicate, and the values are presented as percent means ± S.D considering E44 invasion as 100%. Increase or decrease in cell association/invasion was compared to E44, *p < 0.05 by Student's t test.
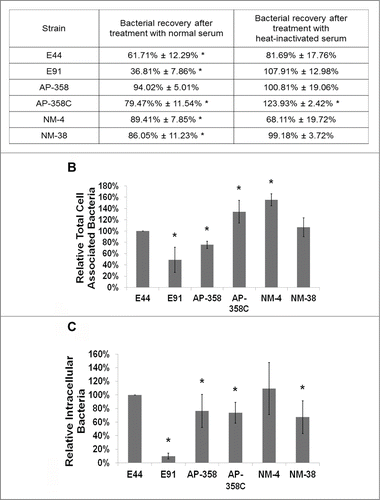
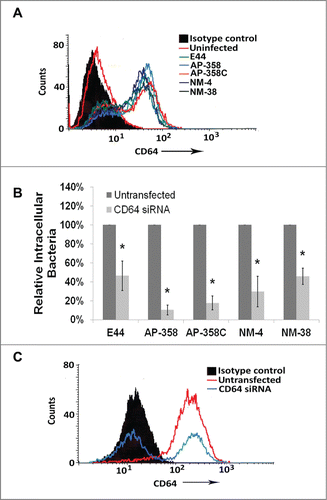
The next step in the pathogenesis of meningitis would be bacterial multiplication in immune cells, especially in macrophages, to reach high-grade bacteremia. Therefore, C57BL/6 mouse bone marrow-derived macrophages (BMDMs) were infected with APEC and NMEC strains to compare their association and invasion of the cells. The total cell-associated (∼4.18 × 105 to 7.53 × 105 CFU per well) and invasion (∼7.46 × 102 to 8.3 × 103 CFU per well) frequencies were comparable to the levels exhibited by E44 (). Although E91 was moderately associated with the cells (∼3.36 × 105 CFU per well), it invaded BMDMs poorly (∼8.75 × 101 CFU per well). Next, we examined whether APEC and NMEC strains also require CD64 on the surface of macrophages for efficient interaction and invasion. Flow cytometry analysis of CD64 expression in response to infection of RAW 264.7 macrophages with the different strains was performed. All the strains induced similar levels of CD64 expression on the cell surface upon infection (). To confirm whether CD64 acts as a receptor for APEC and NMEC strains, RAW 264.7 macrophages were transfected with CD64 siRNA and subjected to invasion assays. All the strains invaded 50% or lower in siRNA-transfected macrophages compared to the invasion of untransfected cells (). Analysis of CD64 silencing by flow cytometry indicated that siRNA-mediated silencing of CD64 expression was efficient (). These data confirm that APEC and NMEC strains require CD64 for efficient interaction with and invasion of macrophages, similar to E44.
Figure 3. APEC and NMEC strains require CD64 for macrophage invasion, similar to E44. RAW 264.7 macrophages were infected with the different APEC (AP) and NMEC (NM) strains, and the infected macrophages were subjected to flow cytometry analysis to determine the surface expression of CD64 in response to infection (A). RAW 264.7 macrophages were transfected with siRNA to CD64, and the cells were allowed to recover for 24 h before performing invasion assays with the strains (B). The effect of CD64 silencing by the siRNA was concurrently verified by flow cytometry (C). The invasion assays were performed at least 3 times in triplicate, and the values are presented as percent means ± S.D considering the bacterial invasion of untransfected cells as 100%. Decrease in the invasion of the strains in the presence of siRNA was statistically significant compared to untransfected cells, *p < 0.05 by Student's t-test.
Invasion of HCMEC/D3 by APEC and NMEC strains requires Ecgp96 expression
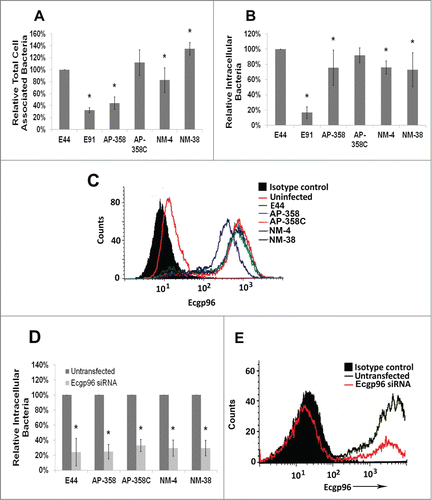
E. coli must traverse the blood-brain barrier to cause meningitis in neonates. Therefore, using HCMEC/D3, a very well-established in vitro model of the BBB,Citation23 we examined whether APEC and NMEC strains also invade HCMEC/D3 in a similar fashion to that of E44. Cell association and invasion experiments showed that the binding efficiencies of these strains vary (6.72 × 105 to 2.14 × 106 CFU per well); however, invasion frequencies were similar in HCMEC/D3 (2.48 × 102 to 1.24 × 103 CFU per well) while E91 poorly invaded the cells (6.45 × 101 CFU per well) (). Previous studies have shown that OmpA of E. coli K1 interacts with Ecgp96 for binding and invading HCMEC/D3. Since APEC and other NMEC strains behave similarly to E44, we speculated that these strains also use Ecgp96 as a receptor to invade HCMEC. As shown in , infection with APEC and NMEC strains increased the expression of Ecgp96 on the surface of HCMEC/D3. The levels of Ecgp96 expression were similar to that of E44 induced levels in these cells. Subsequently, to examine the effect of Ecgp96 on the invasion of APEC and NMEC strains, Ecgp96 siRNA-transfected HCMEC/D3 were used for invasion assays. Lack of this OmpA receptor in HCMEC/D3 significantly prevented the invasion of these strains (). Efficient knockdown of gp96 was concurrently verified by flow cytometry (). These results indicate that Ecgp96 acts as a receptor for tested APEC and NMEC strains in HCMEC/D3.
Figure 4. APEC and NMEC strains manipulate Ecgp96 for brain endothelial cell invasion. Cell association (A) and invasion (B) patterns of APEC (AP) and NMEC (NM) strains in HCMEC/D3 were performed. APEC and NMEC infected HCMEC/D3 were subjected to flow cytometry to examine the surface expression of Ecgp96 in response to infection (C). HCMEC/D3 were transfected with siRNA to Ecgp96, and the cells were allowed to recover for 24 h before performing invasion assays with the strains. The percent reduction in invasion of each strain in the presence of siRNA was calculated relative to their invasion taken as 100% in the absence of siRNA (D). Efficient Ecgp96 silencing was confirmed by flow cytometry (E). The cell association/invasion experiments were performed at least 3 times in triplicate and the bacteria bound or invaded are presented as percent means ± S.D taking the total cell association or invasion of E44 as 100%. Increase or decrease in cell association/invasion of the strains was statistically significant compared to untransfected HCMEC/D3, *p < 0.05 by Student's t-test.
Pathogenicity of APEC and NMEC strains in a newborn mouse model of meningitis
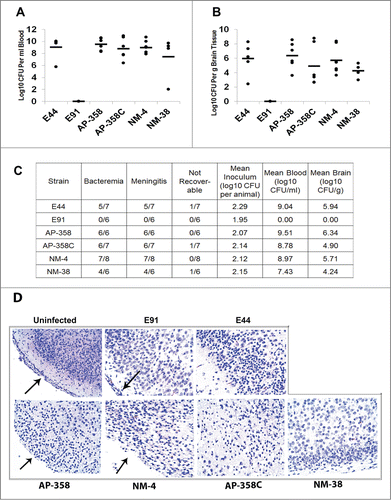
Newborn rat or mouse models of meningitis are well-established using the E44 strain. Although several APEC and NMEC strains have been tested in the rat model, none has been tested in the mouse model of meningitis. Herein, 3-day-old mice were infected with APEC strains (358 and 358C), NMEC strains (4 and 38), E44, or E91 by intraperitoneal injection and then examined for the blood and brain bacterial count. The occurrence of meningitis was evaluated by positive CSF cultures from these animals. These four strains exhibited comparable bacterial load to that of E44 in the newborn mouse model, as indicated by a lack of statistically significant differences in the blood () and brain () bacterial counts by ANOVA. Moreover, all the strains caused meningitis between 19 and 48 h postinfection (). In contrast, E91 did not show any bacteremia and consequently no meningitis. H&E staining of the brain sections obtained from the infected mice showed that all the strains damaged the meningeal portion of the brain, while the brain morphology of E91-infected pups was intact and typical of a normal, uninfected brain with distinct cortex and meninges (). However, brain damage patterns were indistinguishable from each strain in H&E except that meningeal layer was completely lost in APEC and NMEC strains similar to E44. Consequently, we performed immunofluorescence staining for neutrophil infiltration and apoptosis in corresponding brain sections to determine the patterns unique for each strain.
Figure 5. APEC (AP) strains cause meningitis similar to that of NMEC (NM) strains. Three-day-old mice were infected with 100 CFU of each of the strains via the intraperitoneal route and the bacterial load in blood (A) and brain (B) were enumerated. Excluding E91, statistical analysis by one-way ANOVA indicates no significant difference in blood and brain bacterial loads among the APEC and NMEC strains when compared to E44. No colonies were recovered from E91 infected pups. The summary of the animal experiments are tabulated (C). H&E staining of the brain sections was performed to assess the pathological changes (D). Arrows indicate the presence or loss of meninges.
Induction of apoptosis and neutrophil infiltration in the brains of infected mice is strain specific
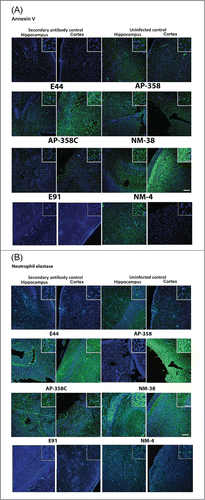
Since H&E staining showed gross pathology of the brains isolated from infected mice, sections stained with annexin V for apoptosis and neutrophil elastase for neutrophil infiltration and evaluated by a confocal microscope. Both E44 and NMEC 38 induced apoptosis of neuronal cells extensively in the cortex region and moderately in the hippocampal region (). APEC strains induced less apoptosis in both the cortex and hippocampus regions but recruited comparatively more neutrophils into the cortex. Of note, very few neutrophils were observed in the hippocampus of the brains of pups infected with E44 and NMEC 38. These results suggest that the mortality of the newborn mice infected with APEC and NMEC strains could be due to slightly different pathological patterns although all the pups showed positive CSF cultures. By contrast, E91-infected brains showed intact morphology and lack of neutrophil infiltration or apoptosis, an obvious reflection of the lack of ability of this strain to cause meningitis.
Figure 6. Patterns of neuronal apoptosis and neutrophil infiltration in the brains of infected newborn mice were distinct for each APEC (AP) or NMEC (NM) strain. Sections adjacent to the ones used for H&E staining, were subjected to immunofluorescence staining with Annexin V (for apoptosis) (A) and neutrophil elastase (for neutrophil infiltration) (B) in the cortex and hippocampus regions. Insets show magnified areas in the sections. Scale bar = 100 µm.
Discussion
Much attention has been directed toward tackling zoonotic infections, a worldwide health concern. The zoonotic potential of APEC strains has been attributed to the presence of common virulence traits that they share with NMEC or uropathogenic E. coli (UPEC). Newborns are specifically susceptible to ExPEC infection, due their immature immune system, and most likely acquire the infection before, during, or after delivery.Citation24 Although ExPEC infections can be easily treatable with antibiotics previously, increased antibiotic resistance of these strains renders current treatments ineffective.Citation25 Developing new strategies to prevent ExPEC infections needs complete understanding of virulence and zoonotic risk as well as constant surveillance of antibiotic resistance patterns of these strains in animals and humans. Therefore, potential zoonotic traits of APEC strains pose a higher risk for neonates since the mortality rates from neonatal sepsis/meningitis are 10% in developed countries and up to 58% in developing countries.Citation2 Since there is limited understanding of the possible infection modalities of APEC strains, we examined the possibility of them sharing a common infection pattern with the prototypic neonatal meningitis strain E44 using in vitro culture models and a newborn mouse model of meningitis.
The pathogenic mechanisms of E44 have been unraveled in recent years. As an ExPEC strain, E44 initially avoids complement attack, even at low counts of 103 CFU, after which it invades macrophages.Citation6 The bacterium uses its major virulence factor OmpA to bind to specific N-glycans on the macrophage surface receptor CD64.Citation13 Once inside the macrophages, bacteria inhibit macrophage apoptosis, multiply, and are released into the bloodstream in high numbers to eventually reach the BBB.Citation12 The bacteria then use OmpA to bind Ecgp96 on the BBB to invade the brain endothelium and disseminate into the central nervous system.Citation10 Our current study shows that representative APEC strains also follow a strikingly similar pattern of infection, along with NMEC clinical isolates. Interestingly, although all the strains used in this study cause meningitis, each of them seems to exhibit a distinct pattern of infection in vitro as well as pathological infliction in the brain. Tivendale et al., identified the pattern of genes associated with conserved virulence regions of APEC plasmids and found that they shared virulence traits with E44, although APEC strains possess additional virulence factors, which could impact the pathogenic mechanisms of these strains.Citation16 Furthermore, while the majority of the APEC and NMEC strains highlighted in the current study had OmpA sequences that are 100% identical to the OmpA from E44, 2 APEC strains (APEC 59 and APEC 79) had slight differences. Specifically, APEC 59 OmpA had 2 amino acid changes and APEC 79 OmpA had 3 amino acid changes and 4 amino acid insertions, compared to E44 OmpA, in the extracellular loop 3 (Fig. S4). Furthermore, APEC 79 OmpA had 4 amino acid changes compared to E44 OmpA in the extracellular loops 1 and 2. Finally, APEC 79 OmpA also had 2 amino acid changes compared to E44 OmpA in the C-terminus. The amino acid changes in APEC 59 and APEC 79 compared to E44 do not have any significant impact on the invasive ability of these strains.
Multiple epidemiological studies have suggested that poultry is a source of human ExPEC infection due to their presence in the intestines of poultry and poultry meat from retail markets, and these strains are often genetically similar to those found to be responsible for human infections.Citation26-28 Large plasmids present in ExPEC strains appears to contribute to their zoonotic potential. In particular, the ST95 multi locus sequence type (MLST) is widely prevalent in APEC strains from North America and Europe.Citation29,30 The data in the present study show that the APEC/NMEC strains show variable invasion patterns, regardless of the MLST or the presence or absence of plasmids (). Although the invasion of some of these strains is reduced compared to E44, these strains have been shown to cause meningitis in a rat model. Our previous studies have demonstrated that efficient multiplication of E44 in macrophages is critical for eventually crossing the BBB. Consistent with this observation, we show here that APEC 358, APEC 358C, NMEC 4, and NMEC 38 invade as well as or more than E44 in RAW 264.7 cells, BMDMs, or HCMEC/D3.
The only striking difference between E44 and the other strains is that E44 harbors the cnf1 gene (cytotoxic necrotizing factor 1) while the other strains do not (). However, the bacterial burden in the brain was similar for E44, APEC, and NMEC strains. Interestingly, we have observed that a cnf1 deletion mutant of E44 (Δcnf1) invades more than the wild-type strains in macrophages and concurrently invades the brain in higher numbers. The Δcnf1 strain complemented with a plasmid containing cnf1, however, behaves similarly to E44 (unpublished data). Since E44 is a prototypical E. coli strain for studying neonatal meningitis, the expression of CNF1 in E44 may be an exclusive virulence trait that the strain uses to adapt to the host, and the relevance of this is currently unknown. Nevertheless, our results show that OmpA-specific receptors in macrophages and HCMEC/D3 seem to be critical for the invasion of these strains in the respective cell types. The antibodies raised against recombinant OmpA recognized only the C-terminal portion, which is localized in the periplasm.Citation31 Moreover, OmpA is known for improper folding in neutral pH solutions,Citation32 and therefore neither the antibodies nor the recombinant protein could be used for blocking experiments. Thus, our efforts to prove the direct role of OmpA per se were unsuccessful. We are currently in the process of creating ompA mutants of some of the APEC and NMEC strains used in this study, and a more comprehensive analysis will be performed using these strains in the future. Nevertheless, based on the requirement of CD64 and Ecgp96 by these strains for invading macrophages and HCMEC/D3 respectively, the presence of OmpA in these strains may seem to dictate their virulence and zoonotic potential, while the presence or absence of CNF1 or other virulence factors might also cause a discernible effect on their virulence. Multi-drug resistance in APEC strains is a common problem and, therefore, poses a bigger health threat if they infect humans. Therefore, small molecule inhibitors of bacterial binding to host cells might serve as alternative therapy for the ever increasing resistance to antibiotics in these strains.
Materials and Methods
Bacterial strains, antibodies, and other reagents
Escherichia coli K1 E44 (OmpA+ E. coli) is a spontaneous rifampicin-resistant mutant of strain NMEC RS218 (serotype O18:K1: H7), which was isolated from the cerebrospinal fluid of a newborn with meningitis, and E91 (OmpA- E. coli) is an ompA deletion mutant of E44.Citation33,34 Bacteria were grown in Luria-Bertani (LB) medium with appropriate antibiotics: E44, 100 µg/ml rifampicin; E91, 12.5 µg/ml tetracycline. Antibodies to CD64 and gp96 were from Santa Cruz Biotechnology (#sc-15364) and Genetex (#GTX103203), respectively. Lipofectamine 2000 (#11668-027), siRNAs specific to CD64 (s65915, #4390771), and gp96 (HSS110955, #1299001), and secondary antibodies tagged to various fluorophores were obtained from Life Technologies. Antibodies to neutrophil elastase (#bs-6982R), and annexin V (#bs-0398R) were purchased from One World Lab. Other bacterial strains are described in . All APEC and NMEC strains used in this study harbor a K1 capsule. To obtain plasmid cured strains, the bacteria were grown in Brain Heart Infusion broth at 42°C, subcultured into fresh broth every 24 hours and plated on MacConkey agar. Then, single colonies were randomly selected for analysis by PCR for plasmid-linked genes. DNA from clones putatively identified as ‘cured’ were then subjected to agarose gel electrophoresis to confirm plasmid loss.
E. coli cell association and invasion assays
RAW 264.7 macrophages were grown to confluence (approximately 105 cells per well) in 24-well plates. They were then incubated with 106 CFU of the strains in experimental medium (DMEM containing 5% heat-inactivated fetal bovine serum) for 60 min at 37°C in a CO2 incubator. HCMEC/D3 were cultured and maintained as described previously.Citation23 For cell association and invasion assays, HCMEC/D3 cultured in 24-well plates were incubated for 90 min with 107 CFU of E. coli. The monolayers were washed with RPMI 1640 3 times and further incubated with gentamicin (100 µg/ml) reconstituted in experimental medium containing for 1 h to kill bound bacteria. The monolayers were washed again and lysed with 0.5% Triton X-100. The intracellular bacteria were determined by plating dilutions on sheep blood agar. To enumerate the total cell-associated bacteria, the experiments were performed without gentamicin.
PCR for ompA sequencing and multiple sequence alignment using Clustal® Omega
Genomic DNA was prepared from bacterial cultures using the ZR Fungal/Bacterial DNA MiniPrep Kit (Zymo Research, Irvine, CA), and amplified by PCR (forward primer: 5′- ATGAAAAAGACAGCTATCGC -3′; reverse primer 5′- TTAAGCCTGCGGCTGAGTTA -3′). PCR amplicons were imaged on a 0.5% agarose-TAE gel with ethidium bromide. PCR amplicons were cleaned using the QIAquick PCR Purification Kit (Qiagen, Valencia, CA) and quantified using the ND-1000 Spectrophotometer (NanoDrop, Wilmington, DE). Purified amplicons were then submitted to Genewiz (Newbury Park, CA) for sequencing. The resultant sequences were analyzed using FinchTV v1.4.0 (Geospiza, Seattle, WA) and aligned using Clustal-Omega v1.2.1 (The European Bioinformatics Institute, www.ebi.ac.uk/Tools/msa/clustalo).
Flow cytometry
To detect the surface expression of CD64 or Ecgp96, RAW 264.7 or HCMEC/D3 cells grown in 6-well plates were washed 3 times with PBS and then detached with TrypLE Express (Life Technologies) from the plates. The cells were fixed using BD Cytofix for 15 min, washed and pre-incubated for 30 min with blocking/wash buffer (PBS+3% normal goat serum) to mask non-specific binding sites. Cells were then incubated with anti-Myc antibody or an isotype-matched control antibody for 1 h at 4°C and washed with buffer. Then FITC-conjugated secondary antibody was added, incubated for 30 min at 4°C and washed with the buffer. The stained cells were then analyzed by 4-color flow cytometry using FACS Calibur Cell Quest Pro software (BD Biosciences, San Jose, CA), and at least 10,000 events were collected for analysis. Results are expressed as bar graphs or histogram overlays with respect to isotype-matched antibody controls. For siRNA studies, 70-80%- confluent RAW 264.7 or HCMEC/D3 in 6-well plates were transfected with 100 pmoles of siRNA to CD64 or Ecgp96, respectively, and the cells were subjected to flow cytometry as mentioned above.
Serum survival assays
The bacterial suspensions (106 CFU/ml) were incubated for 15 min at 37°C with shaking with freshly prepared serum (normal or heat-inactivated) diluted to 40% in gelatin/veronal buffer (Sigma). Aliquots of 10 μl were removed and serially diluted in saline, and dilutions were plated on sheep blood agar. The plates were incubated at 37°C overnight, and the colonies were counted. Percent survival at 15 minutes exposure was calculated based on CFU values obtained from colony counts relative to each strain's values at 0 minutes taken as 100%.
Invasion assays with bone marrow derived macrophages
Bone marrow derived macrophages were isolated from femurs of adult C57BL/6 mice (The Jackson Laboratory, Bar Harbor, ME and Harlan Laboratories, Indianapolis, IN), and seeded in 24-well plates with growth media supplemented with 20% fetal bovine serum, 10 ng/ml macrophage colony-stimulating factor (M-CSF), and 1:1,000 Bac-Off antibiotic solution (Cell Systems Corporation, Kirkland, WA). Growth media was replaced at 3 d post-seeding. Cells were grown to a density of 105 cells per well at approximately 80% confluency at 5 d post-seeding. Invasion assays were performed as previously described, with an increased bacterial invasion time of 2 hours.
Newborn mouse model of meningitis and tissue immunostaining
The Institutional Animal Care and Use Committee of the Saban Research Institute at Children's Hospital Los Angeles (CHLA) approved the animal studies (Protocol #59–14) and followed National Institutes of Health guidelines for the performance of animal experiments. Breeding pairs of C57BL/6 wild-type mice were obtained from Jackson Laboratories and Harlan Laboratories. Three-day-old mouse pups received an intraperitoneal inoculation of 102 colony forming units (CFU) of E. coli K1 or pyrogen-free saline. The pups were monitored up to 48 h postinfection for signs of illness. Pups were sacrificed, cerebrospinal fluid (CSF) was collected aseptically by cisternal puncture, 5 µl of blood was collected by cardiac puncture, and the brains were removed. Animals with traumatic CSF were not considered for the experimental calculations. CSF was directly inoculated into LB broth and incubated overnight at 37°C. The blood samples and one-half of each brain were homogenized and plated on LB agar containing rifampicin (for E44) or blood agar (for APEC and NMEC strains) to determine the bacterial load. The other half of the brain from control and infected pups was embedded in paraffin for tissue sectioning followed by immunostaining. Sections of paraffinized brain from control and infected pups were sequentially rehydrated using xylene, 100% ethanol, 95% ethanol, 50% ethanol, and water. The slides were trypsinized (0.25% trypsin) for 20 min at 37°C for antigen unmasking. The sections were blocked in PBS containing 3% normal goat serum for 30 min at room temperature, stained with 1:100 dilution of neutrophil elastase or annexin V antibody, followed by 1:250 dilution of Alexa 488 secondary antibody and visualized by confocal microscopy.
Statistical analysis
The unpaired Student's t-test was used for statistical analysis of all binding and invasion assays. One-way ANOVA was used for statistical analysis of blood and brain samples from the newborn mouse model. P-values <0.05 were considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_Material.zip
Download Zip (192.8 KB)Acknowledgments
We sincerely thank Lodewijk Spanjaard and Netherlands Reference Laboratory for Bacterial Meningitis for providing the parent NMEC strains used in this study. We also thank G. Esteban Fernandez for assistance with confocal imaging.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by funds from NICHD (NS73115) and NIAID (AI40567) to NVP.
References
- Gaschignard J, Levy C, Romain O, Cohen R, Bingen E, Aujard Y, Boileau P. Neonatal Bacterial Meningitis: 444 Cases in 7 Years. Pediatr Infect Dis J 2007; 30:212-7; http://dx.doi.org/10.1097/INF.0b013e3181fab1e7
- Furyk JS, Swann O, Molyneux E. Systematic review: neonatal meningitis in the developing world. Trop Med Int Health 2011; 16:672-9; PMID:21395927; http://dx.doi.org/10.1111/j.1365-3156.2011.02750.x
- Raymond J, Lopez E, Bonacorsi S, Poyart C, Moriette G, Jarreau PH, Bingen E. Evidence for transmission of Escherichia coli from mother to child in late-onset neonatal infection. Pediatr Infect Dis J 2008; 27:186-8; PMID:18174860
- Manges AR, Johnson JR. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin Infect Dis 2012; 55:712-9; PMID:22615330; http://dx.doi.org/10.1093/cid/cis502
- Manges AR, Smith SP, Lau BJ, Nuval CJ, Eisenberg JN, Dietrich PS, Riley LW. Retail meat consumption and the acquisition of antimicrobial resistant Escherichia coli causing urinary tract infections: a case-control study. Foodborne Pathog Dis 2007; 4:419-31; PMID:18041952; http://dx.doi.org/10.1089/fpd.2007.0026
- Wooster DG, Maruvada R, Blom AM, Prasadarao NV. Logarithmic phase Escherichia coli K1 efficiently avoids serum killing by promoting C4bp-mediated C3b and C4b degradation. Immunology 2006; 117:482-93; PMID:16556262; http://dx.doi.org/10.1111/j.1365-2567.2006.02323.x
- Mittal R, Krishnan S, Gonzalez-Gomez I, Prasadarao NV. Deciphering the Roles of Outer Membrane Protein A Extracellular Loops in the Pathogenesis of Escherichia coli K1 Meningitis. J Biol Chem 2011; 286:2183-93; PMID:21071448; http://dx.doi.org/10.1074/jbc.M110.178236
- Mittal R, Sukumaran SK, Selvaraj SK, Wooster DG, Babu MM, Schreiber AD, Verbeek JS, Prasadarao NV. Fcgamma receptor I α chain (CD64) expression in macrophages is critical for the onset of meningitis by Escherichia coli K1. PLoS Pathog 2010; 6:e1001203; PMID:21124939; http://dx.doi.org/10.1371/journal.ppat.1001203
- Mittal R, Prasadarao NV. gp96 expression in neutrophils is critical for the onset of Escherichia coli K1 (RS218) meningitis. Nat Commun 2011; 2:552; PMID:22109526; http://dx.doi.org/10.1038/ncomms1554
- Prasadarao NV. Identification of Escherichia coli outer membrane protein A receptor on human brain microvascular endothelial cells. Infect Immun 2002; 70:4556-63; PMID:12117968; http://dx.doi.org/10.1128/IAI.70.8.4556-4563.2002
- Sukumaran SK, Shimada H, Prasadarao NV. Entry and intracellular replication of Escherichia coli K1 in macrophages require expression of outer membrane protein A. Infect Immun 2003; 71:5951-61; PMID:14500515; http://dx.doi.org/10.1128/IAI.71.10.5951-5961.2003
- Sukumaran SK, Selvaraj SK, Prasadarao NV. Inhibition of apoptosis by Escherichia coli K1 is accompanied by increased expression of BclXL and blockade of mitochondrial cytochrome c release in macrophages. Infect Immun 2004; 72:6012-22; PMID:15385505; http://dx.doi.org/10.1128/IAI.72.10.6012-6022.2004
- Krishnan S, Liu F, Abrol R, Hodges J, Goddard WA, 3rd, Prasadarao NV. The interaction of N-glycans in Fcgamma receptor I α-chain with Escherichia coli K1 outer membrane protein A for entry into macrophages: experimental and computational analysis. J Biol Chem 2014; 289:30937-49; PMID:25231998; http://dx.doi.org/10.1074/jbc.M114.599407
- Krishnan S, Fernandez GE, Sacks DB, Prasadarao NV. IQGAP1 mediates the disruption of adherens junctions to promote Escherichia coli K1 invasion of brain endothelial cells. Cell Microbiol 2012; 14:1415-33; PMID:22519731; http://dx.doi.org/10.1111/j.1462-5822.2012.01805.x
- Sukumaran SK, Prasadarao NV. Escherichia coli K1 invasion increases human brain microvascular endothelial cell monolayer permeability by disassembling vascular-endothelial cadherins at tight junctions. J Infect Dis 2003; 188:1295-309; PMID:14593586; http://dx.doi.org/10.1086/379042
- Tivendale KA, Logue CM, Kariyawasam S, Jordan D, Hussein A, Li G, Wannemuehler Y, Nolan LK. Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect Immun 2010; 78:3412-9; PMID:20515929; http://dx.doi.org/10.1128/IAI.00347-10
- Kim KS. Mechanisms of microbial traversal of the blood-brain barrier. Nat Rev Microbiol 2008; 6:625-34; PMID:18604221; http://dx.doi.org/10.1038/nrmicro1952
- Logue CM, Doetkott C, Mangiamele P, Wannemuehler YM, Johnson TJ, Tivendale KA, Li G, Sherwood JS, Nolan LK. Genotypic and phenotypic traits that distinguish neonatal meningitis-associated Escherichia coli from fecal E. coli isolates of healthy human hosts. Appl Environ Microbiol 2012; 78:5824-30; PMID:22706051; http://dx.doi.org/10.1128/AEM.07869-11
- Johnson TJ, Siek KE, Johnson SJ, Nolan LK. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J Bacteriol 2006; 188:745-58; PMID:16385064; http://dx.doi.org/10.1128/JB.188.2.745-758.2006
- Mittal R, Gonzalez-Gomez I, Panigrahy A, Goth K, Bonnet R, Prasadarao NV. IL-10 administration reduces PGE-2 levels and promotes CR3-mediated clearance of Escherichia coli K1 by phagocytes in meningitis. J Exp Med 2010; 207:1307-19; PMID:20498022; http://dx.doi.org/10.1084/jem.20092265
- Prasadarao NV, Wass CA, Kim KS. Endothelial cell GlcNAc β 1-4GlcNAc epitopes for outer membrane protein A enhance traversal of Escherichia coli across the blood-brain barrier. Infect Immun 1996; 64:154-60; PMID:8557333
- Massad G, Arceneaux JEL, Byers BR. Acquisition of Iron from Host Sources by Mesophilic Aeromonas Species. J Gen Microbiol 1991; 137:237-41; PMID:1826735; http://dx.doi.org/10.1099/00221287-137-2-237
- Weksler BB, Subileau EA, Perriere N, Charneau P, Holloway K, Leveque M, Tricoire-Leignel H, Nicotra A, Bourdoulous S, Turowski P, Male DK, Roux F, Greenwood J, Romero IA, Couraud PO. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J 2005; 19:1872-4; PMID:16141364
- Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect 2003; 5:449-56; PMID:12738001; http://dx.doi.org/10.1016/S1286-4579(03)00049-2
- Pitout JD. Extraintestinal Pathogenic Escherichia coli: A Combination of Virulence with Antibiotic Resistance. Front Microbiol 2012; 3:9; PMID:22294983; http://dx.doi.org/10.3389/fmicb.2012.00009
- Johnson TJ, Kariyawasam S, Wannemuehler Y, Mangiamele P, Johnson SJ, Doetkott C, Skyberg JA, Lynne AM, Johnson JR, Nolan LK. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J Bacteriol 2007; 189:3228-36; PMID:17293413; http://dx.doi.org/10.1128/JB.01726-06
- Johnson TJ, Wannemuehler Y, Johnson SJ, Stell AL, Doetkott C, Johnson JR, Kim KS, Spanjaard L, Nolan LK. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl Environ Microbiol 2008; 74:7043-50; PMID:18820066; http://dx.doi.org/10.1128/AEM.01395-08
- Johnson TJ, Logue CM, Wannemuehler Y, Kariyawasam S, Doetkott C, DebRoy C, White DG, Nolan LK. Examination of the source and extended virulence genotypes of Escherichia coli contaminating retail poultry meat. Foodborne Pathog Dis 2009; 6:657-67; PMID:19580453; http://dx.doi.org/10.1089/fpd.2009.0266
- Skyberg JA, Johnson TJ, Johnson JR, Clabots C, Logue CM, Nolan LK. Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect Immun 2006; 74:6287-92; PMID:16954398; http://dx.doi.org/10.1128/IAI.00363-06
- Johnson TJ, Jordan D, Kariyawasam S, Stell AL, Bell NP, Wannemuehler YM, Alarcon CF, Li G, Tivendale KA, Logue CM, Nolan LK. Sequence analysis and characterization of a transferable hybrid plasmid encoding multidrug resistance and enabling zoonotic potential for extraintestinal Escherichia coli. Infect Immun 2010; 78:1931-42; PMID:20160015; http://dx.doi.org/10.1128/IAI.01174-09
- Singh SP, Williams YU, Miller S, Nikaido H. The C-terminal domain of Salmonella enterica serovar typhimurium OmpA is an immunodominant antigen in mice but appears to be only partially exposed on the bacterial cell surface. Infect Immun 2003; 71:3937-46; PMID:12819080; http://dx.doi.org/10.1128/IAI.71.7.3937-3946.2003
- Kleinschmidt JH. Folding kinetics of the outer membrane proteins OmpA and FomA into phospholipid bilayers. Chem Phys Lipids 2006; 141:30-47; PMID:16581049; http://dx.doi.org/10.1016/j.chemphyslip.2006.02.004
- Weiser JN, Gotschlich EC. Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect Immun 1991; 59:2252-8; PMID:1646768
- Prasadarao NV, Wass CA, Weiser JN, Stins MF, Huang SH, Kim KS. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect Immun 1996; 64:146-53; PMID:8557332
