Abstract
Enterotoxigenic Escherichia coli (ETEC) serotype O169:H41 has been an extremely destructive epidemic ETEC type worldwide. The strain harbors a large unstable plasmid that is regarded as responsible for its virulence, although its etiology has remained unknown. To examine its genetic background specifically on the unstable retention and responsibility in the unique adherence to epithelial cells and enterotoxin production, the complete sequence of a plasmid, pEntYN10, purified from the serotype strain was determined. The length is 145,082 bp; its GC content is 46.15%. It contains 182 CDSs, which include 3 colonization factors (CFs), an enterotoxin, and large number of insertion sequences. The repertory of plasmid stability genes was extraordinarily scant. Uniquely, results showed that 3 CFs, CS6, CS8 (CFA/III)-like, and K88 (F4)-like were encoded redundantly in the plasmid with unique variations among previously known subtypes. These three CFs preserved their respective gene structures similarly to those of other ETEC strains reported previously with unique sequence variations respectively. It is particularly interesting that the K88-like gene cluster of pEntYN10 had 2 paralogous copies of faeG, which encodes the major component of fimbrial structure. It remains to be verified how the unique variations found in the CFs respectively affect the affinity to infected cells, host range, and virulence of the ETEC strain.
Introduction
Each year, enterotoxigenic Escherichia coli (ETEC) cause nearly one billion cases of diarrheal diseases worldwide. The pathogens often pollute foods and drinking water and which cause endemic prevalence mainly in infants in developing countries and which infect travelers from developed countries.Citation1 Infection gives rise to a severe health hazards in infected areas, resulting in numerous deaths among children.Citation1,2 Etiologically, ETEC harbors one or more cell adhesion factors to colonize on epithelial cells of intestinal surfaces of hosts (colonization factors, CFs). The CFs comprise fimbrial or afimbrial structures on surfaces of bacilli, which facilitate binding to epithelial cells.Citation3,4 Bacilli binding to host cells proliferate on the surface and secrete heat-stable enterotoxin (ST) and/or heat-labile enterotoxin (LT), which disturb the intestinal secretory state, thereby causing watery diarrhea of infected patients.Citation5,6 Such virulence factors of ETEC are generally encoded on plasmids, which can drive the complicated horizontal evolution of pathogenicity of the species.Citation7
Actually, ETEC strains have been defined by possession of their enterotoxin, although their genetic diversity is broad and various serotype strains have been determined as ETEC families from symptomatic patients or foodborne disease outbreaks.Citation3 In spite of low variety in the definitive enterotoxins, CFs of ETEC strains represent wide variation, which explains their diverse antigenicity and structural characteristics.Citation8 In fact, ETEC isolates generally have one or more previously identified CFs. Although adhesive structural molecules of more than 25 types have been identified as CFs, the variation of CFs is not regarded as being completely identified.Citation4 Plasmids encoding CFs and enterotoxins also represent broad diversity including numerous insertion sequences (ISs) and transposons. Host bacilli losing or curing their plasmids lose their fimbrial structure on surface and adhesive activities to host cells generally. This result implies that the virulence of ETEC is closely associated with the retention of its plasmid.Citation9,10
E. coli O169:H41 is an ST-producing ETEC isolates which causes foodborne diarrheic disease.Citation11,12 It was found first as an etiological strain of foodborne case in Japan. Subsequently, its global dissemination has been reported from various countries.Citation13-17 The serotype exhibited variation in ribotypes and outer membrane protein binding patterns, showing its diversity from different outbreaks.Citation18 One unique characteristic of the strain was its atypical adherence manner to epithelial cell lines such as HEp-2, which resembles that of enteroaggregative E. coli (EAggEC).Citation18 It is considered that CFs and enterotoxin of the serotype strains are also encoded in plasmids because the phenotype disappears easily after its passage in liquid medium.Citation18 This disappearance suggests that its virulence plasmid is unstable in vitro and that it is potentially cured out of host infection stage smoothly.
For the specific examination of its unique cell adhesion and unstable retention from the genetic perspective in this study, the complete sequence of a plasmid, pEntYN10, purified from the serotype strain YN10 was determined. Putative CDSs were annotated. In addition, CF-encoding genes were compared with previously reported sequences to assess their potential as virulence factors.
Results
General structure of pEntYN10
To ascertain the genetic features related to its instability and virulence of the host strain ETEC YN10, the plasmid pEntYN10 was purified directly; its sequence was determined. Using the next-generation sequencer FLX system, 17,583,280 bp of 50,934 reads (average length = 345.22 bp, GC content = 46.65%) were obtained. From the read data, 45 contigs longer than 500 bp (N50 = 12,474 bp) were assembled. They were finally combined by gap-filling analysis into single large circular sequences of 145,082 bp. The GC content was 46.15%. On the plasmid, 182 CDSs were estimated by annotation (a circular map is presented in ; gene list on Table S1). This plasmid codes repA1 and repA2 replication genes of incFII group, which means that it belongs to the RepFII plasmid family. The F-transfer pili genes are drastically truncated in the plasmid with the result that only genes traI and traX remain. This truncation of the transfer system was structurally similar to pETEC_80 ().Citation19 Although its gene components are similar to the plasmid of the family sequenced previously,Citation19-23 pEntYN10 is marked by its large size, high percentage of ISs, small number of stability genes, and various repertories of adhesion genes, as described below.
Figure 1. Circular representation of pEntYN10. The innermost circle represents G + C content (% of 1,450 bp window length). Greater and less than 50% are colored respectively as purple and green. The outer circles represent annotated CDSs with their strand orientation (outer – forward; inner – reverse). Colors show their putative categories of function as follows: virulence and cell adhesion, red; IS, pink; replication and plasmid stability, green; other functions, blue. Virulence and cell adhesion related CDSs were assigned by their names.
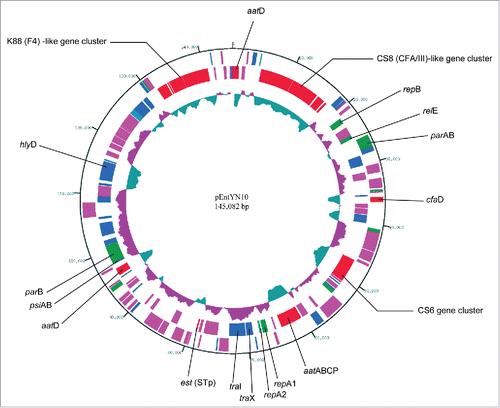
Table 1. Characteristics of RepFIIA plasmids from human ETEC strains
Plasmid stability genes on pEntYN10
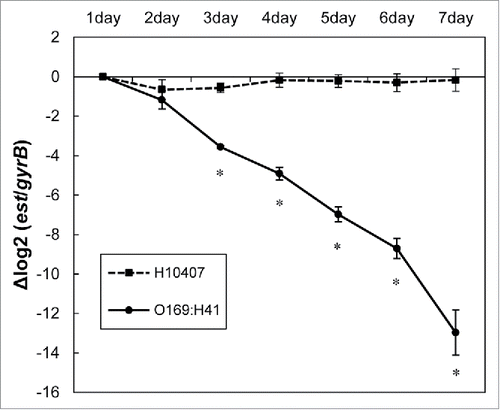
Instability of pEntYN10 in vitro has been noticed in its passage process and has been discussed in the previous paper.Citation12 Actually, it was observed that the plasmid was lost during passages more drastically than a referential ETEC strain H10407 along with in vitro passages (). The entire sequence of pEntYN10 revealed that well-known stability genes on pEntYN10 were fewer than those of other sequenced plasmids (). RepFII plasmids generally possess stbAB and psiAB in their backbones,Citation7 although pEntYN10 lacks a stbAB operon. In addition to the fragile backbone, other stability genes such as hok/mok/sok system and sopAB were also not encoded on pEntYN10. Putative stability related genes of pEntYN10 are restricted to genes harboring partition domains (CDS no. 28, 29, and 124), one toxin/antitoxin relE family (CDS no. 29 and 30), and 4 genes possessing resolvase domain (CDS no. 52, 69, 116, and 159) (Table S1).
Figure 2. Stability of pEntYN10 during passages was verified during 7 d by ΔΔCt comparison between genes on plasmid (est)/chromosome (gyrB) using realtime PCR. The Y-axis represents the difference of Ct value (ΔCt) between est and gyrB of respective strains. Asterisks denote statistically significant difference (*P < 0.001) by Student's t-test between ETEC strain O169:H41 (YN10) and positive control (H10407).
Potential virulent genes encoded on pEntYN10
An outstanding feature of pEntYN10 was that 3 CFs, CS6, CS8 (CFA/III)-like, and K88 (F4)-like, coincided in the single plasmid. These three CFs preserved their respective gene structures and low GC contents as those of other ETEC strains reported previously (),Citation24-27 with unique sequence variations (). Adhesion to HEp-2 cells with an aggregative pattern was unique to YN10 and disappeared in a derivative strain lacking the plasmid after passages (), which suggests that the plasmid correlated to the property of adhesion to host cells.
Figure 3. Linear comparison of adhesion related gene clusters between pEntYN10 and other plasmids of ETEC strains reported previously. The gene clusters were retrieved from the GenBank database: (A) CS6 [accession ID:U04846]; (B) CFA/III [accession ID:AB049751]; (C) K88 gene clusters for comparison were retrieved from the complete sequence of plasmid pUMNK88 [accession ID:CP002730]. The length scales are shown in respective bars. Genes coding fimbrial structure proteins are shown as greyed. They were analyzed in detail for this study.
![Figure 3. Linear comparison of adhesion related gene clusters between pEntYN10 and other plasmids of ETEC strains reported previously. The gene clusters were retrieved from the GenBank database: (A) CS6 [accession ID:U04846]; (B) CFA/III [accession ID:AB049751]; (C) K88 gene clusters for comparison were retrieved from the complete sequence of plasmid pUMNK88 [accession ID:CP002730]. The length scales are shown in respective bars. Genes coding fimbrial structure proteins are shown as greyed. They were analyzed in detail for this study.](/cms/asset/1c7eb716-4029-49ee-a5f1-0c54c8782bc5/kvir_a_1094606_f0003_b.gif)
Figure 4. Light micrograph of HEp-2 cells infected by ETEC strains (A). Scale bars, 10 μm. The number of adhered bacteria was counted quantitatively for each strain (B). Asterisks denote statistically significant difference (*P < 0.001) by Student's t-test between ETEC strain O169:H41 (YN10) and other strains.
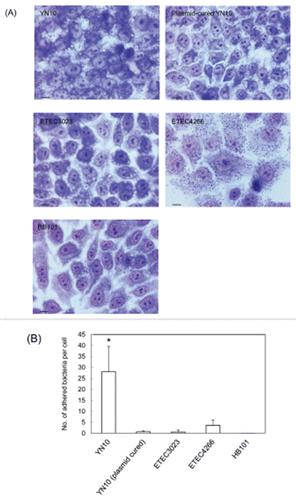
Table 2. Predicted CDSs on pEntYN10 related to cell adhesion
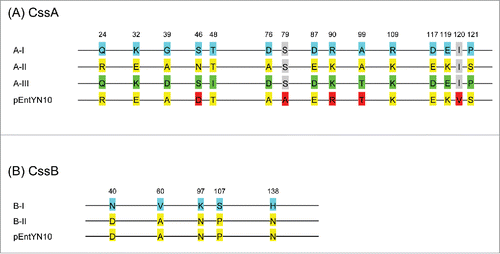
CS6 genes have high levels of sequence identity with previously reported genes (). CS6-coding genes cssA and cssB are classifiable respectively into 3 (A-I, A-II, and A-III) and 2 subtypes (B-I and B-II).Citation28 The CS6 variant of pEntYN10 was found to be unique but similar to A-II/B-II by amino acid sequence homology ().
Figure 5. Sequence variation of CssA and CssB subunit of CS6. (A) CssA. Blue labels are referential amino acids for the type AI, yellow for type AII, and green for type AIII. Red labels differ from the reference amino acids. (B) CssB. Blue labels are referential amino acids for type BI, and yellow labels are for the type BII. Numbers refer to amino acid residues of the reference sequences.
Similarity of amino acid sequences between CS8 on pEntYN10 and previously reported ETEC strains was lower than those of CS6 (). The CS8 components reportedly form a long rod-like fimbrial structure on the cell surface of ETEC, to attach specifically to human intestinal cells.Citation29,30 The main structural protein is CofA, which possesses structural homology with the main fimbrial component of V. cholerae, TcpA.Citation31 The amino acid sequence of CofA of pEntYN10 was similar to that of pSH1134, a plasmid of an ETEC strain 260-1 (). Recently, 5 residues in this protein were reported as highly conserved to construct the filamentous structure.Citation31,32 These residues of CofA in pEntYN10 were also preserved (), which supports the functional conservation.
Figure 6. Comparison of CofA between pEntYN10 and ETEC 260-1 [BAB62897] and TcpA from V. cholerae O1 [AAK20785]. (A) A phylogenetic tree based on Neighbor Joining method. (B) Amino acid sequence alignment of the 3 proteins by CLUSTAL 2.1. Highly conserved residues responsible for the stable structure of pili of V. cholerae (Met26, Glu30, Arg51, Leu98, and Glu108) are shaded.
![Figure 6. Comparison of CofA between pEntYN10 and ETEC 260-1 [BAB62897] and TcpA from V. cholerae O1 [AAK20785]. (A) A phylogenetic tree based on Neighbor Joining method. (B) Amino acid sequence alignment of the 3 proteins by CLUSTAL 2.1. Highly conserved residues responsible for the stable structure of pili of V. cholerae (Met26, Glu30, Arg51, Leu98, and Glu108) are shaded.](/cms/asset/e2e803b0-f702-45c8-9623-76b6467cc704/kvir_a_1094606_f0006_b.gif)
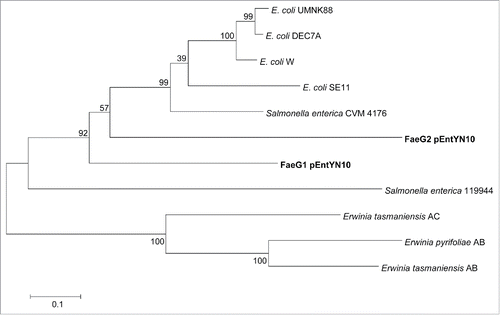
The K88 gene cluster also encodes fimbrial structure components.Citation33 It is particularly interesting that the gene cluster of pEntYN10 had 2 paralogous copies of faeG, which encodes the major component of the K88 fimbrial structure (). Phylogenetic relation of FaeG sequences exhibited that these 2 paralogous genes, faeG1 and faeG2, were separated from other E. coli strains and they are more similar to that of Salmonella enterica serovar Infantis (). These genes were topologically close with long branches, which implies their deep genetic divergence.
Discussion
Recent developments in genomic analysis using next generation sequencers have yielded an extremely large number of draft sequences from widely diverse species. They were registered in databases, expanding the plentiful data resources. Such enormous amounts of information have driven a paradigm shift in various research fields, especially those of comparative genomics, genetic diversity, and molecular evolution.Citation34 In spite of the powerful strategy, it is noteworthy that the current state of sequence techniques providing massive amounts of short reads has hindered the scrutiny of the structural arrangements of genomes, and has presented an obstacle to the specific examination of the genetic nature of virulent plasmids harboring complicated structures and active dynamics because of their many mobile elements. For instance, a virulent plasmid pCoo has an atypical structure integrated with 2 backbones of replication units (RepFIIA and RepI1).Citation21 Although such a unique structure can provide insights into the evolutionary dynamics of pathogenicity of ETEC, it is difficult to ascertain that structure from a simple accumulation of short read sequences. This study revealed the complete sequence of a plasmid of an ETEC strain, pEntYN10, which might help to estimate not only phenotypic features of the strain but also its evolutionary background.
A curious feature of pEntYN10 is its small number of stability-related genes (). The instability was concordantly verified in vitro, in contrast to stable retention of well-known ETEC strain H10407 plasmids (). Instability in vitro might provide benefits of an infectious habitat for the host bacilli. It is noteworthy that instability of pEntYN10 was observed only in vitro curture media. Indeed, plasmid-positive O169:H41 strains were actually isolated from clinical feces,Citation18 perhaps because of the higher stability of the plasmid in vivo. Plasmid stability in vivo should be elucidated. Some putative stability-related genes found in pEntYN10 such as a relE family gene or resolvase genes might be a clue to approach it.
In addition to instability, results showed that the tra region of pEntYN10 is largely truncated, implying that the plasmid lacks a notorious strategy, the F-transfer system, to transfer itself horizontally to other bacilli because of a lack of an essential gene: traA. Similar truncation was reported in pETEC948 and pETEC_80 (), although that truncation was probably compensated by other coexisting plasmids in their host strains. O169 clinical strains reportedly possess no other coexisting plasmids,Citation18 therefore, it remains to be seen whether the plasmid has the potential to be transferred horizontally in natural environments. Strong attachment to epithelial cells provided by the plasmid might enhance the survival fitness of host bacilli to compensate for the defects sufficiently as an independent replicon.
In our study, cell attachment pattern of ETEC strain YN10 in vitro was extraordinarily unique (). Functional redundancy of as many as 3 CFs on pEntYN10 may reinforce its adhesion to host cells in infectious stages more widely and tightly, which can enlarge their habitats as a pathogen. The redundancy may also affect its epidemiological features and pathogenicity of ETEC O169:H41.Citation13-17 Each role of CFs that might contribute to pathogenicity and virulence should be scrutinized one by one using recombinant plasmids to uncover it.
In addition, 3 CFs encoded in pEntYN10 possessed unreported variations. The most unique features were found in K88-like fimbrial components. pEntYN10 encodes 2 copies of structural gene homologs tandemly: faeG1 and faeG2 ( (C)). Such duplication and their unique amino acid sequences might contribute to adhesion to human cells, although the adhesion targets of K88 fimbriae of ETEC have been regarded as restricted to porcine.Citation35 The two paralogous genes have mutually diverted sequences. They are more similar to orthologous faeG of S. Infantis isolated from human patientsCitation36 than those of ETEC isolates (). That fact implies that the K88-like genes of the plasmid might enable O169:H41 strains to attach not to porcine cells but to human cells. However, gene structures and protein sequences of CS6 and CS8 on pEntYN10 were highly conserved, which suggests their functional conservation, although the parental strain YN1 was not reacted with the anti-CS8 MAbs.Citation18 How the unique variations found in the CFs respectively affect the affinity to the infected cells, host range, and virulence should be verified. Infection experiments using animal models might also provide more credible insights in their roles, although careful attention must be devoted to the difficulty of examining the possibility of its broad host range driven by the variety of CFs.
In conclusion, the complete sequence of a plasmid of ETEC O169:H41 showed its unique components, implying association with its phenotypic characteristics such as instability in vitro and atypical adhesion pattern to host cells. The CF variety might shed light on the evolutionary strategy and genetic diversity of ETEC pathogenicity.
Materials and Methods
Bacterial strain
An ETEC YN10 was a derivative of the original strain ETEC O169:H41 YN1, isolated from a diarrheic patient during an outbreak foodborne disease of Osaka City, Japan in 1991.Citation12 Our previous studyCitation18 revealed that the ETEC O169:H41 strains did not agglutinate human or bovine red blood cells in a mannose-resistant manner. The strains showed positive reaction in dot-blot tests using anti-CS6 antibodies. Although CS6 is a structural protein of CFA/IV, no fimbrial structure was identified.Citation18
Instability test of plasmid pEntYN10
YN10 and a control strain H10407 were passaged for 7 d in 10 ml of TSB broth at 37°C. At each passage, 1 μl of each culture incubated for 24 h was transferred into fresh broth. Remaining cultured bacilli were lysed to purify the genomic and plasmid DNA, using Gentra Puregene Tissue Kit (Qiagen Inc., Valencia, CA). The copy numbers of plasmids were quantified using real-time PCR with SYBR Green targeting ST producing gene (est) using primers (STp-f: 5′- CTTTCCCCTCTTTTAGTCAGTCAACT- 3′ and STp-r: 5′- GCAGTAAAATGTGTTGTTCATATTTTCTG- 3′).Citation37 For chromosomal control, gyrB gene was also quantified according to a previous report.Citation38
DNA sequencing
To purify the plasmid pEntYN10, the host strain YN10 was grown in Luria-Bertani broth at 37°C with vigorous aeration. The plasmid was purified by agarose extraction for separation from chromosomal DNA (NucleoBond BAC100; Macherey Nagel, GmbH and Co. KG, Germany), from cultured bacilli within at least 3 times passage from the original isolation.
The draft sequences of the plasmid were obtained using a genome sequencer (FLX System; 454 Life Sciences, Roche Applied Science, Branford, CT). Linkage of each contig estimated by assembly software GS De Novo Assembler (ver. 2.5.3) was verified using the gap filling method with PCR and direct sequencing of the amplified products.
Annotation
To check coding sequences (CDSs) on the plasmid, an automated annotation system, Microbial Genome Annotation Pipeline (MiGAP), was subjected to the complete sequence of the plasmid at first (http://www.migap.org/index.php/en).Citation39 Subsequently, all annotated CDSs were curated further by search of the homologous protein sequences in GenBank using BLASTP. To avoid systematic sequencing errors by 454 FLX sequencer, putative frameshift errors caused by more than 5 polynucleotide regions were checked using Sanger sequencing. The entire sequence and annotation information of the plasmid was registered at DDBJ [accession no: AP014654].
Tissue culture adhesion tests
Adhesion tests of YN10 and other E. coli strains ETEC3023, ETEC4266 (isolated from diarrheal patients admitted to the Infectious Diseases Hospital, Kolkata, IndiaCitation40), and a laboratory strain HB101 for negative control to HEp-2 cells were conducted by infection with cultured bacilli (MOI = 1.0 × 10) on monolayers of HEp-2 cells in 0.5 mL of Basal Eagle's medium as a previous study.Citation18 Plasmid-cured YN10 was cloned from parental YN10 passaged during 10 d in TSB. After 3 h incubation at 37°C, the monolayers were washed 3 times with PBS(-). Then 0.5 mL of the medium was added. After more 3 h incubation, the monolayers were washed again 3 times as in the previous step. The cells were fixed with absolute methanol and were stained with Giemsa.
To compare the adhesion patterns of respective strains quantitatively, the adhesion tests were operated 5 times and the number of bacteria adhering to 60 epithelial cells was counted and its average was calculated.
Pairwise alignment
Pairwise alignment of amino acid sequences of orthologous CS genes were conducted using the EMBOSS needle program at the website of EMBL-EBI (http://www.ebi.ac.uk/Tools/psa/).
Construction of phylogenetic trees
Phylogenetic trees based on amino acid sequences were constructed using Neighbor-Joining method with a Poisson correction model and a 500 replicate bootstrap analysis using MEGA 6.06.Citation41 Bootstrap consensus trees were calculated. Then the average bootstrap values were included on the trees.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Supplemental_Table.zip
Download Zip (23.1 KB)Funding
This study was partially supported by a grant from the Ministry of Health, Labor, and Welfare of Japan (H24-Syokuhin-Ippan-010).
References
- Qadri F, Svennerholm AM, Faruque AS, Sack RB. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev 2005; 18:465–83; PMID:16020685; http://dx.doi.org/10.1128/CMR.18.3.465-483.2005
- Wenneras C, Erling V. Prevalence of enterotoxigenic Escherichia coli -associated diarrhoea and carrier state in the developing world. J Health Popul Nutr 2004; 22:370–82; PMID:15663170; http://www.jstor.org/stable/23499155
- Levine MM. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J Infect Dis 1987; 155:377–89; PMID:3543152; http://dx.doi.org/10.1093/infdis/155.3.377
- Gaastra W, Svennerholm AM. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol 1996; 4:444–52; PMID:8950814; http://dx.doi.org/10.1016/0966-842X(96)10068-8
- Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev 1998; 11:142–201; PMID:9457432
- Fleckenstein JM, Hardwidge PR, Munson GP, Rasko DA, Sommerfelt H, Steinsland H. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect 2010; 12:89–98; PMID:19883790; http://dx.doi.org/10.1016/j.micinf.2009.10.002
- Johnson TJ, Nolan LK. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev 2009; 73:750–74; PMID:19946140; http://dx.doi.org/10.1128/MMBR.00015-09
- Sahl JW, Steinsland H, Redman JC, Angiuoli SV, Nataro JP, Sommerfelt H, Rasko DA. A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect Immun 2011; 79:950–60; PMID:21078854; http://dx.doi.org/10.1128/IAI.00932-10
- Schifferli DM, Beachey EH, Taylor RK. The 987P fimbrial gene cluster of enterotoxigenic Escherichia coli is plasmid encoded. Infect Immun 1990; 58:149–56; PMID:1967167
- Casey TA, Moon HW. Genetic characterization and virulence of enterotoxigenic Escherichia coli mutants which have lost virulence genes in vivo. Infect Immun 1990; 58:4156–8; PMID:2254037
- Ando K, Itaya T, Aoki A, Saito A, Masaki H, Tokumaru Y. An outbreak of food poisoning caused by enterotoxigenic Escherichia coli O169:H41. Jpn J Food Microbiol 1993; 10:77–81; PMID:24270144; http://doi.org/10.7883/yoken.66.530
- Nishikawa Y, Hanaoka M, Ogasawara J, Moyer NP, Kimura T. Heat-stable enterotoxin-producing Escherichia coli O169:H41 in Japan. Emerg Infect Dis 1995; 1:61; PMID:8903162; http://dx.doi.org/10.3201/eid0102.950206
- Beatty ME, Bopp CA, Wells JG, Greene KD, Puhr ND, Mintz ED. Enterotoxin-producing Escherichia coli O169:H41, United States. Emerg Infect Dis 2004; 10:518–21; PMID:15109427; http://dx.doi.org/10.3201/eid1003.030268
- Cho SH, Kim J, Oh KH, Hu JK, Seo J, Oh SS, Hur MJ, Choi YH, Youn SK, Chung GT, et al. Outbreak of enterotoxigenic Escherichia coli O169 enteritis in schoolchildren associated with consumption of kimchi, Republic of Korea, 2012. Epidemiol Infect 2014; 142:616–23; PMID:23800632; http://dx.doi.org/10.1017/S0950268813001477
- Devasia RA, Jones TF, Ward J, Stafford L, Hardin H, Bopp C, Beatty M, Mintz E, Schaffner W. Endemically acquired foodborne outbreak of enterotoxin-producing Escherichia coli serotype O169:H41. Am J Med 2006; 119:168 e7-10; PMID:16443428; http://dx.doi.org/10.1016/j.amjmed.2005.07.063
- Hamada K, Tsuji H, Shimada K. Outbreaks of heat stable enterotoxin-producing Escherichia coli O169 in the Kinki district in Japan: epidemiological analysis by pulsed-field gel electrophoresis. Jpn J Infect Dis 1999; 52:165–7; PMID:10592898
- Harada T, Itoh K, Yamaguchi Y, Hirai Y, Kanki M, Kawatsu K, Seto K, Taguchi M, Kumeda Y. A foodborne outbreak of gastrointestinal illness caused by enterotoxigenic Escherichia coli serotype O169:H41 in Osaka, Japan. Jpn J Infect Dis 2013; 66:530–3; PMID:24270144; http://dx.doi.org/10.7883/yoken.66.530
- Nishikawa Y, Helander A, Ogasawara J, Moyer NP, Hanaoka M, Hase A, Yasukawa A. Epidemiology and properties of heat-stable enterotoxin-producing Escherichia coli serotype O169:H41. Epidemiol Infect 1998; 121:31–42; PMID:9747753; http://dx.doi.org/10.1017/S0950268898001046
- Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, et al. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 2008; 190:6881–93; PMID:18676672; http://dx.doi.org/10.1128/JB.00619-08
- Crossman LC, Chaudhuri RR, Beatson SA, Wells TJ, Desvaux M, Cunningham AF, Petty NK, Mahon V, Brinkley C, Hobman JL, et al. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J Bacteriol 2010; 192:5822–31; PMID:20802035; http://dx.doi.org/10.1128/JB.00710-10
- Froehlich B, Parkhill J, Sanders M, Quail MA, Scott JR. The pCoo plasmid of enterotoxigenic Escherichia coli is a mosaic cointegrate. J Bacteriol 2005; 187:6509–16; PMID:16159784; http://dx.doi.org/10.1128/JB.187.18.6509-6516.2005
- Ochi S, Shimizu T, Ohtani K, Ichinose Y, Arimitsu H, Tsukamoto K, Kato M, Tsuji T. Nucleotide sequence analysis of the enterotoxigenic Escherichia coli Ent plasmid. DNA Res 2009; 16:299–309; PMID:19767599; http://dx.doi.org/10.1093/dnares/dsp015
- Wajima T, Sabui S, Kano S, Ramamurthy T, Chatterjee NS, Hamabata T. Entire sequence of the colonization factor coli surface antigen 6-encoding plasmid pCss165 from an enterotoxigenic Escherichia coli clinical isolate. Plasmid 2013; 70:343–52; PMID:23933356; http://dx.doi.org/10.1016/j.plasmid.2013.07.006
- Taniguchi T, Akeda Y, Haba A, Yasuda Y, Yamamoto K, Honda T, Tochikubo K. Gene cluster for assembly of pilus colonization factor antigen III of enterotoxigenic Escherichia coli. Infect Immun 2001; 69:5864–73; PMID:11500465; http://dx.doi.org/10.1128/IAI.69.9.5864-5873.2001
- Verdonck F, Cox E, Schepers E, Imberechts H, Joensuu J, Goddeeris BM. Conserved regions in the sequence of the F4 (K88) fimbrial adhesin FaeG suggest a donor strand mechanism in F4 assembly. Vet Microbiol 2004; 102:215–25; PMID:15327796; http://dx.doi.org/10.1016/j.vetmic.2004.06.002
- Wolf MK, de Haan LA, Cassels FJ, Willshaw GA, Warren R, Boedeker EC, Gaastra W. The CS6 colonization factor of human enterotoxigenic Escherichia coli contains two heterologous major subunits. FEMS Microbiol Lett 1997; 148:35–42; PMID:9066107; http://dx.doi.org/10.1111/j.1574-6968.1997.tb10263.x
- Shepard SM, Danzeisen JL, Isaacson RE, Seemann T, Achtman M, Johnson TJ. Genome sequences and phylogenetic analysis of K88- and F18-positive porcine enterotoxigenic Escherichia coli. J Bacteriol 2012; 194:395–405; PMID:22081385; http://dx.doi.org/10.1128/JB.06225-11
- Sabui S, Ghosal A, Dutta S, Ghosh A, Ramamurthy T, Nataro JP, Hamabata T, Chatterjee NS. Allelic variation in colonization factor CS6 of enterotoxigenic Escherichia coli isolated from patients with acute diarrhoea and controls. J Med Microbiol 2010; 59:770–9; PMID:20299505; http://dx.doi.org/10.1099/jmm.0.017582-0
- Honda T, Arita M, Miwatani T. Characterization of new hydrophobic pili of human enterotoxigenic Escherichia coli: a possible new colonization factor. Infect Immun 1984; 43:959–65; PMID:6199307
- Shinagawa H, Taniguchi T, Yamaguchi O, Yamamoto K, Honda T. Cloning of the genes that control formation of the fimbrial colonization factor antigen III (CFA/III) from an enterotoxigenic Escherichia coli. Microbiol Immunol 1993; 37:689–94; PMID:7903787; http://dx.doi.org/10.1111/j.1348-0421.1993.tb01693.x
- Fukakusa S, Kawahara K, Nakamura S, Iwashita T, Baba S, Nishimura M, Kobayashi Y, Honda T, Iida T, Taniguchi T, et al. Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing. Acta Crystallogr D Biol Crystallogr 2012; 68:1418–29; PMID:22993096; http://dx.doi.org/10.1107/S0907444912034464
- Li J, Lim MS, Li S, Brock M, Pique ME, Woods VL, Jr, Craig L. Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen/deuterium exchange mass spectrometry. Structure 2008; 16:137–48; PMID:18184591; http://dx.doi.org/10.1016/j.str.2007.10.027
- Mol O, Oudega B. Molecular and structural aspects of fimbriae biosynthesis and assembly in Escherichia coli. FEMS Microbiol Rev 1996; 19:25–52; PMID:8916554; http://dx.doi.org/10.1111/j.1574-6976.1996.tb00252.x
- Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods 2008; 5:16–8; PMID:18165802; http://dx.doi.org/10.1038/nmeth1156
- Blanco J, Blanco M, Garabal JI, Gonzalez EA. Enterotoxins, colonization factors and serotypes of enterotoxigenic Escherichia coli from humans and animals. Microbiologia 1991; 7:57–73; PMID:1684712
- Aviv G, Tsyba K, Steck N, Salmon-Divon M, Cornelius A, Rahav G, Grassl GA, Gal-Mor O. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ Microbiol 2014; 16:977–94; PMID:24320043; http://dx.doi.org/10.1111/1462-2920.12351
- Hidaka A, Hokyo T, Arikawa K, Fujihara S, Ogasawara J, Hase A, Hara-Kudo Y, Nishikawa Y. Multiplex real-time PCR for exhaustive detection of diarrhoeagenic Escherichia coli. J Appl Microbiol 2009; 106:410–20; PMID:19200309; http://dx.doi.org/10.1111/j.1365-2672.2008.04043.x
- Taylor DE, Rooker M, Keelan M, Ng LK, Martin I, Perna NT, Burland NT, Blattner FR. Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J Bacteriol 2002; 184:4690–8; PMID:12169592; http://dx.doi.org/10.1128/JB.184.17.4690-4698.2002
- Sugawara H, Ohyama A, Mori H, Kurokawa K. Microbial genome annotation pipeline (MiGAP) for diverse users. The 20th International Conference on Genome Informatics (GIW2009). Yokohama, Japan, 2009.
- Ghosal A, Bhowmick R, Nandy RK, Ramamurthy T, Chatterjee NS. PCR-based identification of common colonization factor antigens of enterotoxigenic Escherichia coli. J Clin Microbiol 2007; 45:3068–3071; PMID:17596357; http://dx.doi.org/10.1128/JCM.00646-07
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30:2725–9; PMID:24132122; http://dx.doi.org/10.1093/molbev/mst197