Abstract
Pathogenicity of the saprobe Aspergillus fumigatus strictly depends on nutrient acquisition during infection, as fungal growth determines colonisation and invasion of a susceptible host. Primary metabolism has to be considered as a valid target for antimycotic therapy, based on the fact that several fungal anabolic pathways are not conserved in higher eukaryotes. To test whether fungal proliferation during invasive aspergillosis relies on endogenous biosynthesis of aromatic amino acids, defined auxotrophic mutants of A. fumigatus were generated and assessed for their infectious capacities in neutropenic mice and found to be strongly attenuated in virulence. Moreover, essentiality of the complete biosynthetic pathway could be demonstrated, corroborated by conditional gene expression in infected animals and inhibitor studies. This brief report not only validates the aromatic amino acid biosynthesis pathway of A. fumigatus to be a promising antifungal target but furthermore demonstrates feasibility of conditional gene expression in a murine infection model of aspergillosis.
Introduction
It is well established that microbial pathogenicity is a multi-factorial trait that comprises specific, host-damaging virulence factors but also general, non-specific cellular characteristics that support colonisation and infection of a susceptible host. Among these, metabolic versatility is crucial, based on the fact that pathogenic microorganisms need to grow and survive within the surrounding tissue and might have to colonize diverse niches inside the host.Citation1 Accordingly, successful exploitation of the host microenvironment by an invading pathogen determines proliferation and consequently pathogenesis, and biosynthetic routes in the context of available substrates define a fundamental aspect of the pathogen-host interplay.
Fungal metabolism has to be considered as a valid target for antimycotic therapy, based on the fact that several fungal anabolic pathways are not well conserved or even absent in higher eukaryotes. For the ubiquitous mold Aspergillus fumigatus, the main causative agent of aspergillosis, several of such pathogen-specific biosynthetic reactions have been described.Citation2 As a primary saprobe, A. fumigatus thrives by degradation of the growth substrate via osmotrophy, based on abundant secretion of hydrolytic enzymatic activities that decompose organic polymers. Break-down products of this extracellular digestion are then taken up to be channelled into the primary fungal metabolism. During infection, this feeding mode likely contributes to growth and, therefore, virulence as well as to dissemination: degradation of the primarily colonized pulmonary tissue supports angioinvasion and infection of secondary organs, eventually resulting in systemic disease progression. Distinct virulence factors are hard to define for A. fumigatus, and comparative genome analyses indicate the absence of prime candidates that may serve as specific mediators of virulence but support the multi-factorial nature of its pathogenicity.Citation3 The versatile metabolism of A. fumigatus has to be regarded as an essential virulence determinant that has evolved to support growth in its prime ecological niche but which also promotes its pathogenic potential.Citation4 By assessing the virulence potential of corresponding mutant strains in suitable infection models, some of the underlying pathways could be identified as such disease determinants, like biosynthesis of para-aminobenzoic acid, uracil/uridine, lysine, or isoleucine.Citation5-9
We here explore the contribution of aromatic amino acid biosynthesis to growth and virulence of A. fumigatus during invasive aspergillosis. This biosynthetic route feeds from 2 intermediates of carbon metabolism and results in chorismic acid as the last common compound.Citation10 From this so-called shikimate pathway,Citation11 branched biosynthesis of tyrosine and phenylalanine and of tryptophan is accomplished (). This anabolic pathway was found to be highly preserved among algae, bacteria, fungi, higher plants, and apicomplexa,Citation12,13 but absent in animals, making it a promising target for therapeutic substances. Moreover, the intermediate chorismic acid and the product tryptophan serve as precursors for ubiquinone and nicotinamide, respectively, which are substantially involved in the respiratory chain and cellular redox potential. A known inhibitor of the shikimate pathway is N(phosphonomethyl)glycine (synonym: glyphosate), which has been established as a broad-spectrum systemic herbicide for decades. In fungi, however, any interference with biosynthetic routes that results in imbalances of amino acid homeostasis may be counteracted by the so-called cross-pathway control (CPC) regulatory system that acts in A. fumigatus by its transcriptional activator CpcA on a variety of target genes to increase their transcription and, therefore, gene product levels.Citation14
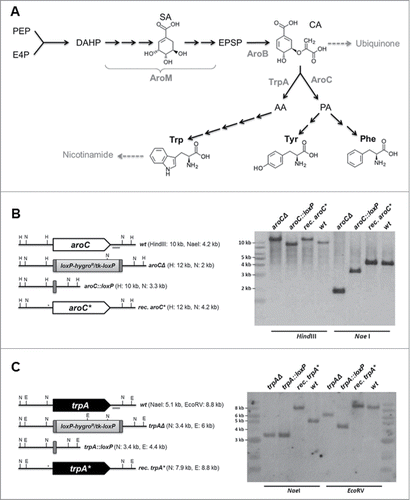
Figure 1. Targeting of genes encoding key enzymes of fungal aromatic amino acids biosynthesis. (A) Schematic outline of aromatic amino acid biosynthesis in Aspergillus. Starting reagents erythrose-4-phosphate (E4P) and phosphoenolpyruvate (PEP) as well as intermediates chorismic acid (CA), prephenic acid (PA), and anthanilate (AA) are indicated, together with encoding genes of key enzymatic activities, such as aroM (multifunctional enzyme, encoded by A. fumigatus annotated locus AfuA_1G13740), aroB (chorismate synthase, AfuA_1G06940), aroC (chorismate mutase, AfuA_5G13130), and trpA (anthranilate synthase I, AfuA_6G12580). (B) and (C) Schematic outlines of the genomic situation for the targeted gene loci aroC and trpA together with Southern hybridization analyses indicating successful deletion, marker excision, and reconstitution.
To date the role of aromatic amino acid biosynthesis in the context of aspergillosis has not been addressed. Our data collected on this topic demonstrate validity of the approach of targeting fungal primary metabolic routes of for identifying A. fumigatus virulence determinants and, furthermore, establish a conditional expression system in the context of infection by this human-pathogenic mold.
Results and Discussion
In order to generate defined auxotrophic mutants of A. fumigatus that would be unable to synthesize aromatic amino acids, genes encoding key enzymes of the pathways were deleted in a suitable recipient strain (AfS35, akuA::loxP) by gene replacement after homologous recombination followed by marker excision, validation by Southern hybridization (), and phenotyping. First, the Aspergillus chorismate mutase-encoding gene aroCCitation15 (AfuA_5G13130) was deleted, resulting in a strain (AfS155) unable to convert chorismate to prephenate and therefore being auxotroph for both tyrosine and phenylalanine, as demonstrated by growth tests on respective solid media (). Interestingly, this deletant germinated poorly in liquid medium containing all necessary supplements (): microscopic inspection of conidia inoculated in liquid minimal medium supplemented with tyrosine and phenylalanine revealed that only 30% of the aroCΔ deletant spores had germinated after prolonged incubation time of 18 hours at 37°C, in contrast to the wild-type strain and the reconstituted aroC+ control strain, which both showed germination rates of about 50%. Moreover, hyphal extension appeared significantly reduced for the aroCΔ strain in liquid culture. This phenotype of impaired supplementation of the Tyr-/Phe- auxotrophy indicates an altered uptake capacity in this three-dimensional and presumably microaerophilic environment.
Figure 2. A. fumigatus strains that are auxotrophic for phenylalanine/tyrosine or tryptophan are severely attenuated in virulence. (A) Strains deleted for aroC or trpA are auxotroph for tyrosine and phenylalanine or tryptophan, respectively, as demonstrated by their growth phenotype on supplemented media. 1 × 103 conidia of deletion strains AfS155 (aroCΔ) or AfS157 (trpAΔ) and their reconstituted derivatives AfS156 (aroC*) or AfS158 (trpA*) were spotted on Aspergillus minimal medium (AMM) supplemented with phenylalanine (F), tyrosine (Y), tryptophan (W), or anthranilate (AA) and growth was monitored after 3 days of incubation at 37°C. (B) Germination of an aroCΔ deletion strain is significantly impaired in liquid culture, as assessed by microscopic inspection of inoculated conidia in supplemented culture medium after 18 hours incubation at 37°C: while germ tubes and elongated hyphae became evident for the wild-type progenitor strain or the trpAΔ mutant, could almost none of these structures be seen for the tyrosine/phenylanine auxotroph. (C) Inhibitor studies indicate that deletion of aroC or trpA does not significantly interfere with respiratory capacities of A. fumigatus. Shown are dilution series of indicated strains on culture medium supplemented with the inhibitors CCCP and sodium azide, respectively, after 3 days of growth at 37°C. (D) Virulence of aromatic amino acid biosynthesis deletants in neutropenic mice after pulmonary (left, cohorts of n=15) or intravenous (right, n=5) inoculation with 2 × 105 and 1 × 105 conidia, respectively. Kaplan-Meier plots clearly indicate a significant attenuation of each deletion mutant that is further corroborated by low competitive indices (CI) as determined from mixed pulmonary inoculations applying each deletant together with the progenitor strain AfS35 (middle panel).
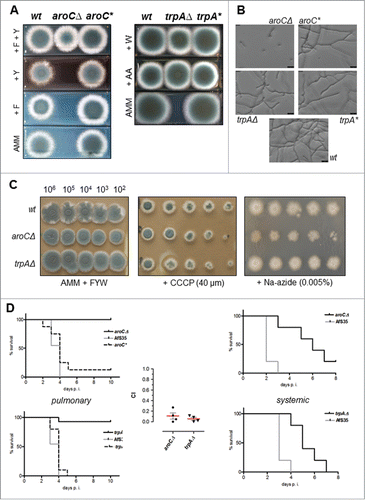
Targeting of the anthranilate synthase component I-encoding gene trpA (AfuA_6G12580) of A. fumigatus strain AfS35 resulted in a deletant (AfS157) that grew only in the presence of externally supplied tryptophan or, alternatively, the intermediate anthranilate (). In contrast to the aroCΔ deletant AfS155 could the auxotrophy of AfS157 (trpAΔ) be rescued by tryptophan supplementation in liquid culture as revealed by microscopic inspection of inoculated conidia (). As aromatic amino acids are precursors for ubiquinone and nicotinamide biosynthesis, compounds that are substantially involved in the respiratory chain and redox potential, we tested both auxotrophic strains for any phenotypes related to their respiratory condition (). Propagation of AfS155 and AfS157 in the presence of respiration disturbing agents such as CCCP, which inhibits oxidative phosphorylation, or sodium azide, a mitochondrial inhibitor, revealed none or only minor deficiencies of either mutant strain under adequate supplementation.
When testing the auxotrophic strains for virulence in an established neutropenic mouse model invasive aspergillosis (), a significant attenuation of the aroCΔ mutant became apparent in a pulmonary infection series (p < 0.001) and for a systemic one (p = 0.0047). This demonstrates that the strict auxotrophic requirements of this mutant could not sufficiently be supplemented in the respective host environment. Similarly, the trpAΔ mutant turned out to be essentially avirulent (p = 0.001) or severely attenuated (p = 0.0047) when tested in the according disease models for pulmonary or systemic aspergillosis, respectively, indicating the nutritional shortage at the respective infection sites. The severe attenuation of virulence displayed by the auxotrophs could further be demonstrated by quantification of competitive indices from mixed inoculation experiments with conidia from each deletion strain together with the wild-type-like progenitor strain AfS35 (). When monitoring the infectious process after the systemic inoculation by fungal burden determination and histological inspection we could surprisingly not detect any fungal elements in the renal tissue (data not shown), indicating that the auxotrophic strains do not invade or proliferate in the kidneys during the course of this infection model and that the observed delayed mortality of the animals is more likely the result of events in the peripheral vascular system.
Having obtained these promising results, we made an effort to generate a mutant strain that would be auxotroph for all 3 aromatic amino acids, hypothesizing that such an auxotroph would be robustly avirulent. For this purpose, deleting the trpA gene in the aroCΔ mutant as well as aroC targeting in the trpAΔ strain was attempted, but no correct double mutant could be identified after colony purification from primary transformants. Targeting the aroM gene (AfuA_1G13740), which encodes a multifunctional component of the Aspergillus shikimate pathway,Citation16 led to the same outcome, indicating that the biosynthetic pathway is per se indispensable for the growth of the fungus. This was eventually supported by a comprehensive heterokaryon rescue analysisCitation17 of conidial suspensions harvested from the primary presumed aroMΔ transformants, from which no viable descendants could be regrown on supplemented selective medium (data not shown).
To substantiate this phenotypical observation further, we made use of a conditional expression approach to place a shikimate pathway gene under control of a tunable promoter. In the course of such a conditional promoter replacement (CPR),Citation18,19 the coding sequence of the aroB gene (AfuA_1G06940) encoding the unique chorismate synthase enzyme was fused to a tetracycline-dependent expression module, transcription of which is active only in the presence of this drug or its derivative doxycycline ().Citation20 After gene replacement, the resulting strain AfS159 [Tet-ON::aroB <ptrA>] was validated by Southern hybridization () and tested for its conditional auxotrophy, that could not be complemented by the presence of aromatic amino acids (), and eventually assessed for virulence (). Inoculating a cohort of neutropenic mice by intranasal instillation of 2 × 105 conidia of strain AfS159 and supplementing the drinking water of this control group with 0.2% doxycycline resulted in pulmonary aspergillosis, while the majority of animals from a cohort lacking doxycycline survived and did not display signs of disease (p = 0.0012). Numbers of colony forming units retrieved from the lungs of respective animals corroborated these observations, with fungal burdens being in the range of wild-type levels in the presence of doxycycline and elimination of the Tet-ON::aroB strain when the antibiotic was omitted. Control groups of immune-competent or neutropenic animals that had been inoculated with the wild-type A. fumigatus progenitor AfS35 and given doxycycline did not differ in their response compared to animals not receiving the antibiotic, ruling out an influence of this compound on the course of infection. Histopathological analysis revealed that infections under doxycycline supplementations resulted in extensive fungal invasion in the murine lung tissue while organs from animals of the -Dox cohort showed only minor pathological signs due to restricted or absent growth of the fungal pathogen (). Molecular analysis of the fungal isolates recovered from the succumbed animals of the -Dox group surprisingly revealed a recombination event in the expression moduleCitation21 that had placed the aroB coding sequence behind the full length gpdA promoter element to result in constitutive expression (). Further in vitro analysis of this phenomenon by plating AfS159 conidia on culture medium without doxycycline and counting the numbers of colonies arising from these plates revealed that in a fraction of approximately 0.1% this recombination would occur. This finding validates essentiality of aroB expression in vivo for supporting pulmonary aspergillosis in susceptible animals. Survival times of neutropenic mice also strongly correlated with the presence of doxycycline when inoculated systemically with the conditional Tet-ON::aroB expression strain (): While all animals of the cohort receiving this inducer via the drinking water succumbed to the infection within 4 days, mice kept in the absence of doxycycline displayed a significantly (p = 0.002) longer survival time.
Figure 3. The shikimate pathway is essential for growth of A. fumigatus. (A) Schematic outline of the conditional promoter replacement for aroB by the Tet-ON module to achieve doxycycline-dependent transcription. The recombination event between the gpdA promoter and the identical sequence of the minimal promoter (pmin) resulting in constitutive expression of the gene under Tet-ON control is indicated. (B) Schematic outline together with molecular analyses of the aroB genomic locus after Tet-ON conditional promoter replacement and recombination in the Tet-ON module by Southern analysis with the indicated probe after EcoRV digestion and diagnostic PCR using oligonucleotides that anneal at the specified positions, respectively. (C) Growth of the conditional promoter replacement strain AfS159 [Tet-ON::aroB<ptrA>] depends on the presence of doxycycline, as validated on supplemented medium containing 1 μg·ml-1 of this antibiotic, whereas supplementation of aromatic amino acids does not rescue the growth defect that is caused by silenced aroB transcription in the absence of doxycycline. (D) In vivo repression of aroB gene expression during pulmonary (upper panel, n=11) and systemic (lower panel, n=5) inoculation of susceptible mice is achieved in the absence of doxycycline, demonstrated by significant virulence attenuation of the Tet-ON::aroB strain in the respective cohort. In mice of the -Dox cohort that succumbed to pulmonary aspergillosis, a recombination event in the Tet-ON::aroB cassette had caused constitutive expression of the otherwise silenced gene to reconstitute full virulence. Histopathological inspections revealed hyphal proliferation and invasion in the murine lungs under doxycycline supplementation but no mycelial elements in its absence.
![Figure 3. The shikimate pathway is essential for growth of A. fumigatus. (A) Schematic outline of the conditional promoter replacement for aroB by the Tet-ON module to achieve doxycycline-dependent transcription. The recombination event between the gpdA promoter and the identical sequence of the minimal promoter (pmin) resulting in constitutive expression of the gene under Tet-ON control is indicated. (B) Schematic outline together with molecular analyses of the aroB genomic locus after Tet-ON conditional promoter replacement and recombination in the Tet-ON module by Southern analysis with the indicated probe after EcoRV digestion and diagnostic PCR using oligonucleotides that anneal at the specified positions, respectively. (C) Growth of the conditional promoter replacement strain AfS159 [Tet-ON::aroB<ptrA>] depends on the presence of doxycycline, as validated on supplemented medium containing 1 μg·ml-1 of this antibiotic, whereas supplementation of aromatic amino acids does not rescue the growth defect that is caused by silenced aroB transcription in the absence of doxycycline. (D) In vivo repression of aroB gene expression during pulmonary (upper panel, n=11) and systemic (lower panel, n=5) inoculation of susceptible mice is achieved in the absence of doxycycline, demonstrated by significant virulence attenuation of the Tet-ON::aroB strain in the respective cohort. In mice of the -Dox cohort that succumbed to pulmonary aspergillosis, a recombination event in the Tet-ON::aroB cassette had caused constitutive expression of the otherwise silenced gene to reconstitute full virulence. Histopathological inspections revealed hyphal proliferation and invasion in the murine lungs under doxycycline supplementation but no mycelial elements in its absence.](/cms/asset/480efb73-4406-4744-a7d8-00343e69eefd/kvir_a_1109766_f0003_oc.gif)
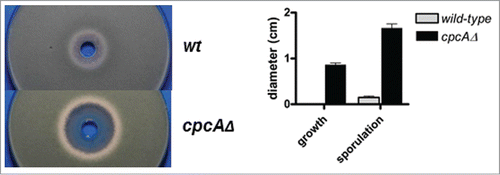
In a further step we aimed at validating our findings by inhibitor studies. The herbicide glyphosate is a well-established inhibitor of the shikimate pathway. To test whether this compound would act as A. fumigatus growth inhibitor by interference with the common aromatic amino acid biosynthesis route and to assess any influence of the CPC system, a fungal wild-type isolate and its congenic cpcAΔ deletion mutant AfS01Citation14 were examined on solid minimal medium in an agar diffusion test (). A clear and prominent halo of growth inhibition surrounded by a zone of reduced sporulation became evident in the presence of the herbicide but only for the CPC deletant, which demonstrates the compensatory capability of the cross-pathway control upon amino acid starvation.
Figure 4. Growth inhibition studies with the shikimate pathway inhibitor glyphosate demonstrate susceptibility of A. fumigatus in a CpcA-dependent manner. Shown are inoculated culture plates (left), to which the herbicide was applied in a centrical reservoir at a concentration of 150 mg/ml together with a quantitative analysis (right) from 3 independent experimental setups presenting the halos' diameters corrected for the punch hole.
In this study we provide proof-of-concept for a conditional expression system suitable for in vivo gene silencing with the aim to validate the aromatic amino acid biosynthesis pathway as an antifungal drug target. The necessity for novel and effective antifungal compounds emerges from the currently limited number of established targets, i.e. the fungal cell wall, plasma membrane, and ergosterol biosynthesis, which are targeted by echinocandins, polyenes, or azoles, respectively. Fungal-specific genes and their products are prime candidates for interfering with pathogenesis, and metabolism represents a fundamental aspect of fungal physiology that is expedient to interfere with. However, recent studies on the physiology of A. fumigatus have revealed a high degree of versatility as well as redundancy with respect to nutrient utilization,Citation22 arising from the osmotrophic lifestyle of this prime saprobe, which hampers identification of distinct factors supporting in vivo growth. Unique enzymes of primary metabolism that may even be absent in the mammalian host have to be considered as highly promising candidates. In this study we have successfully identified the biosynthesis of tyrosine and phenylalanine or tryptophan as such valid targets for antifungal therapy during aspergillosis. Given the extensive knowledge available for each enzyme that is encoded by the targeted genes aroCCitation15,23 and trpA,Citation24 respectively, studies with inhibitors interfering with these biosynthetic reactions are the logical next steps to define potential new classes of antifungal substances that might be exploited for prophylactic purposes. This might be especially promising given the fact that the aroCΔ mutant strain turned out to be avirulent upon pulmonary inoculation, which is the natural route of infection for invasive aspergilosis. In particular, structural insights that have been gained for the conserved chorismate mutase enzymeCitation25 could initiate modeling and screening approaches in a straight-forward manner. Moreover, the fact that the aroC gene appears not to be under transcriptional regulation of the CPC system in AspergillusCitation15 makes its gene product a promising target for therapeutic intervention, because compensatory mechanisms of the cross-pathway control to counteract any starvation condition would not be effective at this branch enzyme of aromatic amino acid biosynthesis.
While each single mutant hindered in biosynthesis of tyrosine/phenylalanine or tryptophan was viable in the presence of the corresponding amino acids, any deletant presumably auxotrophic for all 3 aromatic amino acids turned out to be unable to grow under otherwise adequate supplementation. One possible explanation for this unexpected phenotype lies in insufficient transport capacities for all 3 collective supplements. As a consequence, the shikimate pathway became apparent as an essential biosynthetic route. Such pathways serve as prime candidates in drug target prioritization studies,Citation26 and essentiality of corresponding genes can be validated by parasexual geneticsCitation27 or conditional expression from controllable promoters.Citation28 For the latter, several heterologous as well as one homologous system have been described for A. fumigatus, making use of metabolic genes or a tetracycline-depending system.Citation18,19,Citation29 While the former appear as restricted in their applicability, the latter has been validated only for in vitro studies to date.Citation20 Our results demonstrate applicability of the Tet-ON system in susceptible mice inoculated with A. fumigatus, although with the caveat of recombination in the conditional expression module that would result in constitutively high expression of the target gene to cause contra-productive effects. Thus, genotyping of strains rescued from succumbed animals is mandatory in infection series that deploy the original Tet-ON system. Advanced modules as recently validated might also be the utilised.Citation21 Successful in vivo application of a conditional promoter replacement strain of A. fumigatus employing the Tet-ON system extends the molecular toolbox to scrutinize virulence attributes of this opportunistic fungal pathogen.
Conclusions
The fact that aromatic amino acid biosynthesis might be targeted to interfere with aspergillosis may stimulate the identification of antifungal substances from the pool of already available compounds directed against this well-characterized anabolic route. Furthermore, we provide proof-of-concept for the Tet-ON expression system to be suitable for identification of essential A. fumigatus genes and for in vivo validation during pulmonary as well as systemic infection studies, opening the perspective for comprehensive and complementary studies to identify novel drug targets of A. fumigatus.
Methods
Strains and culture media
A non-homologous recombination-deficient derivative of the A. fumigatus clinical isolate D141Citation30 served as reference recipient. Fungal strains were generally cultured at 37°C in/on nitrate-based minimal medium supplemented when required with 4 mM tyrosine. In case of selection for the presence of the hygromycin B resistance marker, 50 µg/ml of this antibiotic were applied.
Generation of recombinant fungal strains
Cloning of gene replacement cassettes was essentially carried out as described following standard protocols of recombinant DNA technology by fusing homologous arms to a recyclable selection marker module.Citation31 For gene targeting, reconstitution, or promoter replacement, recipient strains were transformed by induced protoplast fusion and selection of primary transformants was carried out in the presence of respective antifungal substances. After colony purification, recombined isolates were screened by diagnostic PCR to become eventually validated by Southern analyses.
Models of infection
Infection studies in neutropenic murine models of pulmonary or systemic aspergillosis were carried out as describedCitation32 with female BALB/c mice (from Charles Rivers Breeding Laboratories, Sulzfeld, Germany) and inoculation doses of 2 × 105 conidiospores suspended in 40 µl of saline for intranasal instillation or of 1 × 105 conidia in a 50 µl suspension for intravenous injection into the lateral tail vein, respectively. For competitive index (CI) assessment,Citation6 mice were intranasally inoculated with a 1:1 mixture of 2 × 104 conidia from AfS35 and the respective deletion strain to become sacrificed after 4 days. Lungs were explanted and aliquots from homogenized tissue were spread onto media containing or lacking the respective amino acid in order to differentiate between the wild-type progenitor and its auxotrophic deletant. Statistical analyses for comparison of survival curves among cohorts of infected mice were performed using the GraphPad Prism 6 software to be compared by the Log-rank (Mantel-Cox) test. Histological cryo-sectioning was performed on 4% formaldehyde-fixed lungs, staining procedures with hemotoxylin and eosin together with Grocott's Methenamine Silver were carried out according to standard protocols.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Michaela Dümig for excellent technical assistance, Dr. Tina Schäfer for precious help, Prof. Dr. Christian Bogdan for critical review of the manuscript, and all other members of the departments for support and discussions.
Funding
This work was financially supported by the German Research Foundation (KR2294/1 and KR2294/3–1), the European Science Foundation by its Fuminomics Research Networking Program (06-RNP-132), the University of Würzburg, the Free State of Bavaria, the Microbiology Institute at the University Hospital of Erlangen, the University of Erlangen-Nürnberg (ELAN-12–08–17–1), as well as the German Federal Ministry of Education and Research (FKZ 031A408A) by funding the AspMetNet consortium during the first call of the Infect-ERA research co-ordination action.
References
- Rohmer L, Hocquet D, Miller SI. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol 2011; 19:341-8; PMID:21600774; http://dx.doi.org/10.1016/j.tim.2011.04.003
- Andersen MR. Elucidation of primary metabolic pathways in Aspergillus species: Orphaned research in characterizing orphan genes. Brief Funct Genomics 2014; 13(6):451-5; PMID:25114096
- Tekaia F, Latgé JP. Aspergillus fumigatus: saprophyte or pathogen? Curr Opin Microbiol 2005; 8:385-92; PMID:16019255; http://dx.doi.org/10.1016/j.mib.2005.06.017
- Rhodes JC. Aspergillus fumigatus: growth and virulence. Med Mycol 2006; 44 Suppl 1:S77-81; PMID:17050423; http://dx.doi.org/10.1080/13693780600779419
- d'Enfert C, Diaquin M, Delit A, Wuscher N, Debeaupuis JP, Huerre M, Latgé JP. Attenuated virulence of uridine-uracil auxotrophs of Aspergillus fumigatus. Infect Immun 1996; 64:4401-5; PMID:8926121
- Brown JS, Aufauvre-Brown A, Brown J, Jennings JM, Arst H, Jr., Holden DW. Signature-tagged and directed mutagenesis identify PABA synthetase as essential for Aspergillus fumigatus pathogenicity. Mol Microbiol 2000; 36:1371-80; PMID:10931287; http://dx.doi.org/10.1046/j.1365-2958.2000.01953.x
- Liebmann B, Mühleisen TW, Müller M, Hecht M, Weidner G, Braun A, Brock M, Brakhage AA. Deletion of the Aspergillus fumigatus lysine biosynthesis gene lysF encoding homoaconitase leads to attenuated virulence in a low-dose mouse infection model of invasive aspergillosis. Arch Microbiol 2004; 181:378-83; PMID:15052376; http://dx.doi.org/10.1007/s00203-004-0667-3
- Schöbel F, Jacobsen ID, Brock M. Evaluation of lysine biosynthesis as an antifungal drug target: biochemical characterization of Aspergillus fumigatus homocitrate synthase and virulence studies. Eukaryot Cell 2010; 9:878-93; PMID:20363898; http://dx.doi.org/10.1128/EC.00020-10
- Oliver JD, Kaye SJ, Tuckwell D, Johns AE, Macdonald DA, Livermore J, Warn PA, Birch M, Bromley MJ. The Aspergillus fumigatus dihydroxyacid dehydratase Ilv3A/IlvC is required for full virulence. PLoS One 2012; 7:e43559; PMID:23028460; http://dx.doi.org/10.1371/journal.pone.0043559
- Gibson F. The elusive branch-point compound of aromatic amino acid biosynthesis. Trends Biochem Sci 1999; 24:36-8; PMID:10087921; http://dx.doi.org/10.1016/S0968-0004(98)01330-9
- Herrmann KM, Weaver LM. The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol 1999; 50:473-503; PMID:15012217; http://dx.doi.org/10.1146/annurev.arplant.50.1.473
- Bentley R. The shikimate pathway - a metabolic tree with many branches. Crit Rev Biochem Mol Biol 1990; 25:307-84; PMID:2279393; http://dx.doi.org/10.3109/10409239009090615
- Roberts F, Roberts CW, Johnson JJ, Kyle DE, Krell T, Coggins JR, Coombs GH, Milhous WK, Tzipori S, Ferguson DJ, et al. Evidence for the shikimate pathway in apicomplexan parasites. Nature 1998; 393:801-5; PMID:9655396; http://dx.doi.org/10.1038/30718
- Krappmann S, Bignell EM, Reichard U, Rogers T, Haynes K, Braus GH. The Aspergillus fumigatus transcriptional activator CpcA contributes significantly to the virulence of this fungal pathogen. Mol Microbiol 2004; 52:785-99; PMID:15101984; http://dx.doi.org/10.1111/j.1365-2958.2004.04015.x
- Krappmann S, Helmstaedt K, Gerstberger T, Eckert S, Hoffmann B, Hoppert M, Schnappauf G, Braus GH. The aroC gene of Aspergillus nidulans codes for a monofunctional, allosterically regulated chorismate mutase. J Biol Chem 1999; 274:22275-82; PMID:10428795; http://dx.doi.org/10.1074/jbc.274.32.22275
- Charles IG, Keyte JW, Brammar WJ, Smith M, Hawkins AR. The isolation and nucleotide sequence of the complex AROM locus of Aspergillus nidulans. Nucleic Acids Res 1986; 14:2201-13; PMID:3515316; http://dx.doi.org/10.1093/nar/14.5.2201
- Osmani AH, Oakley BR, Osmani SA. Identification and analysis of essential Aspergillus nidulans genes using the heterokaryon rescue technique. Nat Protoc 2006; 1:2517-26; PMID:17406500; http://dx.doi.org/10.1038/nprot.2006.406
- Hu W, Sillaots S, Lemieux S, Davison J, Kauffman S, Breton A, Linteau A, Xin C, Bowman J, Becker J, et al. Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog 2007; 3:e24; PMID:17352532; http://dx.doi.org/10.1371/journal.ppat.0030024
- Romero B, Turner G, Olivas I, Laborda F, De Lucas JR. The Aspergillus nidulans alcA promoter drives tightly regulated conditional gene expression in Aspergillus fumigatus permitting validation of essential genes in this human pathogen. Fungal Genet Biol 2003; 40:103-14; PMID:14516763; http://dx.doi.org/10.1016/S1087-1845(03)00090-2
- Dichtl K, Helmschrott C, Dirr F, Wagener J. Deciphering cell wall integrity signalling in Aspergillus fumigatus: identification and functional characterization of cell wall stress sensors and relevant Rho GTPases. Mol Microbiol 2012; 83:506-19; PMID:22220813; http://dx.doi.org/10.1111/j.1365-2958.2011.07946.x
- Helmschrott C, Sasse A, Samantaray S, Krappmann S, Wagener J. Upgrading fungal gene expression on demand: improved systems for doxycycline-dependent silencing in Aspergillus fumigatus. Appl Environ Microbiol 2013; 79:1751; PMID:23275515; http://dx.doi.org/10.1128/AEM.03626-12
- Amich J, Krappmann S. Deciphering metabolic traits of the fungal pathogen Aspergillus fumigatus: redundancy vs. essentiality. Front Microbiol 2012; 3:414; PMID:23264772
- Romero RM, Roberts MF, Phillipson JD. Chorismate mutase in microorganisms and plants. Phytochemistry 1995; 40:1015-25; http://dx.doi.org/10.1016/0031-9422(95)00408-Y
- Romero RM, Roberts MF, Phillipson JD. Anthranilate synthase in microorganisms and plants. Phytochemistry 1995; 39:263-76; PMID:7495530; http://dx.doi.org/10.1016/0031-9422(95)00010-5
- Sträter N, Schnappauf G, Braus G, Lipscomb WN. Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures. Structure 1997; 5:1437-52; http://dx.doi.org/10.1016/S0969-2126(97)00294-3
- Lu Y, Deng J, Rhodes JC, Lu H, Lu LJ. Predicting essential genes for identifying potential drug targets in Aspergillus fumigatus. Comput Biol Chem 2014; 50:29-40; PMID:24569026; http://dx.doi.org/10.1016/j.compbiolchem.2014.01.011
- Firon A, Beauvais A, Latgé JP, Couve E, Grosjean-Cournoyer MC, d'Enfert C. Characterization of essential genes by parasexual genetics in the human fungal pathogen Aspergillus fumigatus: impact of genomic rearrangements associated with electroporation of DNA. Genetics 2002; 161:1077-87; PMID:12136012
- Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol 2003; 50:167-81; PMID:14507372; http://dx.doi.org/10.1046/j.1365-2958.2003.03697.x
- Vogt K, Bhabhra R, Rhodes JC, Askew DS. Doxycycline-regulated gene expression in the opportunistic fungal pathogen Aspergillus fumigatus. BMC Microbiol 2005; 5:1; PMID:15649330; http://dx.doi.org/10.1186/1471-2180-5-1
- Krappmann S, Sasse C, Braus GH. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end joining-deficient genetic background. Eukaryot Cell 2006; 5:212-5; PMID:16400185; http://dx.doi.org/10.1128/EC.5.1.212-215.2006
- Krappmann S, Bayram Ö, Braus GH. Deletion and allelic exchange of the Aspergillus fumigatus veA locus via a novel recyclable marker module. Eukaryot Cell 2005; 4:1298-307; PMID:16002655; http://dx.doi.org/10.1128/EC.4.7.1298-1307.2005
- Amich J, Schafferer L, Haas H, Krappmann S. Regulation of sulphur assimilation is essential for virulence and affects iron homeostasis of the human-pathogenic mould Aspergillus fumigatus. PLoS Pathog 2013; 9:e1003573; PMID:24009505; http://dx.doi.org/10.1371/journal.ppat.1003573
