Abstract
Pmps (Polymorphic Membrane Proteins) are a group of membrane bound surface exposed chlamydial proteins that have been characterized as autotransporter adhesins and are important in the initial phase of chlamydial infection. These proteins all contain conserved GGA (I, L, V) and FxxN tetrapeptide motifs in the N-terminal portion of each protein. All chlamydial species express Pmps. Even in the chlamydia-related bacteria Waddlia chondrophila, a Pmp-like adhesin has been identified, demonstrating the importance of Pmps in Chlamydiales biology. Chlamydial species vary in the number of pmp genes and their differentially regulated expression during the infectious cycle or in response to stress. Studies have also demonstrated that Pmps are able to induce innate immune functional responses in infected cells, including production of IL-8, IL-6 and MCP-1, by activating the transcription factor NF-κB. Human serum studies have indicated that although anti-Pmp specific antibodies are produced in response to a chlamydial infection, the response is variable depending on the Pmp protein. In C. trachomatis, PmpB, PmpC, PmpD and PmpI were the proteins eliciting the strongest immune response among adolescents with and without pelvic inflammatory disease (PID). In contrast, PmpA and PmpE elicited the weakest antibody response. Interestingly, there seems to be a gender bias for Pmp recognition with a stronger anti-Pmp reactivity in male patients. Furthermore, anti-PmpA antibodies might contribute to adverse pregnancy outcomes, at least among women with PID. In vitro studies indicated that dendritic cells infected with C. muridarum were able to present PmpG and PmpF on their MHC class II receptors and T cells were able to recognize the MHC class-II bound peptides. In addition, vaccination with PmpEFGH and Major Outer Membrane Protein (MOMP) significantly protected mice against a genital tract C. muridarum infection, suggesting that Pmps may be an important component of a multi-subunit chlamydial vaccine. Thus, Pmps might be important not only for the pathogenesis of chlamydial infection, but also as potential candidate vaccine proteins.
Introduction
The Chlamydia genus encompasses 11 species: C. trachomatis (human sexually transmitted disease and eye infection), C. muridarum (causes disease in mice and hamsters), C. suis (infects pigs), C. pneumoniae (responsible for human respiratory infections), C. pecorum (common pathogen in livestock), C. psittaci (prevalent in birds and causing pneumonia in humans), C. abortus (causes abortion in mammals), C. felis (species found in cats), C. caviae (species causing infection in guinea pigs), C. avium (comprising strains from pigeons and psittacine birds) and C. gallinacea (strains from poultry).Citation1-3 C. trachomatis can cause sexually transmitted diseases, which can lead to ectopic pregnancies, pelvic inflammatory disease (PID), tubal infertility, and miscarriage.Citation4-11 C. trachomatis is of particular importance to human health because the infection is mostly asymptomatic and induces inflammatory responses that can lead to immunopathological sequelae. The World Health Organization estimates that since 2008 there are over 100 million new sexually transmitted cases due to C. trachomatis infection.Citation12 C. trachomatis can also cause trachoma (ocular disease) that can lead to scarring and blindness.Citation13,14 In fact, trachoma is the leading cause of blindness worldwide.
Chlamydia exists as either the infectious, non-replicating extracellular elementary body (EB) or the reticulate body (RB), which is noninfectious, replicating and strictly intracellular.Citation15 For such an obligate intracellular bacteria, one of the most important steps for infecting eukaryotic cells is the attachment to host's cells mediated by adhesin proteins. Several adhesins have been identified, including OmcB,Citation16 and polymorphic membrane proteins.
In this review we will discuss the main characteristics of polymorphic membrane proteins (Pmps), which are autotransporter-like immunogenic surface-exposed proteins that have been found to play an important role not only as adhesins, but also as potent antigenic proteins involved in the immunopathogenesis of chlamydial infections. We will review the regulation of Pmps' and describe their diversity in the different chlamydial species with a particular focus on C. trachomatis and C. pneumoniae (). We will also discuss their functional properties as adhesins as well as their role in pathogenesis, especially by triggering cytokine production and inducing inflammation and pathological lesions. Finally, their utilization as potential chlamydial vaccine candidates will also be presented.
Table 1. Summary of Pmp research
Structure and Regulation of PMPs
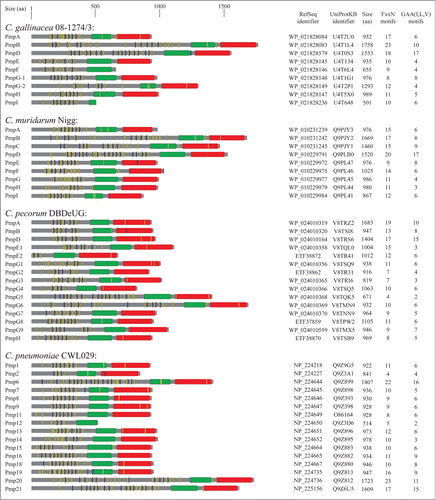
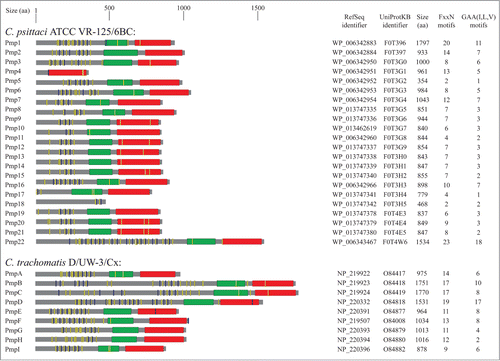
Pmps are a group of membrane bound proteins present in all chlamydial species. These proteins are grouped together by the fact that all exhibit conserved GGA(I, L, V) and FxxN tetrapeptide motifs in their N-terminal portion.Citation17 Pmps were first identified in C. psittaciCitation18,19 and subsequent studies have shown that all the other members of the Chlamydia genus also encoded Pmp proteins.Citation20-24 However, chlamydial species vary in the number of pmp genes. depicts the main characteristics of Pmp proteins in reference strains C. abortus S26/323, C. avium 10DC88,Citation2 C. caviae GPIC,Citation22 C. felis Fe/C-56,Citation20 C. gallinacea 08-1274/32, C. muridarum Nigg,Citation21 C. pecorum DBDeUG,Citation25 C. pneumoniae CWL029,Citation26 C. psittaci ATCC VR-125/6BC,Citation27 and C. trachomatis D/UW-3/Cx.Citation28 Alternative names of Pmp in the Chlamydia genus are shown in Figure S1.
Figure 1. Schematic representation of Pmp proteins in Chlamidiaceae. The UniProt identifier, RefSeq identifier and the amino acid length are shown for each Pmp protein. Conserved domains were detected using MOTIF search (http://www.genome.jp/tools/motif/): FxxN motifs (yellow), GAA (I,L,V) motifs (blue), central PMP_M region (green) and autotransporter β-domain (red). Proteins containing frameshift mutations were omitted in the figure.
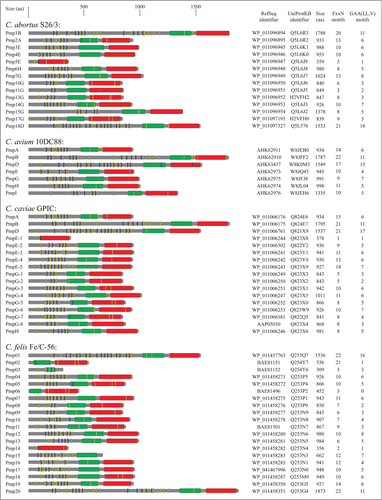
Studies have demonstrated that C. trachomatis (serovar A/HAR13 and D/UW-3) and C. muridarum (strain Nigg) both contain 9 pmp genesCitation21,28,Citation29 whereas 17 and 18 genes encode for Pmps in C. caviae and C. abortus, respectively.Citation21,23,Citation28,29 Of note, a pmp-like encoding gene has also been identified in the genome of the Chlamydia-related bacteria Waddlia chondrophila.Citation30
Numerous studies have indicated that Pmps function as autotransporters.Citation18,31-33 Autotransporter proteins are characterized by 3 functional domains, which include a cleavable N-terminal signal for translocation to the membrane, a passenger domain for surface localization or secretion and a C-terminal β-barrel translocator sequence for outer membrane translocation.Citation17,34 Pmp proteins share many of the characteristics of classical autotransporters including an N-terminal Sec-dependent leader sequence, a C-terminal β-barrel sequence and a passenger domain.Citation35-38 Autotransporter proteins are widespread in Gram-negative bacteria and contribute to virulence of many pathogens.Citation34,36,Citation39 These virulence factors include among others secreted proteases (Neisseria gonorrhoeae, Haemophilus influenzae and Shigella flexneri), adhesins (Yersinia enterocolitica, pathogenic Escherichia coli, Salmonella enterica and Bordetella pertussis) and cytotoxins (Bordetella pertussis and Helicobacter pylori).Citation38,40
Bacteria from the Chlamydia genus possess a large number of autotransporter genes compared to other gram-negative bacteria, suggesting an important role of these chlamydial autotransporter proteins in pathogenesis. C. pneumoniae contains 21 Pmps (5 of which contain frameshift mutations), which account for 17.5% of its coding capacity.Citation41 In addition to MOMP and outer membrane protein 2 (Omp2), Pmps represent the major proteins in C. pneumoniae outer membrane complex.Citation32 These Pmps are characterized by subtype G containing Pmp1-13 and subtype E encompassing Pmp15-18. The remaining 4 subtypes (Pmp14, 19, 20, 21) are represented by a single gene.Citation26,42 Studies using 2D-PAGE and RT-qPCR have demonstrated that all pmp genes are transcribed and expressed during the C. pneumoniae developmental cycleCitation43,44 and encode for proteins of various sizes.Citation43,44 Pmp6, pmp20 and pmp21 genes encode for proteins that are between 1408 and 1724 amino acids long whereas pmp12, 3, 4, 5, 10 and 17 genes transcribe proteins that are shorter due to either truncation of the C-terminal (i.e. Pmp12) or frameshift mutation (i.e., pmp3, 4, 5, 10, 17).Citation45 Pmp proteins can also be processed into smaller fragments as in the case of Pmp6, 20 and the PmpD ortholog Pmp21.Citation32 Studies investigating C. pneumoniae Pmp processing suggest that the EBs produce at least 3 different forms of Pmp21 (N, M and C) and that the N-Pmp21 and M-Pmp21 forms are expressed on the chlamydial surface.Citation32,33,Citation46
The C. trachomatis genome encodes for 9 Pmp proteins termed PmpA-I that are further subdivided into A (PmpA), B (PmpB, C) D (PmpD), E (PmpE, F) G (PmpG, I) and H (PmpH) subtypes.Citation42 Similar to C. pneumoniae, a high percentage of coding capacity (13.6%) is dedicated to C. trachomatis pmp genesCitation28,41,Citation42 suggesting that these genes play an important role in Chlamydia biology and pathogenesis.Citation47,48 Proteins between subtypes share variable similarities in terms of amino acid sequences. Indeed, C. trachomatis (serovar E) PmpG and PmpI (subtype G) are 25% similar and PmpB and PmpC (subtype B) share 43% homology.Citation42 There is also interspecies and inter-serovar Pmp homology. C. trachomatis subtype PmpD and C. pneumoniae Pmp21 share 33% homology at the protein levelCitation42 and PmpD is the second highest conserved Pmp demonstrating 99.2% homology among C. trachomatis serovars.Citation49 These similarities suggest common functional roles across chlamydial serovars and species for Pmps. However, there is some controversy on whether all 9 of the C. trachomatis Pmps are expressed. Early investigations detected only PmpB, D, E, F, G and H,Citation50-52 whereas later studies demonstrated that all C. trachomatis pmp genes were transcribed.Citation17,53 C. trachomatis pmp genes are located in 2 clusters with pmpA-C and pmpE-I comprising each cluster and pmpD being genetically isolated.Citation28 Furthermore, pmpABC, pmpFE and pmpGH are co-transcribed in vitro indicating that these genes are organized in operons.Citation54 Of all of the C. trachomatis Pmps, PmpA, D and I are the most conserved, with PmpA having 99.9%, PmpD 99.1% and PmpI 99.2% amino acid sequence similarity.Citation54 All 9 pmps have been shown to be transcribed in vitro in various C. trachomatis strains including serovars D, E, and L2Citation17,31,Citation55 and are all translocated to the bacterial surface.Citation17 However, C. trachomatis pmp paralogues exhibit variable transcription patterns. Expression studies conducted by Kiselev and colleagues demonstrated that the pmpD gene from C. trachomatis serovar L2 was upregulated between 16 and 24 hours after infection, which is the time RBs differentiate into EBs.Citation31 Studies by Carrasco et al. and Nunes and colleagues demonstrated that pmpA transcription peaked at 12 and 18 hours post infection.Citation54,55 Similarly, pmpI was transcribed at 18 hours post infection. PmpBC, pmpEF, and pmpGH were all co-transcribed at later time points (32 and 48 hours post infection).Citation54
There is evidence that Pmps are differently regulated in response to stress. Penicillin, a β-lactam antibiotic, is known to disrupt the growth of Chlamydia by blocking the conversion of RBs into EBs and induce so-called aberrant bodies.Citation56,57 Further studies have demonstrated that penicillin also modulates the transcription and protein processing of several Pmps. Carrasco and colleagues demonstrated that when exposed to penicillin, transcription of pmpB, C, E, F, G, and H was downregulated, whereas expression of pmpA, D and I was mostly unaffected.Citation54 The fact that pmpA, pmpD and pmpI were transcribed even under stress conditions such as penicillin suggests a critical role for these Pmps for the survival of the organism in a hostile environment and may be important for pathogenesis. Similarly, Kiselev et al. demonstrated that in the presence of penicillin, the cleavage and secretion of the passenger domain of PmpD was suppressed in C. trachomatis L2 serovar. This suppression was mediated by the inhibition of membrane proteases by penicillin, such as signal peptidases I, which cleave the autotransporter protein.Citation31 These data suggest that penicillin may inhibit Pmp transcription and post-translation processing in different chlamydial serovars and strains.
Studies on PmpD have suggested that Pmps undergo an extensive processing before their translocation to the chlamydial surface and exist as oligomers with a flower-like structure.Citation58 Werhl et al. has suggested a multistep process where the full length PmpD is exported to the periplasmic space via the Sec-machinery. Then, after the signal sequence is cleaved, the C-terminal portion forms a β- barrel and the N-terminal passenger domain is exported and cleaved, resulting in an N-terminal surface exposed protein.Citation33 Several investigators have generated antibodies against the N-terminal portion of all C. trachomatis Pmps including PmpD,Citation17,47 further supporting the hypothesis that Pmps are present on the surface of Chlamydia and that the N-terminal portions of these proteins are accessible to antibodies. However, in a study by Kiselev et al., anti-PmpD polyclonal antibodies only recognized PmpD on the surface of RBs and not EBsCitation31 suggesting only RBs are able to express Pmps on their outer surface membrane. In addition, more recent studies showed that PmpD might also be expressed on EBs but at a lower level.Citation59 Noteworthy, Molleken and colleagues demonstrated that C. pneumoniae Pmp21 is located on the surface of both EBs and RBs during infectionCitation60 indicating that the preferential expression of PmpD of C. trachomatis on RBs is not a common feature shared by all chlamydial Pmps.
Functional Properties as Adhesins
In 2004, Wehrl et al. investigated the ability of a polyclonal rabbit serum raised against N-Pmp21 to inhibit C. pneumoniae infection in vitro.Citation33 A year later, Finco and colleagues demonstrated a similar result using a mouse kidney cell line (LLC-MK2) and polyclonal mouse sera to Pmp2 and Pmp10.Citation61 These studies highlighted the importance of Pmps in the initial phases of infection. Furthermore, their role as adhesins was suggested by the fact that Anaplasma phagocytophilum express adhesins that contain the GGA(I, L, V) and FxxN motifsCitation62 which are also present in chlamydial Pmps.
Molleken et al. demonstrated that the C. pneumoniae Pmp21 adhesion to cell surface receptors required at least 2 copies of the Pmp repeats: either FxxN+FxxN or FxxN+GGA(I, L, A). Thus, scrambling one of the Pmp21-derived synthetic peptide FxxN motifs did not modulate the adhesive property of Pmp21, whereas deleting the 2 tetrapeptide FxxN sequences resulted in significant reduction or complete loss of adhesion capacity.Citation60 They proposed that these repeats either indirectly modulate the protein conformation implicated in adhesion or directly modulate adhesin interaction with the eukaryotic receptors. Moreover, a more recent study identified EGFR (epidermal growth factor receptor) as the receptor for Pmp21.Citation63 depicts the localization of GGA (I, L, V) and FxxN motifs of Pmp proteins in reference strains of the Chlamydia genus.
C. trachomatis PmpD has also been shown to be a surface exposed and neutralizing target for anti-PmpD antibodies. Neutralizing assays were performed on HaK (hamster kidney) cells using 3 major C. trachomatis serogroups representing ocular serovars (A, Ba, C), noninvasive genital serovars (D, E, F, G, K) and genital invasive serovar (L2). Results from this study indicated that serovars Ba, D, E and L2 were more efficiently neutralized than serovars A, C, F, G and K. The α-PmpD serum failed to neutralize C. muridarum, C. pneumoniae and C. caviae indicating that PmpD is a specific strain-dependent neutralizing target.Citation47 Interestingly, this study also demonstrated that antibody to MOMP and LPS blocked the ability of PmpD anti-serum to neutralize the chlamydial infection in HaK cells. MOMP and LPS are 2 of the most abundant immunodominant antigens on the EB surface.Citation64 This could imply that during an infection, MOMP and LPS may act as decoys for neutralizing antibodies by blocking their binding to Pmps, which are important in the initial phase of infection. A 2014 study conducted by Becker et al. investigated the ability of all C. trachomatis serovar E Pmps to act as adhesins in 2 human cells lines. The study demonstrated that PmpA-I mediated adhesion to human epithelial (HeLa) and endothelial (Hep-2) cells suggesting all C. trachomatis Pmps are involved in the chlamydial infection process and implicating the Pmps as a group of virulence factors.Citation59 There is also evidence that Pmps may exist as 2 distinct forms and are variably expressed depending on whether they are on EBs or RBs. Western blot analysis and scanning electron microscopy studies have indicated that PmpD is more abundant on RBs than EBs and the RBs have a more homogeneous distribution whereas the PmpD on the EB exhibited a more polarized localization.Citation58
Furthermore, PmpD might not only function as a surface protein, but also undergoes multiple proteolytic processing resulting in a soluble protein containing 3 fragments (p111, p73 and p30) that is restricted to infected cells.Citation58 The authors of this study hypothesized that since the PmpD fragments are similar to Neisseria secreted IgA protease H. pylori VacA secreted toxins, the p11, p73 or p30 proteins may interact with immune cells to induce inflammatory cascades that are important in pathogenicity.Citation58 Recently, a study conducted by Kari and colleagues used a serovar D pmpD null mutant, to define the role of PmpD in the pathogenesis of chlamydial infection. Although the study demonstrated that pmpD is not an essential chlamydial gene, pmpD mutants did exhibit abnormal morphology or ultrastructural phenotype and were significantly deficient in host-cell attachment in human epithelial cells during early host-cell interactions (70% reduction of the infection ability). When they performed the same experiment using a mouse cell line, the pmpD mutant and wild-type C. trachomatis EBs were equally competent in their ability to infect mouse cells. Although mouse and human chlamydial strains share 72% PmpD sequence similarity, the possible subtle differences in the proteins structure and configuration between the mouse and human PmpD may account for different infection capabilities and tropism. Using a macaque model of ocular infection, the authors also demonstrated that chlamydial burden was significantly reduced in monkeys infected with the pmpD mutant compared to control monkeys infected with wild-type C. trachomatis during the first 14 days of infection. Interestingly, at later time points (2 weeks post-infection), there were no differences in terms of ocular bacterial burden.Citation65 These data suggest that PmpD plays a critical role in chlamydial pathogenesis at the early stages of infection.
Human Pathogenesis
Human studies have demonstrated that although C. trachomatis-infected patients develop antibodies to Pmps, not all Pmps are recognized equally. Tan et al. showed that PmpB, C, D and I were the most frequently recognized and elicited a stronger humoral immune response, whereas PmpA and E were the lowest recognized in sera that included women with pelvic inflammatory disease (PID), adolescent young females and male patients. This finding suggests that these Pmps may be more frequently exposed or abundantly expressed on the C. trachomatis surface.Citation48 Interestingly, there was a gender bias for Pmp recognition. Serum from male patients tended to have a stronger anti-Pmp reactivity compared with serum from female patients. Furthermore, PmpB was the most recognized Pmp in sera from female patients (82% of serum) whereas in male sera PmpD was the most recognized Pmp (92% of serum). There was also a difference between adolescent female patients and women that exhibited PID. Antibodies specific for PmpB and PmpI were more prevalent in the PID group suggesting that these Pmps might contribute to the development of chronic inflammation. Another study investigating human sera in women with PID suggested that PmpA might have a role in adverse pregnancy outcomes.Citation66 Taylor et al. demonstrated that women who had PID and also tested positive for anti-PmpA specific antibodies had a significantly lower chance of getting pregnant, less likely chance to have a live birth, and were more likely to have an upper genital tract infection than women who did not test positive for anti-PmpA antibodies. Furthermore, women who were positive for PmpI also were more likely to have upper genital tract infections.Citation66
C. trachomatis includes 3 human biovars composed of different serovars. Serovars Ab, B, Ba cause trachoma. Serovar D-K cause urethritis, PID, ectopic pregnancy and neonatal pneumonia and serovars L1, L2 and L3 are able to cause lymphogranuloma venereum (LGV). Gomes et al. demonstrated that only sera from serovars D, E and G-infected patients reacted to recombinant PmpC whereas all infected patients reacted to MOMP.Citation67 Nunes and colleagues performed a similar study where sera from 39 adolescents were tested for immunoreactivity to recombinant PmpD and PmpF. All sera from patients that were positive for chlamydial serovars Ba, E, F and K also reacted to recombinant PmpD protein. However, patients that were infected with strains D, Ia, J and G did not have immunoreactive sera to PmpD. Surprisingly, no sera from any of the infected patients reacted to recombinant PmpF.Citation55 This suggests that the expression of Pmps in different chlamydial serovars is variably regulated and may induce differential immune responses with specific serovars. It may also indicate that PmpC and PmpD elicit an especially robust humoral immune response in adolescent females.
Immunology
Studies have also demonstrated that Pmps are able to activate innate immune functional responses in infected cells. A study by Niessner et al. tested the ability of 15 C. pneumoniae Pmps to induce cytokine production in a human cell line. The study revealed that only Pmp20 and Pmp21 were able to induce IL-6 and MCP-1 (monocyte chemoattractant protein-1) secretion in human umbilical vein endothelial cells (HUVECs) in a dose-dependent manner. When the 2 Pmps were co-cultured in HUVECs there was no increase in cytokine production compared to a single Pmp HUVEC culture suggesting the 2 Pmps were not synergistic in their ability to elicit IL-6 and MCP-1 secretion. Furthermore, the study revealed that Pmp20 and Pmp21 induce their effect on endothelial cell cytokine production by activation of NF-κB (nuclear factor kappa B) pathway, which is a major pathway for inflammatory activation.Citation68 Another study investigating the ability of a C. trachomatis Pmp to induce cytokine production demonstrated that recombinant PmpD incubated with a human monocyte cell line (THP-1) induced robust IL-8 production in a dose dependent manner.Citation33
Vaccine Studies
The asymptomatic nature of a chlamydial infection and its subsequent long-term consequences such as ectopic pregnancy, preterm delivery and infertility makes a chlamydial vaccine paramount.Citation11 The first chlamydial vaccine studies utilized avirulent intact live C. trachomatis as a prophylactic for trachoma infection. However, even though in some cases the vaccine provided considerable protection from infection and pathology, some vaccinated individuals developed more severe disease after subsequent C. trachomatis infection.Citation69,70 Vaccines containing live organisms are optimal because they contain not only native antigens that are recognized by antigen presenting cells but also negate the need for adjuvants. However, the drawbacks include the need for cold storage and the possibility of avirulent strains converting back to a strain that is able to cause disease.Citation71 Subsequent chlamydial vaccine studies have utilized multiple strategies including subunit antigenic determinants, recombinant proteins and plasmid DNA in conjunction with adjuvants or other delivery vehicles to increase immunogenicity.Citation11,72 Even though chlamydial major outer membrane protein (MOMP) is the most investigated vaccine candidate in animal and nonhuman primate studies, the protection elicited has been suboptimalCitation73-75 necessitating further research for more efficacious vaccine candidates.
Immunity against Chlamydia requires CD4+ T cells and to a lesser degree CD8+ T cells that recognize specific chlamydial antigens on MHC molecules and the production of INF-γ. One of the major difficulties in developing an effective chlamydial vaccine is identifying the MHC-bound chlamydial protein epitopes that are recognized by T cells. A 2008 study conducted by Grotenbreg et al. investigated specific chlamydial epitopes that are able to activate CD8+ T cells. They discovered that MHC tetramers corresponding to PmpI-D were able to induce the expansion of purified CD8+ T cells from mice previously infected with C. trachomatis LGV serovar L2.Citation76 A study conducted by Finco and colleagues demonstrated that chlamydial proteins including MOMP contain T cell and B cell epitopes or both.Citation77-79 Other chlamydial proteins have also been proposed as candidates for a chlamydial vaccine including Omp2 (outer membrane protein 2),Citation80 HPS60 (chlamydial shock protein 60),Citation81 CPAF (chlamydial protease-like activity factor),Citation82,83 Cap1 (class I accessible protein 1)Citation84 and CrpA (cysteine-rich protein A).Citation81
Pmps generally contain only T cell epitopesCitation85 and have also been proposed as vaccine candidates. Indeed, several studies have investigated the efficacy of various Pmps to protect against Chlamydia infection and activate T cells.Citation86-91 Using affinity chromatography and tandem mass spectrometry, Karunakaran et al. demonstrated that dendritic cells infected with C. muridarum presented PmpG and F on their MHC class II receptors. Furthermore, purified Chlamydia-specific CD4+ T cells produced high levels of INF-γ after co-culture with the previously C. muridarum-infected dendritic cells suggesting T cell recognition of the MHC class-II bound Pmps.Citation86 In another study, mice receiving adoptively transferred dendritic cells that were previously pulsed with PmpG and F were partially protected against intranasal and genital C. muridarum infection.Citation88 A study conducted by Mygind and colleagues discovered that splenic cells from C. pneumoniae nasally infected mice produced significant amounts of INF-γ after incubation with recombinant Pmp8, Pmp20 and Pmp21 and the INF-γ production was shown to be mediated by CD4+ T cells. Recombinant Pmp6, 9, 10, 11 were recognized inconsistently over time but with low responses.Citation87 Yu and colleagues investigated the ability of PmpG, PmpE/F and MOMP in conjunction with either CpG-ODN, AbISCO-100 or DDA/TDB adjuvants to protect mice from C. muridarum genital infection. The results demonstrated that a mutlisubunit vaccine containing PmpG + PmpE/F + MOMP + DDA/TDB exhibited the highest degree of protection. In addition, compared to CpG and AbISCO, DDA/TDB + PmpG induced the strongest INF-γ responses by CD4+ T cells.Citation89 A later study by Yu et al. showed that vaccination with PmpE, F, G and H significantly protected mice against a genital tract C. muridarum infection as measured by cervicovaginal shedding. Furthermore, PmpG persisted on splenic antigen presenting cells for at least 6 months. Interestingly, splenocyte culture that were restimulated with PmpG, F, E and H from C. muridarum genitally infected mice demonstrated robust INF-γ production in response to PmpG and H, low production after PmpF restimulation and no production after PmpE restimulation.Citation91 In fact, studies by various researchers have also demonstrated the immunodominance of PmpG in the murine model.Citation92-94 A more recent study by Yu et al. investigated protection elicited by a multisubunit vaccine containing PmpEFGH + Th1 polarizing adjuvant DDA/MPL in a genital tract infection. They demonstrated that a vaccine containing PmpEFGH + MOMP + DDA/MPL was superior in protecting against a C. muridarum genital infection compared to PmpG, MOMP or PmpEFGH measured by cervicovaginal shedding. They also showed that PmpE and H have low stimulatory capabilities to induce IFN-γ production in CD4+ T cells and therefore may not be suited for use in a chlamydial multisubunit vaccine.Citation90 C. trachomatis PmpG has also been used as an epitope in conjunction with vault nanoparticles. These studies demonstrated that a PmpG-vault nanoparticle vaccine incubated with a human monocyte cell line (THP-1) activated the protease caspase-1 and IL-1β production. In addition, mice immunized with the PmpG/nanoparticle vaccine induced PmpG-specific CD4+ T cells that were able to be restimulated with PmpG in vitro.Citation95
Whereas PmpEFGH + MOMP and adjuvants or PmpG + nanoparticles appear to be promising vaccine candidates for protection against C. trachomatis, PmpD may be an interesting candidate for protection against C. abortus which can cause ovine enzootic abortion and poses a risk to pregnant women.Citation6 Mice immunized with recombinant PmpD in a Vibrio cholerae ghost delivery system elicited a robust antigen-specific IFN-γ, IgA and IgG2c immune response after genital C. abortus challenge. Length of vaginal shedding and number of inclusion forming units recovered following C. abortus challenge was also significantly decreased.Citation96 C. psittaci is an avian chlamydial strain that is capable of causing pneumonia, encephalitis and even death in humans.Citation97 A recent study demonstrated that chickens inoculated with a recombinant herpesvirus of turkeys (HTV) expressing C. psittaci PmpD displayed increased PmpD-specific antibodies and significantly elevated CD4+ T levels.Citation98 Collectively, these studies suggest that Pmps may be good candidates for a chlamydial vaccine because they are outer membrane proteins that are recognized by antigen presenting cells, degraded, processed and presented to CD4+ T cells. Additionally, these chlamydial proteins have been shown to activate T cells and induce cytokine and antibody production that are important in the resolution of a chlamydial infection.
A recent study utilizing nanoparticles demonstrated that robust protection against C. trachomatis was attained in the genital tract of mice after immunization with UV-inactivated C. trachomatis bound to nanoparticles carrying adjuvants. This study highlights the importance of the site of vaccination (vaginal mucosa) and the requirement of multiple vaccinations in order to develop an effective protection against the infection.Citation99
Conclusion
Pmps are a family of membrane bound surface exposed proteins that are expressed in all chlamydial species and other Chlamydiales, including Waddlia chondrophila. Since divergence of Chlamydiaceae and Waddliaceae families occurred more than 1 billion years ago, Pmps have thus a long evolutionary history, assuming that pmp genes have not been transferred horizontally. Pmps have been characterized as autotransporter adhesin proteins that are important in the initial phase of infection and have been shown to induce cytokine production in infected cells. Furthermore, Pmps may be involved in human pathology and infertility. Utilized as a component of a subunit vaccine in conjunction with effective adjuvants and/or delivery systems, these outer membrane proteins may contribute to the development of an effective chlamydial vaccine that elicits a protective Th1-mediated immune response that does not induce adverse immunopathologies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
KVIR_A_1111509_Supplementary.docx
Download MS Word (63.3 KB)Acknowledgments
We thank Anne Ammerdorffer for critical review of the manuscript.
Funding
This work was supported by the Department of Obstetrics and Gynecology, Maternity, Lausanne, Switzerland and by the SNSF grant number 310030_156169/1 attributed to David Baud. David Baud is also supported by the “Fondation Leenaards” through the “Bourse pour la relève académique,” by the “Fondation Divesa” and by the “Loterie Romande.”
References
- Everett KD. Chlamydia and Chlamydiales: more than meets the eye. Vet Microbiol 2000; 75:109–26; PMID:10889402; http://dx.doi.org/10.1016/S0378-1135(00)00213-3
- Sachse K, Laroucau K, Riege K, Wehner S, Dilcher M, Creasy HH, Weidmann M, Myers G, Vorimore F, Vicari N, et al. Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp. nov. and Chlamydia gallinacea sp. nov. Syst Appl Microbiol 2014; 37:79–88; PMID:24461712; http://dx.doi.org/10.1016/j.syapm.2013.12.004
- Wheelhouse N, Longbottom D. Endemic and emerging chlamydial infections of animals and their zoonotic implications. Transbound Emerg Dis 2012; 59:283–91; PMID:22099945; http://dx.doi.org/10.1111/j.1865-1682.2011.01274.x
- Baud D, Goy G, Jaton K, Osterheld MC, Blumer S, Borel N, Vial Y, Hohlfeld P, Pospischil A, Greub G. Role of Chlamydia trachomatis in miscarriage. Emerg Infect Dis 2011; 17:1630–5; PMID:21888787; http://dx.doi.org/10.3201/eid1709.100865
- Baud D, Goy G, Vasilevsky S, Osterheld MC, Roth-Kleiner M, Croxatto A, Greub G. Roles of bovine Waddlia chondrophila and Chlamydia trachomatis in human preterm birth. New Microbes New Infect 2015; 3:41–5; PMID:25755892; http://dx.doi.org/10.1016/j.nmni.2014.11.004
- Baud D, Greub G. Intracellular bacteria and adverse pregnancy outcomes. Clin Microbiol Infect 2011; 17:1312–22; PMID:21884294; http://dx.doi.org/10.1111/j.1469-0691.2011.03604.x
- Baud D, Regan L, Greub G. Emerging role of Chlamydia and Chlamydia-like organisms in adverse pregnancy outcomes. Curr Opin Infect Dis 2008; 21:70–6; PMID:18192789; http://dx.doi.org/10.1097/QCO.0b013e3282f3e6a5
- Hornung S, Thuong BC, Gyger J, Kebbi-Beghdadi C, Vasilevsky S, Greub G, Baud D. Role of Chlamydia trachomatis and emerging Chlamydia-related bacteria in ectopic pregnancy in Vietnam. Epidemiol Infect 2015; (12):2635–8; http://dx.doi.org/10.1017/S0950268814003616
- Karaer A, Mert I, Cavkaytar S, Batioglu S. Serological investigation of the role of selected sexually transmitted infections in the aetiology of ectopic pregnancy. Eur J Contracept Reprod Health Care 2013; 18:68–74; PMID:23256948; http://dx.doi.org/10.3109/13625187.2012.744818
- Kavanagh K, Wallace LA, Robertson C, Wilson P, Scoular A. Estimation of the risk of tubal factor infertility associated with genital chlamydial infection in women: a statistical modelling study. Int J Epidemiol 2013; 42:493–503; PMID:23505256; http://dx.doi.org/10.1093/ije/dyt011
- Vasilevsky S, Greub G, Nardelli-Haefliger D, Baud D. Genital Chlamydia trachomatis: understanding the roles of innate and adaptive immunity in vaccine research. Clin Microbiol Rev 2014; 27:346–70; PMID:24696438; http://dx.doi.org/10.1128/CMR.00105-13
- World Health Organization. Global incidence and prevalence of selected curable sexually transmitted infections. 2008
- Burton MJ, Mabey DC. The global burden of trachoma: a review. PLoS Negl Trop Dis 2009; 3:e460; PMID:19859534; http://dx.doi.org/10.1371/journal.pntd.0000460
- Hu VH, Holland MJ, Burton MJ. Trachoma: protective and pathogenic ocular immune responses to Chlamydia trachomatis. PLoS Negl Trop Dis 2013; 7:e2020; PMID:23457650; http://dx.doi.org/10.1371/journal.pntd.0002020
- Wyrick PB. Chlamydia trachomatis persistence in vitro: an overview. J Infect Dis 2010; 201 Suppl 2:S88–95; PMID:20470046; http://dx.doi.org/10.1086/652394
- Fadel S, Eley A. Chlamydia trachomatis OmcB protein is a surface-exposed glycosaminoglycan-dependent adhesin. J Med Microbiol 2007; 56:15–22; PMID:17172511; http://dx.doi.org/10.1099/jmm.0.46801-0
- Tan C, Hsia RC, Shou H, Carrasco JA, Rank RG, Bavoil PM. Variable expression of surface-exposed polymorphic membrane proteins in in vitro-grown Chlamydia trachomatis. Cell Microbiol 2010; 12:174–87; PMID:19811502; http://dx.doi.org/10.1111/j.1462-5822.2009.01389.x
- Longbottom D, Russell M, Dunbar SM, Jones GE, Herring AJ. Molecular cloning and characterization of the genes coding for the highly immunogenic cluster of 90-kgdalton envelope proteins from the Chlamydia psittaci subtype that causes abortion in sheep. Infect Immun 1998; 66:1317–24; PMID:9529048
- Longbottom D, Russell M, Jones GE, Lainson FA, Herring AJ. Identification of a multigene family coding for the 90 kDa proteins of the ovine abortion subtype of Chlamydia psittaci. FEMS Microbiol Lett 1996; 142:277–81; PMID:8810511; http://dx.doi.org/10.1111/j.1574-6968.1996.tb08443.x
- Azuma Y, Hirakawa H, Yamashita A, Cai Y, Rahman MA, Suzuki H, Mitaku S, Toh H, Goto S, Murakami T, et al. Genome sequence of the cat pathogen, Chlamydophila felis. DNA Res 2006; 13:15–23; PMID:16766509; http://dx.doi.org/10.1093/dnares/dsi027
- Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res 2000; 28:1397–406; PMID:10684935; http://dx.doi.org/10.1093/nar/28.6.1397
- Read TD, Myers GS, Brunham RC, Nelson WC, Paulsen IT, Heidelberg J, Holtzapple E, Khouri H, Federova NB, Carty HA, et al. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res 2003; 31:2134–47; PMID:12682364; http://dx.doi.org/10.1093/nar/gkg321
- Thomson NR, Yeats C, Bell K, Holden MT, Bentley SD, Livingstone M, Cerdeño-Tárraga AM, Harris B, Doggett J, Ormond D, et al. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res 2005; 15:629–40; PMID:15837807; http://dx.doi.org/10.1101/gr.3684805
- Voigt A, Schofl G, Saluz HP. The Chlamydia psittaci genome: a comparative analysis of intracellular pathogens. PLoS One 2012; 7:e35097; PMID:22506068; http://dx.doi.org/10.1371/journal.pone.0035097
- Bachmann NL, Fraser TA, Bertelli C, Jelocnik M, Gillett A, Funnell O, Flanagan C, Myers GS, Timms P, Polkinghorne A. Comparative genomics of koala, cattle and sheep strains of Chlamydia pecorum. BMC Genomics 2014; 15:667; PMID:25106440; http://dx.doi.org/10.1186/1471-2164-15-667
- Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman RW, Olinger L, Grimwood J, Davis RW, Stephens RS. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet 1999; 21:385–9; PMID:10192388; http://dx.doi.org/10.1038/7716
- Voigt A, Schofl G, Heidrich A, Sachse K, Saluz HP. Full-length de novo sequence of the Chlamydophila psittaci type strain, 6BC. J Bacteriol 2011; 193:2662–3; PMID:21441521; http://dx.doi.org/10.1128/JB.00236-11
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 1998; 282:754–9; PMID:9784136; http://dx.doi.org/10.1126/science.282.5389.754
- Carlson JH, Porcella SF, McClarty G, Caldwell HD. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect Immun 2005; 73:6407–18; PMID:16177312; http://dx.doi.org/10.1128/IAI.73.10.6407-6418.2005
- Kebbi-Beghdadi C, Domröse A, Becker E, Cisse OH, Hegemann JH, Greub G. OmpA family proteins and Pmp-like autotransporter: new adhesins of Waddlia chondrophila. Pathog Dis 2015; 73(6):ftv035; In Press; http://dx.doi.org/10.1093/femspd/ftv035
- Kiselev AO, Stamm WE, Yates JR, Lampe MF. Expression, processing, and localization of PmpD of Chlamydia trachomatis Serovar L2 during the chlamydial developmental cycle. PLoS One 2007; 2:e568; PMID:17593967; http://dx.doi.org/10.1371/journal.pone.0000568
- Vandahl BB, Pedersen AS, Gevaert K, Holm A, Vandekerckhove J, Christiansen G, Birkelund S. The expression, processing and localization of polymorphic membrane proteins in Chlamydia pneumoniae strain CWL029. BMC Microbiol 2002; 2:36; PMID:12453305; http://dx.doi.org/10.1186/1471-2180-2-36
- Wehrl W, Brinkmann V, Jungblut PR, Meyer TF, Szczepek AJ. From the inside out - processing of the Chlamydial autotransporter PmpD and its role in bacterial adhesion and activation of human host cells. Mol Microbiol 2004; 51:319–34; PMID:14756775; http://dx.doi.org/10.1046/j.1365-2958.2003.03838.x
- Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev 2004; 68:692–744; PMID:15590781; http://dx.doi.org/10.1128/MMBR.68.4.692-744.2004
- Dautin N, Barnard TJ, Anderson DE, Bernstein HD. Cleavage of a bacterial autotransporter by an evolutionarily convergent autocatalytic mechanism. EMBO J 2007; 26:1942–52; PMID:17347646; http://dx.doi.org/10.1038/sj.emboj.7601638
- Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol 2007; 61:89–112; PMID:17506669; http://dx.doi.org/10.1146/annurev.micro.61.080706.093233
- Henderson IR, Lam AC. Polymorphic proteins of Chlamydia spp. - autotransporters beyond the Proteobacteria. Trends Microbiol 2001; 9:573–8; PMID:11728862; http://dx.doi.org/10.1016/S0966-842X(01)02234-X
- Henderson IR, Nataro JP. Virulence functions of autotransporter proteins. Infect Immun 2001; 69:1231–43; PMID:11179284; http://dx.doi.org/10.1128/IAI.69.3.1231-1243.2001
- Leyton DL, Rossiter AE, Henderson IR. From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol 2012; 10:213–25; PMID:22337167; http://dx.doi.org/10.1038/nrmicro2733
- Tseng TT, Tyler BM, Setubal JC. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol 2009; 9 Suppl 1:S2; PMID:19278550; http://dx.doi.org/10.1186/1471-2180-9-S1-S2
- Rockey DD, Lenart J, Stephens RS. Genome sequencing and our understanding of chlamydiae. Infect Immun 2000; 68:5473–9; PMID:10992442; http://dx.doi.org/10.1128/IAI.68.10.5473-5479.2000
- Grimwood J, Stephens RS. Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb Comp Genomics 1999; 4:187–201; PMID:10587946; http://dx.doi.org/10.1089/omi.1.1999.4.187
- Grimwood J, Olinger L, Stephens RS. Expression of Chlamydia pneumoniae polymorphic membrane protein family genes. Infect Immun 2001; 69:2383–9; PMID:11254597; http://dx.doi.org/10.1128/IAI.69.4.2383-2389.2001
- Vandahl BB, Birkelund S, Demol H, Hoorelbeke B, Christiansen G, Vandekerckhove J, Gevaert K. Proteome analysis of the Chlamydia pneumoniae elementary body. Electrophoresis 2001; 22:1204–23; PMID:11358148; http://dx.doi.org/10.1002/1522-2683()22:6%3c1204::AID-ELPS1204%3e3.0.CO;2-M
- Christiansen G, Pedersen AS, Hjerno K, Vandahl B, Birkelund S. Potential relevance of Chlamydia pneumoniae surface proteins to an effective vaccine. J Infect Dis 2000; 181 Suppl 3:S528–37; PMID:10839754; http://dx.doi.org/10.1086/315633
- Goodall JC, Yeo G, Huang M, Raggiaschi R, Gaston JS. Identification of Chlamydia trachomatis antigens recognized by human CD4+ T lymphocytes by screening an expression library. Eur J Immunol 2001; 31:1513–22; PMID:11465108; http://dx.doi.org/10.1002/1521-4141(200105)31:5%3c1513::AID-IMMU1513%3e3.0.CO;2-U
- Crane DD, Carlson JH, Fischer ER, Bavoil P, Hsia RC, Tan C, Kuo CC, Caldwell HD. Chlamydia trachomatis polymorphic membrane protein D is a species-common pan-neutralizing antigen. Proc Natl Acad Sci U S A 2006; 103:1894–9; PMID:16446444; http://dx.doi.org/10.1073/pnas.0508983103
- Tan C, Hsia RC, Shou H, Haggerty CL, Ness RB, Gaydos CA, Dean D, Scurlock AM, Wilson DP, Bavoil PM. Chlamydia trachomatis-infected patients display variable antibody profiles against the nine-member polymorphic membrane protein family. Infect Immun 2009; 77:3218–26; PMID:19487469; http://dx.doi.org/10.1128/IAI.01566-08
- Gomes JP, Nunes A, Bruno WJ, Borrego MJ, Florindo C, Dean D. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J Bacteriol 2006; 188:275–86; PMID:16352844; http://dx.doi.org/10.1128/JB.188.1.275-286.2006
- Mygind P, Christiansen G, Persson K, Birkelund S. Detection of Chlamydia trachomatis-specific antibodies in human sera by recombinant major outer-membrane protein polyantigens. J Med Microbiol 2000; 49:457–65; PMID:10798559; http://dx.doi.org/10.1099/0022-1317-49-5-457
- Shaw AC, Gevaert K, Demol H, Hoorelbeke B, Vandekerckhove J, Larsen MR, Roepstorff P, Holm A, Christiansen G, Birkelund S. Comparative proteome analysis of Chlamydia trachomatis serovar A, D and L2. Proteomics 2002; 2:164–86; PMID:11840563; http://dx.doi.org/10.1002/1615-9861(200202)2:2%3c164::AID-PROT164%3e3.0.CO;2-U
- Tanzer RJ, Hatch TP. Characterization of outer membrane proteins in Chlamydia trachomatis LGV serovar L2. J Bacteriol 2001; 183:2686–90; PMID:11274132; http://dx.doi.org/10.1128/JB.183.8.2686-2690.2001
- Skipp P, Robinson J, O'Connor CD, Clarke IN. Shotgun proteomic analysis of Chlamydia trachomatis. Proteomics 2005; 5:1558–73; PMID:15838905; http://dx.doi.org/10.1002/pmic.200401044
- Carrasco JA, Tan C, Rank RG, Hsia RC, Bavoil PM. Altered developmental expression of polymorphic membrane proteins in penicillin-stressed Chlamydia trachomatis. Cell Microbiol 2011; 13:1014–25; PMID:21504531; http://dx.doi.org/10.1111/j.1462-5822.2011.01598.x
- Nunes A, Gomes JP, Mead S, Florindo C, Correia H, Borrego MJ, Dean D. Comparative expression profiling of the Chlamydia trachomatis pmp gene family for clinical and reference strains. PLoS One 2007; 2:e878; PMID:17849007; http://dx.doi.org/10.1371/journal.pone.0000878
- Barbour AG, Amano K, Hackstadt T, Perry L, Caldwell HD. Chlamydia trachomatis has penicillin-binding proteins but not detectable muramic acid. J Bacteriol 1982; 151:420–8; PMID:7085567
- Lee CK, Bowie WR, Alexander ER. In vitro assays of the efficacy of antimicrobial agents in controlling Chlamydia trachomatis propagation. Antimicrob Agents Chemother 1978; 13:441–5; PMID:162541; http://dx.doi.org/10.1128/AAC.13.3.441
- Swanson KA, Taylor LD, Frank SD, Sturdevant GL, Fischer ER, Carlson JH, Whitmire WM, Caldwell HD. Chlamydia trachomatis polymorphic membrane protein D is an oligomeric autotransporter with a higher-order structure. Infect Immun 2009; 77:508–16; PMID:19001072; http://dx.doi.org/10.1128/IAI.01173-08
- Becker E, Hegemann JH. All subtypes of the Pmp adhesin family are implicated in chlamydial virulence and show species-specific function. Microbiologyopen 2014; 3:544–56; PMID:24985494; http://dx.doi.org/10.1002/mbo3.186
- Molleken K, Schmidt E, Hegemann JH. Members of the Pmp protein family of Chlamydia pneumoniae mediate adhesion to human cells via short repetitive peptide motifs. Mol Microbiol 2010; 78:1004–17; PMID:21062373; http://dx.doi.org/10.1111/j.1365-2958.2010.07386.x
- Finco O, Bonci A, Agnusdei M, Scarselli M, Petracca R, Norais N, Ferrari G, Garaguso I, Donati M, Sambri V, et al. Identification of new potential vaccine candidates against Chlamydia pneumoniae by multiple screenings. Vaccine 2005; 23:1178–88; PMID:15629361; http://dx.doi.org/10.1016/j.vaccine.2004.07.045
- Girard V, Mourez M. Adhesion mediated by autotransporters of Gram-negative bacteria: structural and functional features. Res Microbiol 2006; 157:407–16; PMID:16725315; http://dx.doi.org/10.1016/j.resmic.2006.02.001
- Molleken K, Becker E, Hegemann JH. The Chlamydia pneumoniae invasin protein Pmp21 recruits the EGF receptor for host cell entry. PLoS Pathog 2013; 9:e1003325; PMID:23633955; http://dx.doi.org/10.1371/journal.ppat.1003325
- Su H, Watkins NG, Zhang YX, Caldwell HD. Chlamydia trachomatis-host cell interactions: role of the chlamydial major outer membrane protein as an adhesin. Infect Immun 1990; 58:1017–25; PMID:2318528
- Kari L, Southern TR, Downey CJ, Watkins HS, Randall LB, Taylor LD, Sturdevant GL, Whitmire WM, Caldwell HD. Chlamydia trachomatis polymorphic membrane protein D is a virulence factor involved in early host-cell interactions. Infect Immun 2014; 82:2756–62; PMID:24733093; http://dx.doi.org/10.1128/IAI.01686-14
- Taylor BD, Darville T, Tan C, Bavoil PM, Ness RB, Haggerty CL. The role of Chlamydia trachomatis polymorphic membrane proteins in inflammation and sequelae among women with pelvic inflammatory disease. Infect Dis Obstet Gynecol 2011; 2011:989762; PMID:22028586; http://dx.doi.org/10.1155/2011/989762
- Gomes JP, Hsia RC, Mead S, Borrego MJ, Dean D. Immunoreactivity and differential developmental expression of known and putative Chlamydia trachomatis membrane proteins for biologically variant serovars representing distinct disease groups. Microbes Infect 2005; 7:410–20; PMID:15784185; http://dx.doi.org/10.1016/j.micinf.2004.11.014
- Niessner A, Kaun C, Zorn G, Speidl W, Turel Z, Christiansen G, Pedersen AS, Birkelund S, Simon S, Georgopoulos A, et al. Polymorphic membrane protein (PMP) 20 and PMP 21 of Chlamydia pneumoniae induce proinflammatory mediators in human endothelial cells in vitro by activation of the nuclear factor-kappaB pathway. J Infect Dis 2003; 188:108–13; PMID:12825178; http://dx.doi.org/10.1086/375827
- Grayston JT, Woolridge RL, Wang SP, Yen CH, Yang CY, Cheng KH, Chang IH. Field studies of protection from infection by experimental trachoma virus vaccine in preschool-aged children on Taiwan. Proc Soc Exp Biol Med 1963; 112:589–95; PMID:13950005; http://dx.doi.org/10.3181/00379727-112-28112
- Sowa S, Sowa J, Collier LH, Blyth WA. Trachoma vaccine field trials in The Gambia. J Hyg 1969; 67:699–717; PMID:5261212; http://dx.doi.org/10.1017/S0022172400042157
- Detmer A, Glenting J. Live bacterial vaccines - a review and identification of potential hazards. Microb Cell Fact 2006; 5:23; PMID:16796731; http://dx.doi.org/10.1186/1475-2859-5-23
- Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol 2005; 5:149–61; PMID:15688042; http://dx.doi.org/10.1038/nri1551
- Kari L, Whitmire WM, Crane DD, Reveneau N, Carlson JH, Goheen MM, Peterson EM, Pal S, de la Maza LM, Caldwell HD. Chlamydia trachomatis native major outer membrane protein induces partial protection in nonhuman primates: implication for a trachoma transmission-blocking vaccine. J Immunol 2009; 182:8063–70; PMID:19494332; http://dx.doi.org/10.4049/jimmunol.0804375
- Pal S, Peterson EM, Rappuoli R, Ratti G, de la Maza LM. Immunization with the Chlamydia trachomatis major outer membrane protein, using adjuvants developed for human vaccines, can induce partial protection in a mouse model against a genital challenge. Vaccine 2006; 24:766–75; PMID:16199110; http://dx.doi.org/10.1016/j.vaccine.2005.08.074
- Singh SR, Hulett K, Pillai SR, Dennis VA, Oh MK, Scissum-Gunn K. Mucosal immunization with recombinant MOMP genetically linked with modified cholera toxin confers protection against Chlamydia trachomatis infection. Vaccine 2006; 24:1213–24; PMID:16194585; http://dx.doi.org/10.1016/j.vaccine.2005.08.097
- Grotenbreg GM, Roan NR, Guillen E, Meijers R, Wang JH, Bell GW, Starnbach MN, Ploegh HL. Discovery of CD8+ T cell epitopes in Chlamydia trachomatis infection through use of caged class I MHC tetramers. Proc Natl Acad Sci U S A 2008; 105:3831–6; PMID:18245382; http://dx.doi.org/10.1073/pnas.0711504105
- Baehr W, Zhang YX, Joseph T, Su H, Nano FE, Everett KD, Caldwell HD. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc Natl Acad Sci U S A 1988; 85:4000–4; PMID:2453883; http://dx.doi.org/10.1073/pnas.85.11.4000
- Finco O, Frigimelica E, Buricchi F, Petracca R, Galli G, Faenzi E, Meoni E, Bonci A, Agnusdei M, Nardelli F, et al. Approach to discover T- and B-cell antigens of intracellular pathogens applied to the design of Chlamydia trachomatis vaccines. Proc Natl Acad Sci U S A 2011; 108:9969–74; PMID:21628568; http://dx.doi.org/10.1073/pnas.1101756108
- Ortiz L, Demick KP, Petersen JW, Polka M, Rudersdorf RA, Van der Pol B, Jones R, Angevine M, DeMars R. Chlamydia trachomatis major outer membrane protein (MOMP) epitopes that activate HLA class II-restricted T cells from infected humans. J Immunol 1996; 157:4554–67; PMID:8906834
- Eko FO, He Q, Brown T, McMillan L, Ifere GO, Ananaba GA, Lyn D, Lubitz W, Kellar KL, Black CM, et al. A novel recombinant multisubunit vaccine against Chlamydia. J Immunol 2004; 173:3375–82; PMID:15322201; http://dx.doi.org/10.4049/jimmunol.173.5.3375
- Starnbach MN, Loomis WP, Ovendale P, Regan D, Hess B, Alderson MR, Fling SP. An inclusion membrane protein from Chlamydia trachomatis enters the MHC class I pathway and stimulates a CD8+ T cell response. J Immunol 2003; 171:4742–9; PMID:14568950; http://dx.doi.org/10.4049/jimmunol.171.9.4742
- Chaganty BK, Murthy AK, Evani SJ, Li W, Guentzel MN, Chambers JP, Zhong G, Arulanandam BP. Heat denatured enzymatically inactive recombinant chlamydial protease-like activity factor induces robust protective immunity against genital chlamydial challenge. Vaccine 2010; 28:2323–9; PMID:20056182; http://dx.doi.org/10.1016/j.vaccine.2009.12.064
- Li W, Murthy AK, Guentzel MN, Seshu J, Forsthuber TG, Zhong G, Arulanandam BP. Antigen-specific CD4+ T cells produce sufficient IFN-gamma to mediate robust protective immunity against genital Chlamydia muridarum infection. J Immunol 2008; 180:3375–82; PMID:18292563; http://dx.doi.org/10.4049/jimmunol.180.5.3375
- Fling SP, Sutherland RA, Steele LN, Hess B, D'Orazio SE, Maisonneuve J, Lampe MF, Probst P, Starnbach MN. CD8+ T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis. Proc Natl Acad Sci U S A 2001; 98:1160–5; PMID:11158611; http://dx.doi.org/10.1073/pnas.98.3.1160
- Coler RN, Bhatia A, Maisonneuve JF, Probst P, Barth B, Ovendale P, Fang H, Alderson M, Lobet Y, Cohen J, et al. Identification and characterization of novel recombinant vaccine antigens for immunization against genital Chlamydia trachomatis. FEMS Immunol Med Microbiol 2009; 55:258–70; PMID:19281568; http://dx.doi.org/10.1111/j.1574-695X.2008.00527.x
- Karunakaran KP, Rey-Ladino J, Stoynov N, Berg K, Shen C, Jiang X, Gabel BR, Yu H, Foster LJ, Brunham RC. Immunoproteomic discovery of novel T cell antigens from the obligate intracellular pathogen Chlamydia. J Immunol 2008; 180:2459–65; PMID:18250455; http://dx.doi.org/10.4049/jimmunol.180.4.2459
- Mygind T, Vandahl B, Pedersen AS, Christiansen G, Hollsberg P, Birkelund S. Identification of an in vivo CD4+ T cell-mediated response to polymorphic membrane proteins of Chlamydia pneumoniae during experimental infection. FEMS Immunol Med Microbiol 2004; 40:129–37; PMID:14987731; http://dx.doi.org/10.1016/S0928-8244(03)00300-6
- Yu H, Jiang X, Shen C, Karunakaran KP, Brunham RC. Novel Chlamydia muridarum T cell antigens induce protective immunity against lung and genital tract infection in murine models. J Immunol 2009; 182:1602–8; PMID:19155509; http://dx.doi.org/10.4049/jimmunol.182.3.1602
- Yu H, Jiang X, Shen C, Karunakaran KP, Jiang J, Rosin NL, Brunham RC. Chlamydia muridarum T-cell antigens formulated with the adjuvant DDA/TDB induce immunity against infection that correlates with a high frequency of gamma interferon (IFN-gamma)/tumor necrosis factor α and IFN-gamma/interleukin-17 double-positive CD4+ T cells. Infect Immun 2010; 78:2272–82; PMID:20231405; http://dx.doi.org/10.1128/IAI.01374-09
- Yu H, Karunakaran KP, Jiang X, Brunham RC. Evaluation of a multisubunit recombinant polymorphic membrane protein and major outer membrane protein T cell vaccine against Chlamydia muridarum genital infection in three strains of mice. Vaccine 2014; 32:4672–80; PMID:24992718; http://dx.doi.org/10.1016/j.vaccine.2014.06.002
- Yu H, Karunakaran KP, Jiang X, Shen C, Andersen P, Brunham RC. Chlamydia muridarum T cell antigens and adjuvants that induce protective immunity in mice. Infect Immun 2012; 80:1510–8; PMID:22290151; http://dx.doi.org/10.1128/IAI.06338-11
- Johnson RM, Yu H, Kerr MS, Slaven JE, Karunakaran KP, Brunham RC. PmpG303-311, a protective vaccine epitope that elicits persistent cellular immune responses in Chlamydia muridarum-immune mice. Infect Immun 2012; 80:2204–11; PMID:22431650; http://dx.doi.org/10.1128/IAI.06339-11
- Karunakaran KP, Yu H, Jiang X, Chan Q, Moon KM, Foster LJ, Brunham RC. Outer membrane proteins preferentially load MHC class II peptides: Implications for as a Chlamydia trachomatis T cell vaccine. Vaccine 2015; 33(18):2159–66; PMID:25738816
- Li LX, McSorley SJ. B cells enhance antigen-specific CD4 T cell priming and prevent bacteria dissemination following Chlamydia muridarum genital tract infection. PLoS Pathog 2013; 9:e1003707; PMID:24204262; http://dx.doi.org/10.1371/journal.ppat.1003707
- Zhu Y, Jiang J, Said-Sadier N, Boxx G, Champion C, Tetlow A, Kickhoefer VA, Rome LH, Ojcius DM, Kelly KA. Activation of the NLRP3 inflammasome by vault nanoparticles expressing a chlamydial epitope. Vaccine 2015; 33:298–306; PMID:25448112; http://dx.doi.org/10.1016/j.vaccine.2014.11.028
- Pan Q, Pais R, Ohandjo A, He C, He Q, Omosun Y, Igietseme JU, Eko FO. Comparative evaluation of the protective efficacy of two formulations of a recombinant Chlamydia abortus subunit candidate vaccine in a mouse model. Vaccine 2015; 33(15):1865–72
- Zhou J, Qiu C, Cao XA, Lin G. Construction and immunogenicity of recombinant adenovirus expressing the major outer membrane protein (MOMP) of Chlamydophila psittaci in chicks. Vaccine 2007; 25:6367–72; PMID:17640776; http://dx.doi.org/10.1016/j.vaccine.2007.06.031
- Liu S, Sun W, Chu J, Huang X, Wu Z, Yan M, Zhang Q, Zhao P, Igietseme JU, Black CM, et al. Construction of Recombinant HVT Expressing PmpD, and Immunological Evaluation against Chlamydia psittaci and Marek's Disease Virus. PLoS One 2015; 10:e0124992; PMID:25893439; http://dx.doi.org/10.1371/journal.pone.0124992
- Stary G, Olive A, Radovic-Moreno AF, Gondek D, Alvarez D, Basto PA, Perro M, Vrbanac VD, Tager AM, Shi J, et al. VACCINES. A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells. Science 2015; 348:aaa8205; PMID:26089520; http://dx.doi.org/10.1126/science.aaa8205
- Kiselev AO, Skinner MC, Lampe MF. Analysis of pmpD expression and PmpD post-translational processing during the life cycle of Chlamydia trachomatis serovars A, D, and L2. PLoS One 2009; 4:e5191; PMID:19367336; http://dx.doi.org/10.1371/journal.pone.0005191
- Gomes JP, Bruno WJ, Borrego MJ, Dean D. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J Bacteriol 2004; 186:4295–306; PMID:15205432; http://dx.doi.org/10.1128/JB.186.13.4295-4306.2004
- Mygind PH, Christiansen G, Roepstorff P, Birkelund S. Membrane proteins PmpG and PmpH are major constituents of Chlamydia trachomatis L2 outer membrane complex. FEMS Microbiol Lett 2000; 186:163–9; PMID:10802165; http://dx.doi.org/10.1111/j.1574-6968.2000.tb09098.x
- Wheelhouse NM, Sait M, Aitchison K, Livingstone M, Wright F, McLean K, Inglis NF, Smith DG, Longbottom D. Processing of Chlamydia abortus polymorphic membrane protein 18D during the chlamydial developmental cycle. PLoS One 2012; 7:e49190; PMID:23145118; http://dx.doi.org/10.1371/journal.pone.0049190
- Wheelhouse N, Sait M, Wilson K, Aitchison K, McLean K, Smith DG, Longbottom D. Expression patterns of five polymorphic membrane proteins during the Chlamydia abortus developmental cycle. Vet Microbiol 2012; 160:525–9; PMID:22776512; http://dx.doi.org/10.1016/j.vetmic.2012.06.017
- Stemke-Hale K, Kaltenboeck B, DeGraves FJ, Sykes KF, Huang J, Bu CH, Johnston SA. Screening the whole genome of a pathogen in vivo for individual protective antigens. Vaccine 2005; 23:3016–25; PMID:15811648; http://dx.doi.org/10.1016/j.vaccine.2004.12.013