Abstract
Candida parapsilosis is an emerging opportunistic pathogen, second in frequency only to C. albicans and commonly associated with both mucosal and systemic infections. Adhesion to biotic surfaces is a key step for the development of mycoses. The C. parapsilosis genome encodes 5 predicted agglutinin-like sequence proteins and their precise role in the adhesion process still remains to be elucidated. In this study, we focused on the putative adhesin Cpar2_404800, in view of its high homology to the most important adhesion molecule in C. albicans. Two independent lineages of C. parapsilosis CPAR2_404800 heterozygous and null mutants were obtained by site-specific deletion. CPAR2_404800 mutants did not differ from wild-type strain in terms of in vitro growth or in their ability to undergo morphogenesis. However, when compared for adhesion to a biotic surface, CPAR2_404800 null mutants exhibited a marked reduction in their adhesion to buccal epithelial cells (>60% reduction of adhesion index). Reintroduction of one copy of CPAR2_404800 gene in the null background restored wild type phenotype. A murine model of urinary tract infection was used to elucidate the in vivo contribution of CPAR2_404800. A 0.5 and 1 log10 reduction in colony forming unit numbers (per gram) was observed respectively in bladder and kidneys obtained from mice infected with null mutant compared to wild-type infected ones. Taken together, these findings provide the first evidence for a direct role of CPAR2_404800 in C. parapsilosis adhesion to host surfaces and demonstrate its contribution to the pathogenesis of murine urinary candidiasis.
Introduction
Molecular characterization of virulence properties of fungal pathogens through gene knock out strategies have led to a deeper understanding of the complex yeast – pathogen interaction. This has been particularly evident for the most common fungal pathogen of humans, Candida albicans Citation1-3. Nevertheless, this approach has been successfully extended to other clinically relevant non-albicans Candida species. Citation4-6 Among these, C. parapsilosis has emerged as a common opportunistic pathogen, responsible for both mucosal and systemic infection, second in frequency only to C. albicans and commonly associated with catheter-related infections in intensive care units. Citation7-10 The entire genomic sequence of C. parapsilosis (strain CDC 317) has been published in 2009. Citation11 This event permitted the direct analysis of a vast number of putative gene sequences, obtained from a comparative genomic investigation. Some virulence-associated genes in C. parapsilosis have already been identified and characterized. Among these, secreted aspartyl protease (SAP) gene familyCitation12, lipases Citation6,13, fatty acid synthase genesCitation14, biofilm and cell wall regulator (BCR1) geneCitation5 and acid threalase (ATC1) gene. Citation15
The ability to adhere to biotic and/or abiotic surfaces represents an essential trait that enables microorganisms to colonize and eventually infect the host. The stable presence of a microbe within the host, especially on skin or mucosal surfaces, is often related to the strong interaction between molecules that act as receptors and their ligands. This interaction can contribute to the unsuccessful clearance of the microorganism by the host and promote fungal stable colonization of human surfaces. Despite the clinical relevance that C. parapsilosis has gained in the past few decades, little is known on the molecular mechanisms underlying adhesion of this yeast to biotic surfaces.
In silico analysis of the genomic sequence of C. parapsilosis indicated the existence of 5 potential homologues of CaALS genes.Citation11 Homology studies revealed that Cpar2_404800 shared the highest sequence homology with CaAls3p, which is considered one of the most important adhesin in C. albicans.Citation16-20 However, according to the Candida Gene Order Browser database (http://cgob3.ucd.ie/), CPAR2_404800 is synthenic with C. albicans ALS7. The function of this gene, named CpALS7 according to its synteny with C. albicans gene, was investigated by targeted gene deletion, performed with the SAT1-flipper cassette system. Citation2,5,6 The aim of the present study was to evaluate the effect of the deletion of one or both the CpALS7 alleles in 2 independent lineages of mutant strains, each including a heterozygous ALS7/als7Δ and a null als7Δ/als7Δ strain. The panel of mutant isolates was characterized for phenotypic traits such as growth rate, ability to grow in the presence of cell wall perturbing agents, cytotoxicity and ability to adhere to human buccal epithelial cells. Pathogenicity of the mutant collection was also assessed in 2 different experimental models, including intra-hemocelic infection of Galleria mellonella larvae Citation21,22 and a murine model of urinary infection. Citation23
Results
In silico analysis of ALS-like genes and selection of CPAR2_404800 as first target for site-specific mutagenesis
According to the Candida Genome Database (CGD) (http://www.candidagenome.org) annotation, Candida parapsilosis possesses 5 potential homologues of CaALS genes (CPAR2_500660; CPAR2_404770; CPAR2_404800; CPAR2_404790; CPAR2_3j404780).
The protein sequences were aligned using Clustal Omega, analyzed with InterPro 45.0 Citation24 and a phylogenetic tree was obtained with Mega6 program Citation25 (data not shown). A quick analysis of the tandem repeat regions was performed with an on-line tool T-REKS Citation26, confirming the adhesin-like conserved features for all 5 proteins. Among these, CPAR2_404800 (gene length 4152 nucleotides, amino acid number 1383) shared the highest sequence homology with C. albicans ALS3, one the most important adhesion molecules in this species. Citation17,19
Pairwise identity scores, obtained with Clustal Omega and based on the alignment of CaAls and CpAls protein sequences, indicated an identity score of 43.36% between Cpar2_404800 and CaAls3, while the majority of the alignment values were below 35%. Since CPAR2_404800 is synthenic with C. albicans ALS7, according to the Candida Gene Order Browser database (http://cgob3.ucd.ie/) CPAR2_404800 was named CpALS7 and selected as first target for site specific mutagenesis to evaluate its role in C. parapsilosis adhesion to biotic surfaces.
Generation of the CpALS7 null mutant and reconstituted strain with the SAT1 flipper cassette
To evaluate the role of CpALS7, both alleles were deleted in C. parapsilosis wild type strain ATCC 22019 as background, using the SAT1 flipper cassette as disruption strategy. Citation2,5,6 Plasmid p35ALS7 (Table S1), containing the CpALS7 disruption cassette was sequenced and results obtained indicate the absence of mismatched bases (Table S2 and Table S3A).
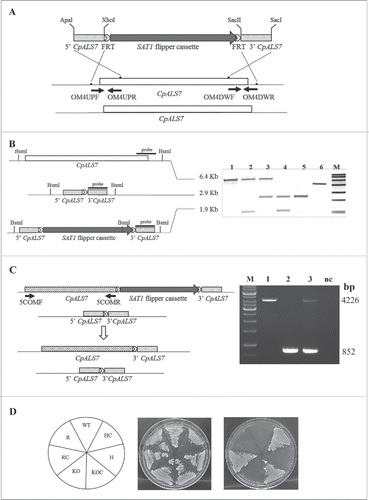
Two separate rounds of transformation, followed by FLP-mediated CaSAT1 resistance marker recycling, gave rise to 2 independent lineages of heterozygous (H) and null strains (KO) lacking one or both copies of CpALS7, respectively (). Genomic DNA from wild type (WT) and the panel of mutant strains was extracted and used to verify the correct integration/excision of the cassette by targeted PCRs (data not shown). Southern blot based restriction analysis with BsmI enzyme was also used to confirm that the appropriate mutagenesis has occurred in each strain, using a 520 bp fragment of the 3′ homology region as a probe. illustrates a scheme of CpALS7 locus in the different strains obtained in this study and shows the Southern blot pattern obtained for each isolate (Lineage b, ). To demonstrate that the phenotype of the null mutant strain was linked to the deletion of CpALS7 only, a functional copy of the gene was reintroduced into the original locus in the null mutant background, using the SAT1 flipper cassette. Transformants were screened by PCR and the correct excision of the cassette was assessed (). In addition, a fragment containing the complete CpALS7 ORF was amplified in the reconstituted strain (R) and the 5′ and 3′ regions of the gene were sequenced, demonstrating complete homology with the wild type sequence (Table S3B). All strains containing SAT1 flipper cassette integrated in genome (HC and KOC strains) were resistant to nurseothricin (NTC) and were maintained on YPD plates supplemented with 100 μg/ml NTC. All the other strains were sensitive to NTC and were maintained on YPD plates ().
Figure 1. CpAls7 disruption strategy based on SAT1 flipper cassette (A). Upstream and downstream homology sequences from C. parapsilosis reference strain ATCC 22019 were amplified and inserted at the ApaI/XhoI and SacII/SacI sites surrounding the SAT1 flipper cassette. The disruption cassette integrated in the CpALS7 allele by homologous recombination. (B). Southern blot hybridization analysis of genomic DNA isolated from the mutants collection and digested with BsmI restriction enzyme. The probe used to verify the correct construction of the mutant collection was amplified by PCR from the downstream homology fragment (schematized by the black bar). M: Roche Dig Labeled Marker VII; 1: wild type (WT); 2: heterozygous with cassette (HC); 3: heterozygous (H); 4: null mutant with cassette (KOC); 5: null mutant (KO); 6: disruption cassette. The expected sizes were 6.4 Kb, 1.9 Kb, 2.9 Kb, and 5.3 Kb for the wild type allele, the allele with the integrated cassette, the deleted allele and the cassette, respectively. (C). The entire coding sequence of CpALS7 was amplified and cloned at the 5′ end of the SAT1 flipper cassette. The reintegration cassette integrated in one of the 2 null mutant alleles by homologous recombination. Primers OM4UPF2 and OM4DWR1 (Table S2) were used to verify the presence of the entire copy of CpALS7 integrated in the correct locus in reconstituted strain (R). Expected fragment lengths: 4.2 Kb for WT and R, 852 bp for KO strains. M: 1Kb DNA ladder (Invitrogen), 1: WT; 2: KO; 3: R; CN: negative control. (D). WT and mutant strains (HC, H, KOC, KO, reconstituted strain with cassette (RC), R) grown for 48 h at 30°C on YPD or nourseothricin (NTC) supplemented YPD plates.
Effect of CpALS7 deletion on C. parapsilosis growth rate on conventional medium or in the presence of cell wall perturbing agents
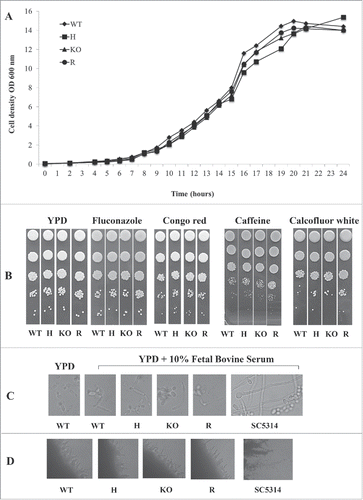
Growth rate in YPD liquid medium was evaluated in the panel of CpALS7 mutant strains. As shown in , panel A, no difference was observed in the growth rate in liquid medium at 37°C. The effect of CpALS7 deletion on cell wall integrity was also evaluated in terms of ability to grow on solid YPD media supplemented with different cell wall perturbing agents, including, congo red, calcofluor white, or other compounds such as fluconazole and caffeine. Citation27-29 In all experiments, stressing agents were tested at sub-fungicidal concentration. At both the temperatures tested (30°C and 37°C), the mutant collection showed a similar growth, compared to the wild type strain in all conditions tested (Fig. S1 and Fig. 2B, respectively).
Figure 2. Phenotypic analysis of C. parapsilosis strains. (A). Growth curve of wild type strain (WT, ATCC22019), CpALS7 heterozygous (H), null (KO) and reconstituted (R) strains in YPD medium at 37°C. (B). Susceptibility to cell wall perturbing agents of the C. parapsilosis strains was evaluated by spot assay, on YPD agar supplemented with the following compounds: fluconazole (0.5 mg/l), congo red (1 mg/l), caffeine (5 mM); calcofluor white (20 mg/l). Approximately 1 × 106 cells and 10 fold dilutions were spotted on different media. Plates were incubated at 30°C (Fig. S1) or 37°C for 48 h, and visually inspected. Experiments were performed in duplicate, with similar results. (C). Production of pseudohyphae by CpALS7 mutant strains. C. albicans SC5314 was also included as positive control. Morphogenesis was induced in YPD broth in presence of 10% FBS. Following 24 h of incubation at 37°C, 10 μl of each culture was directly observed with an optical microscope at 400 × magnification. (D). The ability to produce filaments was also visualized on colony borders grown on spider agar. Photographs were taken following 7-day incubation at 37°C. C. albicans SC5314 represented a positive control for morphogenesis.
These results indicate that the lack of CpALS7 does not alter the ability of C. parapsilosis to grow under these experimental conditions.
Ability to undergo morphogenesis in vitro
The ability to form pseudohyphae, a virulence trait that may be involved in tissue and cell invasion Citation30, was also assessed in the mutant collection. C. parapsilosis is not able to produce true hyphae, as observed in C. albicans, but under inducing conditions budding cells do start to elongate, forming filaments with constrictions at the cell-cell junctions. As shown in , panel C, in the presence of serum, pseudohyphae were observed in C. parapsilosis wild type strain as well as in the mutant strains, indicating that the deletion of CpALS7 did not affect the transition to a filamentous morphology. As expected, C. albicans (control strain SC5314) produced true hyphae. The same result was obtained when morphogenesis was evaluated on solid medium. All C. parapsilosis strains produced filamentation to a similar extent following 7-day incubation at 37°C on spider agar, while C. albicans SC5314 strain gave rise to a more pronounced phenotype ().
Epithelial cell damage assay
The potential cytotoxic activity exerted by CpALS7 mutant panel on the immortalized epithelial cell line A549 was also evaluated. Since C. parapsilosis is considered less pathogenic than C. albicans, a more virulent strain, C. albicans reference strain SC5314, was included in the assay as a positive control.Citation31 No cytotoxic effect was observed with any of the C. parapsilosis strains, at all time points tested, while a progressive increase in cell damage was observed in A549 co-incubated with C. albicans over time (1.7%, 1.8%, 4.3% and 44.9% at 4, 6, 8 and 16 hour incubation respectively). Different MOI were also tested with an increasing number of C. parapsilosis ATCC 22019 yeast cells (data not shown) but no effect was detected, even with a doubled MOI (32 yeast cells per human cell).
Real time –RT PCR analysis
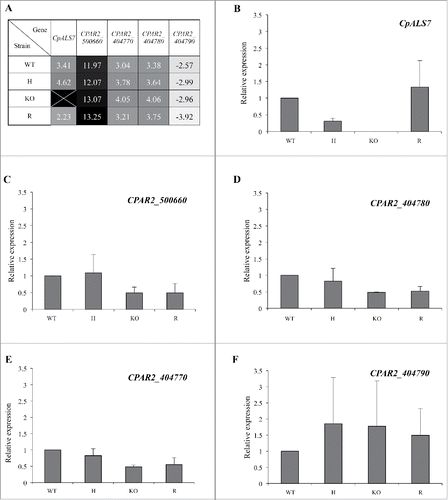
Quantitative expression of CpALS-like genes was determined from late-exponential phase cultures of C. parapsilosis wild type and mutant strains. As evidenced by the heat map depicted in , all 5 genes were expressed under basal growth conditions, even though CPAR2_ 500660 expression was barely detectable in all the strains, with transcripts appearing more than 10 cycles later than the reference gene (actin). Conversely, CPAR2_ 404790 expression was the highest among the ALS-like genes, as indicated by negative values observed for all strains, with transcripts detectable between 2.57 and 3.92 cycles before actin (). As expected, no CpALS7 transcript was observed in the null mutant strain (KO, ), while complemented strain (R) showed the presence of CpALS7 mRNA. No significant changes in other ALS-like gene expression were observed in mutant strains, in these experimental conditions ().
Figure 3. Basal transcriptional profiles of all 5 putative ALS like genes in C. parapsilosis wild type (WT), CpALS7 heterozygous (H), null (KO) and reconstituted (R) mutant strains grown in YPD medium to late exponential-phase. (A). Relative expression heat map of ALS-like genes normalized on actin expression levels. (B-F). Relative expression of each gene in mutant strains normalized on actin and wild type transcripts. At least 3 biological replicates were analyzed. Error bars represent standard error of means of 3 independent experiments.
In vitro adhesion assay to human buccal epithelial cells
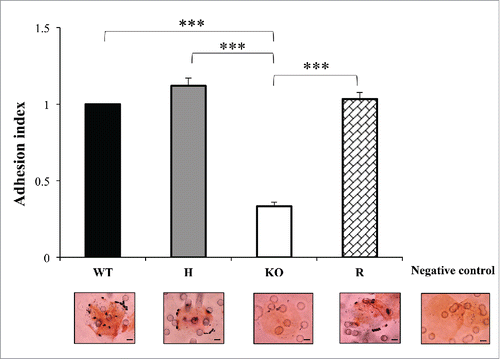
In this study, an adhesion assay was set up to evaluate the adhesion ability of the CpALS7 mutant strains to human buccal epithelial cells (HBECs). Citation32 As illustrated in (data presented for lineage b only), a significant reduction in the adherence to HBECs was observed for both lineages of null mutants, compared to the wild type strain, with more than 60% drop in the adhesion index (0.40 and 0.33 for lineage a and b, respectively; P<0.001). Reintegration of a CpALS7 allele restored the wild type phenotype, confirming the role of CpALS7 in adhesion to HBECs (). In fact, the adhesion properties of the complemented mutant did not significantly differ from wild type ().
Figure 4. The adhesion ability of C. parapsilosis wild type (WT), heterozygous (H), null (KO) and reconstituted (R) mutant strains was tested on human buccal epithelial cells obtained from a healthy donor not colonized with Candida spp. Bars represent adhesion index mean ± standard error of mean. At least 3 biological replicates were used. ***P < 0.001. Micrographs below bars show representative Gram-stained C. parapsilosis blastoconidia from each of the strains adhered to a buccal cell observed at a magnification of 1000 ×. Scale bar denotes 10 μm.
Galleria mellonella intra-hemocelic infection
The pathogenic potential of the strain collection was also assessed in an in vivo infection of the non-conventional host model Galleria mellonella. Dead larvae were differentiated from alive ones by a simple visual analysis: death was associated with a black pigmentation due to a strong melanization and sclerotization of the tissue and with the absence of response to physical pressureCitation22,33 (). Inoculum standardization was established by a preliminary experiment in which different numbers of wild type strain yeast cells were inoculated in groups of larvae (unpublished results). The highest dose (4 × 106 yeast per larva) resulted in a high rate of death within the first 2 days of infection, which could eventually prevent an adequate comparison of C. parapsilosis strains killing ability. An inoculum of 8 × 105 colony forming units (CFUs) per larva resulted in approximately 50% mortality after 2 days of infection. This infecting dose was then chosen to monitor the infection outcome up to 10 days post infection. Healthy larvae were injected with wild type strain and with the CpALS7 mutant strains (lineage b), obtained in this study. All C. parapsilosis strains were able to cause death in G. mellonella, with comparable median survival values: 4 days for wild type; 2 for heterozygous strain; 3.5 for null mutant strain; 2 for CpALS7 reconstituted strain (). The survival curves of larvae infected with C. parapsilosis strains were also compared by the Log-rank (Mantel-Cox) test. No significant differences were observed (P = 0.44) in the median survival rates of infected larvae. After 5 days post infection, 35% larvae infected with wild type and heterozygous strains were still alive. A 30% survival rate was observed for reconstituted strain and 45% for the null mutant. At the end of the time course (10 days), the wild type strain caused 85% mortality in the infected larvae, similar to the heterozygous/reconstituted strain (80% of dead larvae) and null mutant (60%).
Figure 5. Effect of deletion of CpALS7 on C. parapsilosis pathogenicity. (A). Intra-hemocelic infection of Galleria mellonella larvae with CpALS7 wild type (WT) and CpALS7 mutant strains (heterozygous, H; null, KO; and reconstituted, R). Survival curves of G. mellonella infected with CpALS7 wild type and lineage b mutant strains (20 larvae per group) with 8×105 CFUs per larva. Photographs represent typical pigmentation of alive and dead larvae. (B). Murine model of urinary tract infection. Groups of 17 BALB/c mice were transurethrally challenged with approximately 1 × 108 C. parapsilosis cells for each of the indicated strains. Data are expressed as the log10 colony-forming units (CFUs)/g of yeast cells recovered from kidney and urinary bladder homogenates 4 days after the challenge. The log10 CFUs from both kidneys were combined and averaged. A value of 0 was assigned to uninfected organs. Horizontal bars represent median. Log10 counts were compared for statistical significance by non-parametric Wilcoxon rank sum tests. A P value < 0.05 was considered to be statistically significant.
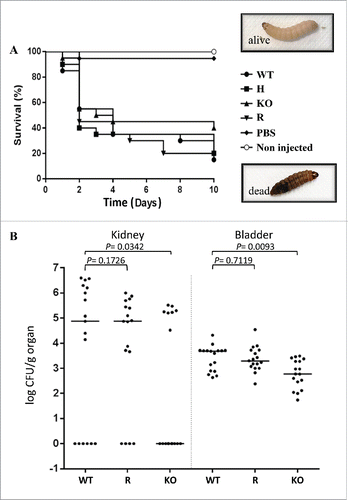
Murine model of urinary infection
Since G. mellonella infection experiments did not show a reduction in virulence for the CpALS7 deleted strain, it was also tested in a mammalian model of urinary infection. Citation23 With the aim to address the effect of CpALS7 deletion on the ability of C. parapsilosis to induce urinary tract infections (UTIs), we compared the pathogenic potential of null mutant, wild type and complemented strains in a mouse model of ascending infection. Preliminary experiments were conducted to test the optimal inoculum useful for BALB/c mice infection via intraurethral catheterization and the time point at which mice would be sacrificed. In particular, 4 different inoculum concentrations (from 106 to 109 yeast cells) and 2 time points (4 and 7 days post infection; PI), were tested. Based on the mean CFUs recovered from kidney and bladders, an inoculum of 108 C. parapsilosis cells and 4 days PI for sacrifice were chosen to conduct the subsequent in vivo experiments (data not shown).
As shown in , the log10 CFUs (per gram of tissue) of C. parapsilosis obtained from mice infected by null mutant was significantly lower with respect to those recovered from wild type strain, 1.1 × 103 vs 4.4 × 103 (P = 0.0093), and 8 × 104 vs 9.7 × 105 (P=0.0342), from bladder and kidneys, respectively. Results obtained by injection of CpALS7 reconstituted strain showed no significant difference compared to wild type strain for bladder and kidneys.
Discussion
Information on virulence and pathogenic traits collected over the past decade for C. albicans, have prepared the path for a better understanding of the molecular mechanisms underlying C. parapsilosis virulence. Colonization and infection are tightly related to adhesion, a common trait shared by most of the pathogenic microorganisms. In fungi, cell wall proteins mediate primary interactions with the host surfaces. Citation16,20,34,35 C. albicans possesses different cell wall proteins, called adhesins, which are usually anchored to the cell wall structure via a GPI anchor residue. The most studied adhesion molecules belong to CaALS gene family, which is composed of 8 members: ALS1-7 and ALS9. Citation16 Different studies, including binding assays, targeted gene deletion, in vitro and in vivo infection models have highlighted the importance of CaALS3 in the adhesion process to host surfaces. Citation18,20,36 C. parapsilosis possesses 5 putative homologues of CaALS, identified by in silico homology studies. Citation11 Actually, little information on the role of ALS genes in C. parapsilosis adhesion to biotic surfaces and pathogenicity is available.
Phenotypic analysis of wild type, heterozygous and null mutants confirmed that the deletion of CpALS7 did not affect the growth rate of the yeast in YPD medium. Moreover, growth and colony morphology of mutant strains were not altered by the presence of cell wall-perturbing agents, with results comparable to those observed for the wild type strain. These findings suggest that deletion of CpALS7 did not cause a marked upheaval in the cell wall arrangement under the experimental conditions used. Further studies, including the use of different stress conditions and a deeper analysis of the cell wall proteome will be required to exclude any effect of CpALS7 deletion in the cell wall organization.
The role of CpALS7 in the adhesion process was assessed in an in vitro model, which involves the co-incubation of yeast cells in the presence of human buccal epithelial cells (HBECs). Even though epithelial monolayers represent continuous layers of cells, resembling the physical structure of a human epithelium, they are not colonized with bacteria. Microbial flora takes an active part in the infectious process, competing with the pathogen and even binding Candida cell adhesins, as demonstrated by Hoyer and colleagues. Citation37 For this reason, we chose adhesion to primary buccal cell as the experimental setting for testing Candida spp. adhesive properties, mimicking the in vivo conditions where the interaction between yeast and host cell microflora occurs as also reported elsewhere. Citation38-40 Despite the extent of experimental variability associated with the use of primary cells, we previously demonstrated that adhesion values obtained in different experiments maintain a common trend, which is conserved even when cells from different donors are used. Citation32 The adhesion assay to HBECs revealed a significant reduction in the adhesion ability of the CpALS7 null mutant. Similar data were observed for the 2 independent lineages of mutants. Moreover, re-integration of a functional copy of CpALS7 in the knock out background restored the adhesion capacity, with values comparable to the wild type ones. These findings demonstrated a role of CpALS7 in the adhesion to buccal epithelial cells, as previously demonstrated for C. albicans Als3. Citation20
Adhesins have also been shown to play a key role in biofilm formation in C. albicans. Citation20 However, the role of CpALS7 in biofilm production by C. parapsilosis could not be fully assessed, since the wild type reference strain ATCC 22019 used to generate mutants failed to produce biofilm at both 30° and 37°C. Citation41
The cytotoxicity of the mutant collection toward the tumoral epithelial cell line A549 was also evaluated, but no cytotoxic effect was observed in cells after incubation with CpALS7 mutant strains, or wild type strain. This finding supports the conclusion of a previous study in which C. parapsilosis did not demonstrate a significant ability to invade and damage immature enterocytes (H4 cells), compared to C. albicans.Citation31 In addition, in other Candida species, a different degree of damage was observed, according to the cell line type used in the assay. For example, a clinical isolate of C. tropicalis was able to reach a 14% cytotoxicity against TCC-SUP cells, 6 % in Caco-2 cell and only 1% with HeLa cell line.Citation31 Viable cells of C. albicans caused a high level of damage to TR-146 cells (oral type cells) 12 h after incubation, with more than 90 % cytotoxicity, while the incubation in the presence of Caco-2 cells (intestinal cells) resulted in a 20 % cytotoxicity. Citation42 In this respect, other immortalized cell lines could be more useful to investigate this aspect of C. parapsilosis pathogenicity.
The pathogenic potential of wild type and mutant strains was first assessed by intra-hemocelic infection of the non-conventional host Galleria mellonella. The use of an invertebrate host offers several advantages, compared to mammalian host, such as the possibility to collect an extensive quantity of data over a short period of time by large screenings, without the use of specialized equipment and with limited bioethical issues. The use of G. mellonella larvae is considered a good choice in the strategy to optimize the infection system and could help reducing the number of further experiments performed on mice. Citation21,22 Survival curves of G. mellonella larvae infected with WT, H and KO strains did not significantly differ. These findings suggested that CpALS7 did not affect the virulence of C. parapsilosis during the dissemination and the invasion of host tissues in a systemic infection. This result can be explained by considering that adhesins are usually involved in the first stages of the infection, when attachment to the host surfaces is critical to prevent microbial eradication by the natural defenses. The direct injection of the pathogen in the hemocelic cavity of the larvae somehow bypassed the colonization process. Optimization of the oral infection by C. parapsilosis in G. mellonella could be considered an alternative approach to compare the pathogenicity of the CpALS7 mutant family.
Alternatively, a murine model of urinary infection was used to test the pathogenic potential of the mutants in a mammalian host. As we recovered CFUs in both kidneys and bladder of mice infected with wild type strain and null mutant we could conclude that both strains are capable of causing bladder infections and to ascend into kidneys. However, a statistically significant reduction in CFUs for BALB/C mice infected with KO strain was observed in comparison with those infected with the WT. This indicates that deletion of CpALS7 results in a reduced ability of the yeast to infect the entire mouse urinary tract and that this gene does influence the pathogenesis of C. parapsilosis in vivo. Nevertheless, other adhesins could be involved in the adhesion to epithelial tissue of bladder and in the migration to kidney, since only an attenuation in the ability of null mutant to cause infection was observed. Whether CpALS7 is involved in other virulence mechanisms beside adhesion remains to be determined.
Overall, this study provides the first evidence for a direct role of CpALS7 in C. parapsilosis adhesion to host surfaces and demonstrated a contribution of this gene to the pathogenesis of murine urinary candidiasis.
Materials and Methods
Strains and growth conditions
Candida parapsilosis strains used in this study are listed in the Table S4. Strains were maintained in 30% glycerol stock frozen at −20°C and −80°C and sub-cultured on YPD agar plates (10 g yeast extract, 20 g peptone, 20 g dextrose, 15 g agar per liter). Yeast strains were routinely grown in YPD liquid medium at 30°C with shaking. For the selection of NTC–resistant transformants, 100 µg ml−1 of NTC (Werner BioAgents, Jena, Germany) was added to YPD agar plates.
Escherichia coli strains used in this study were: E.coli DH5α (genotype F-, endA1, hsdR17 [rk-, mk-], supE44, thi-1, recA1, gyrA96, relA1,Δ[argF-lac]U169,λ-,Φ80dlacZΔM15), kindly provided by Joachim Morschhäuser, and E.coli DH10β (F-, mcrA,Δ[mrr-hsdRMS-mcrBC],Φ80dlacZΔM15,ΔlacX74, endA1, recA1, deoR,Δ[ara,leu]7697, araD139, galU, galK, nupG, rpsL,λ-). Strains were maintained in 15% glycerol stock frozen at −20°C or −80°C and were routinely grown in Luria Bertani (LB) liquid medium or agar plates, under selection pressure when required (100 µg/ml Ampicillin). E. coli cells were grown at 37°C for 16-18 hours with shaking.
Construction of the als7△/als7△ null mutant strain
Candida parapsilosis reference strain ATCC 22019 was selected as parental strain for the generation of 2 independent lineages of mutants, each including a CpALS7 heterozygous strain and a null mutant strain. To generate CpALS7 mutants we used the SAT1-flipper cassette contained in pSFS2 plasmid (Table S1). Citation2 This method relies on the use of a cassette that contains a C. albicans-adapted NTC resistance marker (CaSAT1) under the control of a C. albicans actin promoter (CaACT1) for the selection of integrative transformants. C. albicans adapted FLP gene (CaFLP), whose expression is driven by a C. albicans maltose inducible promoter (CaMAL2), is responsible for cassette excision from the genome. Integration into the target locus occurs with high specificity due to the presence of flanking homologous sequences on both sides of the cassette.
The upstream and downstream CpALS7 homology regions (5′ALS7, 564 bp, from nucleotide −25 to +539, and 3′ALS7, 520 bp, from nucleotide +4006 to +4526) were amplified from ATCC 22019 genome using primers OM3UPF/OM3UPR and OM3DWF/OM3DWR, containing engineered ApaI, XhoI and SacII, SacI restriction sites, respectively (Table S2). The amplified fragments were purified from PCR mixtures with the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA), following the manufacturer's instructions. Plasmid pSFS2 was used as backbone for the first round of ligation, producing plasmid p3ALS7 (Table S1). Plasmidic DNA (pDNA) was isolated with small-scale plasmid DNA isolation (Miniprep) assay. The purified downstream homology region and the pSFS2 plasmid were digested with the combination of SacII/SacI restriction enzymes (New England Biolabs, Ipswich, MA, USA) and then ligated together with T4 DNA ligase (Promega) in order to create plasmid p3ALS7 (Table S3). This plasmid was propagated in E. coli DH10β cells made competent by calcium chloride method. Citation43 p3ALS7 and the purified upstream homology region of CpALS7 were digested with ApaI and XhoI restriction enzymes (New England Biolabs). Ligation of the backbone (p3ALS7) and the insert (5′ALS7) led to the creation of p35ALS7 (Table S1). Plasmid p35ALS7 was sequenced by Eurofins MWG Operon, using M13 forward sequencing primer (−20), 17-mer and M13 reverse sequencing primer (−26), 17-mer and primer But237 (Table S2). Plasmid p35ALS7 was propagated in E. coli DH10β as well and pDNA was isolated. The double digestion with ApaI and SacI produced a linearized fragment of about 5.3 Kb containing the SAT1 flipper disruption cassette flanked by the upstream and downstream CpALS7 sequences.
Construction of the reintegration cassette
To show that the mutant phenotype was caused by the CpALS7 gene deletion, a wild type copy of CpALS7 gene was reintroduced into the als7△/als7△ null mutant strain. p3ALS7 plasmid was used as backbone for the construction of the reintegration cassette. Primers 5COM3F and 5COM3R (Table S2), containing ApaI and SacI sites respectively were used in order to amplify the entire coding sequence of CpALS7 (+4439 bp, comprehensive of −26 upstream and +261 downstream bp) using Q5® High-Fidelity DNA Polymerases by New England Biolabs Inc.. The PCR product and p3ALS7 were digested with ApaI and SacI restriction enzymes and ligated together. The plasmid obtained was named p3RALS7 (Table S1). Since CpALS7 gene contains a restriction site for SacI, it was not possible to perform double digestion of the plasmid. Therefore, p3RALS7 was linearized by ApaI single digestion and directly used to electroporate the null mutant.
Preparation and transformation of C. parapsilosis competent cells
C. parapsilosis strains were transformed by electroporation as previously described, Citation5,6 with some modifications. Overnight cell cultures were diluted 1:100 in 50 ml of fresh YPD broth and incubated at 30°C until the OD600nm reached approximately 1.6-2.0. Cell pellets were suspended in 8 ml of double distilled (DD) H2O, 1 ml of 10 × TE (100 mM Tris-Cl pH7.5, 10mM EDTA 10mM, pH 7.5) and 1 ml of 1 M lithium acetate (pH 7.5) and incubated for 1h at 30°C with gentle shaking. After addition of 100 µl of 1M DTT the incubation was extended for a further 30 min. Candida cultures were washed twice, first with 40 ml of ice-cold DD H2O and then with 25 ml of ice-cold H2O. Finally, cells were washed with 5 ml of 1 M sorbitol and the pellet was suspended in the remaining sorbitol (100 μl) with a tip by gentle swirling. Approximately 1 µg of the purified disruption cassette or the reintegration construct was mixed with 40 µl of C. parapsilosis competent cells, maintained on ice and transferred into a 0.2 cm electroporation cuvette. A negative control, represented by competent cells electroporated with H2O, was added in each experiment. The electroporation system (Gene Pulser, Bio-Rad, Milan, Italy) was set with the following parameters: 2.5 kV, 25 µF, 200 Ω. After the electroporation, 1 ml of YPD supplemented with 1M sorbitol was added to the sample and incubated at 30°C for 4 h to allow cell recovery. 200 µl of the culture and the remaining pellets were then plated on YPD agar supplemented with 100 µg/ml NTC. Negative control pellet was spread on YPD NTC agar plate as well, while 200 µl were plated onto YPD agar plates in order to check cell viability. Plates were grown at 30°C for 2-3 days.
Screening of recombinant colonies of C. parapsilosis
Screening of recombinant colonies of C. parapsilosis was performed by colony PCR. Different primer sets were used to select clones (among heterozygous, null mutants or reintegrated strains, with or without the cassette integrated in the genome) for Southern blot analysis. Primers used are listed in Table S2. Correct construction of the mutant collection was confirmed by Southern blot analysis performed according to the guideline of DIG Application Manual for Filter Hybridization (www.roche-applied-science.com, Roche Diagnostic, Milan, Italy). Total genomic DNA was isolated from WT, HC, H, KOC and KO strains and 1 μg was digested with BsmI restriction enzyme (New England Biolabs). The 3′ALS7 downstream homology region (520 bp) was selected as hybridization probe for Southern blotting experiments.
Sequencing of CpALS7 gene fragments in the complemented strain
CpALS7 gene sequence from CpALS7R strain was sequenced using primers UP4F2/A4REV2 (1405 bp, from nucleotide −416 to +989), CPAG_05056F/A4REV8 (779 bp, from nucleotide +3207 to +3985) and A4FOR9/A4REV9 (442 bp from nucleotide +3785 to +4226) (Table S3B).
Separate pre-mix samples containing CpALS7 gene fragments and appropriate primers were sequenced using the cycle sequencing technology (dideoxy chain termination/cycle sequencing) on ABI 3730XL sequencing apparatus (Eurofins Genomics Ebersberg, Germany)
Growth assays
Growth rate of the mutant collection was analyzed in liquid YPD media. A single colony of each strain was inoculated in 20 ml of YPD broth and incubated overnight at 30°C or 37°C with shaking. Cells were diluted 1:100 in 100 ml of fresh YPD and incubated at 30° or 37°C with shaking. Spectrophotometric readings (OD600nm) were taken every 2 hours for the first 6 hours and then every hour up to 24 h. Cell viability was assessed by plating a diluted aliquot on YPD agar plates. The number of CFU was counted after 24 h incubation at 30°C.
The susceptibility to compounds that interfere with the cell-wall architecture was tested in solid media. C. parapsilosis strains were grown in YPD medium at 30°C or 37°C overnight. Cells were enumerated using a hemocytometer and diluted to a concentration of 107 cells/ml. Serial dilution were set up from 107 cells/ml to 10 cells/ml) in sterile water and 10 μl of each dilution was spotted on YPD agar plates supplemented with cell-wall perturbing agents: 1 mg/l congo red (Sigma Aldrich, Milan, Italy), 0.5 mg/l fluconazole (Sigma Aldrich) 20 mg/l calcofluor white (Sigma Aldrich) and 5 mM caffeine (Merck, Darmstadt, Germany). After incubation at 30°C or 37°C for 24 h or 48 h, plates were photographed.
Filamentation assays
A single colony of each C. parapsilosis strain was inoculated in 10 ml of YPD broth and incubated overnight at 30°C with shaking. C. albicans SC5314 strain was included in the morphogenesis experiments as a positive control. Following incubation, a 106 cells/ml suspension for each strain was prepared in YPD medium supplemented with 10% Fetal Bovine Serum (FBS). Cells were inoculated at the concentration of 6 × 105 cells/ml in 1ml of YPD + 10% FBS in polystyrene 24-well micro titer plates (BD Biosciences) and incubated at 30°C without shaking for 24 h. At the end of the incubation period, 10 μl of each suspension was observed at 400 × magnification in order to evaluate pseudohypha formation.
Morphogenesis was also evaluated on spider agar. From cell suspensions, 10 μl was spotted on the surface of Spider medium (1% nutrient broth, 1% mannitol, 0.2% K2PO4, 2.5% agar, pH 7.2; Citation44). Plates were incubated at 30°C for 7 days and then colonies were observed. This assay was performed in triplicate.
Real Time RT-PCR analysis
Quantitative expression of CpALS genes was determined by real-time reverse transcription (RT)-PCR starting from total RNA of C. parapsilosis strains. Each strain was inoculated in 10 ml of YPD and grown ON at 30°C with shaking. An aliquot (500 μl) of the pre-inoculum was then inoculated in 20 ml of fresh YPD broth and incubated for further 24 h at 30°C. Total RNA was extracted with the Nucleospin RNA (Macherey Nagel, Düren, Germany) according to manufacturer's instructions and stored at −80°C. The quality and quantity of the extracted RNA were spectrophotometrically determined in an UVette® 220-1600 (10 mm path length, 100 μl of sample volume, Eppendorf, Milan, Italy). 1 μg of total RNA in a 20 μl reaction volume was converted into cDNA with random primers, using the Reverse Transcription System kit (Promega), following manufacturer's instructions. Gene expression levels were analyzed by real-time PCR. Primer sequences used for amplification of specific genes are shown in Table S2. Real time PCR mixture (20 µl) contained 1 μl of cDNA, 10 µl of SsoAdvanced™ universal SYBR® Green supermix, 1 µl each of primers (final concentration 0.2 µM) and 7 µl of sterile MilliQ water. Real time PCR was performed in 96 well plates on CFX96 Touch Real-Time PCR Detection System (BioRad) (95°C incubation for 60 s, followed by 40 cycles of 95°C incubation for 5 s and 58°C for 15 s). Each primer pair produced a single amplicon with a uniform melting curve. A standard curve was constructed with a series of purified PCR products and the absolute copy number of amplicons was quantified (amplification efficiency >90%, and <105%). Actin was used as a housekeeping gene for reference (Table S2). The transcription level of detected genes was calculated using the formula of 2−ΔΔCt.
Adhesion assay to human buccal epithelial cells
Adhesion ability of the mutant collection was evaluated on human buccal epithelial cells (HBECs). HBECs were collected from one healthy donor by gently rubbing the inside of the cheeks with sterile swabs, which were then suspended in 5 ml of PBS pH 7.4. Epithelial cells were washed twice in 5 ml of PBS, counted using a hemocytometer, and adjusted to a density of 1.0 × 105 cells/ml in PBS. Yeast cells were grown in 10 ml of YPD broth at 30°C overnight. A volume of 500 μl of the pre-inoculum was then inoculated in 20 ml of fresh YPD broth and incubated for a further 24 h at 30°C. At the end of the incubation period cells were collected, washed twice in PBS, and suspended in 5 ml of PBS. Culture concentration was evaluated using a hemocytometer and a suspension of 1 × 108 cells/ml was prepared. A volume of 200 μl of fungal suspension and an equal volume of HBECs suspension were mixed in sterile glass vials and co-incubated for 45 min at 37°C with gentle shaking. A negative control represented by HBECs and PBS was added in order to check the absence of yeast cells in the oral cavity. Cells were then collected by filtration onto polycarbonate filters (pore diameter of 12 µm, Millipore) and filters were then washed twice in order to remove unbound yeasts cells. Filters were air-dried and then Gram stained. Each filter was examined by a light microscope at 1000 × magnification and number of yeast adherent to 100 HBECs was counted for each C. parapsilosis analyzed strain. The adhesion index was obtained by dividing the mean number of yeast adherent to 100 HBECs by the mean number of wild type yeast adherent to 100 HBECs. The mean adhesion index obtained for 4 independent experiments was then calculated.
LDH assay
A549 human cells were grown in Dulbecco's Modified Eagle Medium (DMEM, Gibco, Monza, Italy) supplemented with 10% FBS (Aurogene, Rome, Italy), 2 mM L-glutamine (Lonza, Basel, Switzerland) and 100 U/ml penicillin-streptomycin (Lonza). Lactate dehydrogenase release (LDH) from cells into medium was monitored as a measure of cell damage using Cyto Tox 96® Non-Radioactive Cytotoxicity Assay (Promega) according to manufacturer's instructions with minor modifications. A549 cells were seeded in a 96 well polystyrene microplate (round bottom) with a density of 10,000 cells/each well. Cells were incubated for 24 h at 37°C, 5% CO2 in order to create a confluent monolayer. WT, H, KO and R strains were inoculated in YNB medium (Becton Dickinson, Milan, Italy) supplemented with 2% dextrose and incubated at 30°C overnight. We used C. albicans SC5314 strain as a positive control and it was inoculated under the same growth conditions used for C. parapsilosis strains. Following incubation, Candida cultures were washed with D-PBS, suspended in assay medium (DMEM supplemented with 2 mM L-glutamine, 0.5 % FBS) and applied to confluent A549 monolayers in a 150 μl volume containing 2.4 × 105 yeast cells. Different plate incubation times were tested (4, 6, 8 or 16 h) at 37°C and 5% CO2. Maximum LDH release was obtained after adding 15 μl of lysis solution in a control set of wells 45 minutes before the end of each incubation period. An aliquot of 50 μl of supernatant was transferred to a 96 well flat-bottom plate and analyzed with a Microplate Reader (Model 550, Bio-Rad) at a wavelength of 490 nm. The percentage of cytotoxicity was then calculated according to the manufacturer's protocol.
Galleria mellonella infection model
The Galleria mellonella larvae (Mous Livebait R.J., The Netherlands) used for experiments were selected to be similar in size (about 0.4 g) and without gray markings. To determine a suitable fungal inoculum a pilot experiment was set up. Different suspensions of yeast cells (4 × 108 cells/ml, 5 × 107 cells/ml and 1 × 107 cells/ml) were used to infect 5 larvae per group by intra-hemocelic infection. 8 × 107 cells/ml was selected as the optimal infection dose in order to monitor the progress of the infection over 10 days. Groups of 20 larvae were injected with WT, H, KO and R (lineage b); 20 larvae were not injected (untreated control) and 20 larvae were injected with an equal volume of sterile PBS. C. parapsilosis strains were grown in YPD medium overnight at 30°C. At the end of the incubation time, cells were collected by centrifugation, washed with sterile PBS (pH 7.4) and counted with a hemocytometer. Larvae were inoculated with 10 μl of a Candida suspension containing 8 × 107 cells/ml by injection in the last pro-leg using a Hamilton syringe (ga26S/51mm/pst2). Prior use and following each injection, the syringe was cleaned with 10 % hypochlorite, 70% ethanol and PBS. Before injection, the last pro-leg was washed as well with a swab soaked with 70% ethanol. Larvae were incubated at room temperature for 10 days and the number of dead larvae was scored daily.
Murine model of urinary infection
In vivo mice experiments were conducted following the ethical protocol approved by the institutional Animal Use Committee (Catholic University of the Sacred Heart, Rome, Italy). For urinary infection, cultures of the CpALS7 null mutant, wild type and reconstituted strains were grown overnight at 30°C in YPD broth (Sigma). Cells were harvested by centrifugation, washed tree times in sterile PBS, and counted by microscopic Bürker chamber. Dilutions were made in order to inoculate 50 µl of physiological solution of yeast cells (108 C. parapsilosis blastoconidia) directly in the bladder of female 10-week-old BALB/c mice (20 to 25 g) by a poly-ethylene catheter (Becton Dickinson). For each strain 17 mice were infected and the number and the vitality of inoculum were confirmed by plate counts in YPD agar (Sigma Aldrich). After 4 days, mice were sacrificed by CO2, and bladder and kidney were excised aseptically, weighed and homogenized in sterile saline by using a Stomacher 80 device (VWR International PBI, Milan, Italy) for kidneys and by a glass pestle for bladders. Organ homogenates were diluted and plated in YPD agar and incubated at 30°C. Following a 48 h incubation, colonies were counted and CFU/g tissue were calculated.
Statistical analysis
Statistical analysis of mean adhesion index data was performed using GraphPad Prism software (version 6.05 for Windows, La Jolla, CA USA). One-way ANOVA followed by Bonferroni's post-hoc test was used to evaluate differences in mean adhesion among different C. parapsilosis strains. Survival curves of G. mellonella larvae were plotted using Kaplan-Meier curve and differences in survival curves of treated larvae were evaluated by the Log-rank (Mantel-Cox) test. Statistical significance for murine experiments was determined by nonparametric Wilcoxon rank sum tests using. A P value <0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Supplementary_Material.zip
Download Zip (470.4 KB)Acknowledgments
We are thankful to Joachim Morschhäuser for providing us with the SAT1 flipper cassette and to Oscar Zaragoza for helpful advices on Galleria mellonella infection. We thank Michela Sali, Università Cattolica del Sacro Cuore, Rome, Italy, for statistical advice. The authors pay tribute to the late Professor Mario Campa for his long-standing inspiration and support.
Funding
This work was supported by the Italian Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR), research grant number RBFR100FLV.
References
- Staab JF, Sundstrom P. URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes. Trends Microbiol 2003; 11:69-73; PMID:12598128; http://dx.doi.org/10.1016/S0966-842X(02)00029-X
- Reuß O, Vik Å, Kolter R, Morschhäuser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 2004; 341:119-27; PMID:15474295; http://dx.doi.org/10.1016/j.gene.2004.06.021
- Xu Q-R, Yan L, Lv Q-Z, Zhou M, Sui X, Cao Y-B, Jiang Y-Y. Molecular genetic techniques for gene manipulation in Candida albicans. Virulence 2014; 5:507-20; PMID:24759671; http://dx.doi.org/10.4161/viru.28893
- Ko BS, Kim J, Kim JH. Production of xylitol from D-xylose by a xylitol dehydrogenase gene-disrupted mutant of Candida tropicalis. Appl Environ Microbiol 2006; 72:4207-13; PMID:16751533; http://dx.doi.org/10.1128/AEM.02699-05
- Ding C, Butler G. Development of a gene knockout system in Candida parapsilosis reveals a conserved role for BCR1 in biofilm formation. Eukaryot Cell 2007; 6:1310-9; PMID:17586721; http://dx.doi.org/10.1128/EC.00136-07
- Gácser A, Trofa D, Schäfer W, Nosanchuk JD. Targeted gene deletion in Candida parapsilosis demonstrates the role of secreted lipase in virulence. J Clin Invest 2007; 117:3049-58; PMID:17853941; http://dx.doi.org/10.1172/JCI32294
- Van Asbeck EC, Clemons KV, Stevens DA. Candida parapsilosis: a review of its epidemiology, pathogenesis, clinical aspects, typing and antimicrobial susceptibility. Crit Rev Microbiol 2009; 35:283-309; PMID:19821642; http://dx.doi.org/10.3109/10408410903213393
- Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. Candida glabrata, Candida parapsilosis and Candida tropicalis: Biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev 2012; 36:288-305; PMID:21569057; http://dx.doi.org/10.1111/j.1574-6976.2011.00278.x
- Pammi M, Holland L, Butler G, Gacser A, Bliss JM. Candida parapsilosis is a significant neonatal pathogen: a systematic review and meta-analysis. Pediatr Infect Dis J 2013; 32:e206-16; PMID:23340551; http://dx.doi.org/10.1097/INF.0b013e3182863a1c
- Klingspor L, Tortorano AM, Peman J, Willinger B, Hamal P, Sendid B, Velegraki A, Kibbler C, Meis JF, Sabino R, et al. Invasive Candida infections in surgical patients in intensive care units: a prospective, multicentre survey initiated by the European Confederation of Medical Mycology (ECMM) (2006-2008). Clin Microbiol Infect 2015; 21:87.e1-10; PMID:25636940; http://dx.doi.org/10.1016/j.cmi.2014.08.011
- Butler G, Rasmussen MD, Lin MF, Santos MAS, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009; 459:657-62; PMID:19465905; http://dx.doi.org/10.1038/nature08064
- Horváth P, Nosanchuk JD, Hamari Z, Vágvölgyi C, Gácser A. The identification of gene duplication and the role of secreted aspartyl proteinase 1 in Candida parapsilosis virulence. J Infect Dis 2012; 205:923-33; PMID:22301631; http://dx.doi.org/10.1093/infdis/jir873
- Trofa D, Soghier L, Long C, Nosanchuk JD, Gacser A, Goldman DL. A rat model of neonatal candidiasis demonstrates the importance of lipases as virulence factors for Candida albicans and Candida parapsilosis. Mycopathologia 2011; 172:169-78; PMID:21667319; http://dx.doi.org/10.1007/s11046-011-9429-3
- Nguyen LN, Cesar GV, Le GTT, Silver DL, Nimrichter L, Nosanchuk JD. Inhibition of Candida parapsilosis fatty acid synthase (Fas2) induces mitochondrial cell death in serum. PLoS Pathog 2012; 8:e1002879; PMID:22952445; http://dx.doi.org/10.1371/journal.ppat.1002879
- Sánchez-Fresneda R, Martínez-Esparza M, Maicas S, Argüelles J-C, Valentín E. In Candida parapsilosis the ATC1 gene encodes for an acid trehalase involved in trehalose hydrolysis, stress resistance and virulence. PLoS One 2014; 9:e99113; PMID:24922533; http://dx.doi.org/10.1371/journal.pone.0099113
- Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol 2001; 9:176-80; PMID:11286882; http://dx.doi.org/10.1016/S0966-842X(01)01984-9
- Zhao X, Oh SH, Cheng G, Green CB, Nuessen JA, Yeater K, Leng RP, Brown AJP, Hoyer LL. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 2004; 150:2415-28; PMID:15256583; http://dx.doi.org/10.1099/mic.0.26943-0
- Oh S-H, Cheng G, Nuessen J A, Jajko R, Yeater KM, Zhao X, Pujol C, Soll DR, Hoyer LL. Functional specificity of Candida albicans Als3p proteins and clade specificity of ALS3 alleles discriminated by the number of copies of the tandem repeat sequence in the central domain. Microbiology 2005; 151:673-81; PMID:15758214; http://dx.doi.org/10.1099/mic.0.27680-0
- Coleman DA, Oh SH, Zhao X, Zhao H, Hutchins JT, Vernachio JH, Patti JM, Hoyer LL. Monoclonal antibodies specific for Candida albicans Als3 that immunolabel fungal cells in vitro and in vivo and block adhesion to host surfaces. J Microbiol Methods 2009; 78:71-8; PMID:19427882; http://dx.doi.org/10.1016/j.mimet.2009.05.002
- Liu Y, Filler SG. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell 2011; 10:168-73; PMID:21115738; http://dx.doi.org/10.1128/EC.00279-10
- García-Rodas R, Casadevall A, Rodríguez-Tudela JL, Cuenca-Estrella M, Zaragoza O. Cryptococcus neoformans capsular enlargement and cellular gigantism during Galleria mellonella infection. PLoS One 2011; 6:1-12; PMID:21915338
- Gago S, García-Rodas R, Cuesta I, Mellado E, Alastruey-Izquierdo A. Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis virulence in the non-conventional host Galleria mellonella. Virulence 2014; 5:278-85; PMID:24193303; http://dx.doi.org/10.4161/viru.26973
- Torelli R, Serror P, Bugli F, Paroni Sterbini F, Florio AR, Stringaro A, Colone M, De Carolis E, Martini C, Giard JC, et al. The PavA-like fibronectin-binding protein of Enterococcus faecalis, EfbA, Is important for virulence in a mouse model of ascending urinary tract infection. J Infect Dis 2012; 206:952-60; PMID:22782954; http://dx.doi.org/10.1093/infdis/jis440
- Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: New developments in the family and domain prediction database. Nucleic Acids Res 2012; 40:306-12; PMID:22096229; http://dx.doi.org/10.1093/nar/gkr948
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30:2725-9; PMID:24132122; http://dx.doi.org/10.1093/molbev/mst197
- Jorda J, Kajava A V. T-REKS: identification of Tandem REpeats in sequences with a K-meanS based algorithm. Bioinformatics 2009; 25:2632-8; PMID:19671691; http://dx.doi.org/10.1093/bioinformatics/btp482
- Heilmann CJ, Sorgo AG, Mohammadi S, Sosinska GJ, de Koster CG, Brul S, de Koning LJ, Klis FM. Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans. Eukaryot Cell 2013; 12:254-64; PMID:23243062; http://dx.doi.org/10.1128/EC.00278-12
- Kopecka M, Gabriel M. The influence of congo red on the cell wall and (1—-3)-beta-D-glucan microfibril biogenesis in Saccharomyces cerevisiae. Arch Microbiol 1992; 158:115-26; PMID:1417414; http://dx.doi.org/10.1007/BF00245214
- Sorgo AG, Heilmann CJ, Dekker HL, Bekker M, Brul S, de Koster CG, de Koning LJ, Klis FM. Effects of fluconazole on the secretome, the wall proteome, and wall integrity of the clinical fungus Candida albicans. Eukaryot Cell 2011; 10:1071-81; PMID:21622905; http://dx.doi.org/10.1128/EC.05011-11
- Abi-Chacra ÉA, Souza LOP, Cruz LP, Braga-Silva LA, Gonçalves DS, Sodré CL, Ribeiro MD, Seabra SH, Figueiredo-Carvalho MHG, Barbedo LS, et al. Phenotypical properties associated with virulence from clinical isolates belonging to the Candida parapsilosis complex. FEMS Yeast Res 2013; 13:831-48; PMID:24103069; http://dx.doi.org/10.1111/1567-1364.12092
- Falgier C, Kegley S, Podgorski H, Heisel T, Storey K, Bendel CM, Gale CA. Candida species differ in their interactions with immature human gastrointestinal epithelial cells. Pediatr Res 2011; 69:384-9; PMID:21283049; http://dx.doi.org/10.1203/PDR.0b013e31821269d5
- Bertini A, De Bernardis F, Hensgens LAM, Sandini S, Senesi S, Tavanti A. Comparison of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis adhesive properties and pathogenicity. Int J Med Microbiol 2013; 303:98-103; PMID:23403338; http://dx.doi.org/10.1016/j.ijmm.2012.12.006
- Jacobsen ID. Galleria mellonella as a model host to study virulence of Candida. Virulence 2014; 5:237-9; PMID:24384470; http://dx.doi.org/10.4161/viru.27434
- Maestre-Reyna M, Diderrich R, Veelders MS, Eulenburg G, Kalugin V, Brückner S, Keller P, Rupp S, Mösch HU, Essen LO. Structural basis for promiscuity and specificity during Candida glabrata invasion of host epithelia. Proc Natl Acad Sci 2012; 109:16864-9; PMID:23035251; http://dx.doi.org/10.1073/pnas.1207653109
- Murciano C, Moyes D, Runglall M, Tobouti P. Evaluation of the role of Candida albicans agglutinin-like sequence (Als) proteins in human oral epithelial cell interactions. PLoS One 2012; 7:e33362; PMID:22428031; http://dx.doi.org/10.1371/journal.pone.0033362
- Lin J, Oh S-H, Jones R, Garnett JA, Salgado PS, Rusnakova S, Matthews SJ, Hoyer LL, Cota E. The peptide-binding cavity is essential for Als3-mediated adhesion of Candida albicans to human cells. J Biol Chem 2014; 289:18401-12; PMID:24802757; http://dx.doi.org/10.1074/jbc.M114.547877
- Hoyer LL, Oh S-H, Jones R, Cota E. A proposed mechanism for the interaction between the Candida albicans Als3 adhesin and streptococcal cell wall proteins. Front Microbiol 2014; 5:564; PMID:25408685; http://dx.doi.org/10.3389/fmicb.2014.00564
- Machado AG, Komiyama EY, Santos SSF Dos, Jorge AOC, Brighenti FL, Koga-Ito CY. In vitro adherence of Candida albicans isolated from patients with chronic periodontitis. J Appl Oral Sci 2011; 19:384-7; PMID:21710096; http://dx.doi.org/10.1590/S1678-77572011005000014
- Silvia S, Annarita S, Silvia A, Marisa C. The MP65 gene is required for cell wall integrity, adherence to epithelial cells and biofilm formation in Candida albicans. BMC Microbiol 2011; 11:106; PMID:21575184; http://dx.doi.org/10.1186/1471-2180-11-106
- Chaves GM, Bates S, Maccallum DM, Odds FC. Candida albicans GRX2, encoding a putative glutaredoxin, is required for virulence in a murine model. Genet Mol Res 2007; 6:1051-63; PMID:18273798
- Tavanti A, Hensgens LAM, Mogavero S, Majoros L, Senesi S, Campa M. Genotypic and phenotypic properties of Candida parapsilosis sensu strictu strains isolated from different geographic regions and body sites. BMC Microbiol 2010; 10:203; PMID:20667137; http://dx.doi.org/10.1186/1471-2180-10-203
- Dalle F, Wächtler B, L'Ollivier C, Holland G, Bannert N, Wilson D, Labruère C, Bonnin A, Hube B. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell Microbiol 2010; 12:248-71; PMID:19863559; http://dx.doi.org/10.1111/j.1462-5822.2009.01394.x
- Sambrook J, Russell DW. Preparation and transformation of competent E. coli using calcium chloride. CSH pdb.prot 3932; 2006; PMID:22485377
- Hall RA, Bates S, Lenardon MD, Maccallum DM, Wagener J, Lowman DW, Kruppa MD, Williams DL, Odds FC, Brown AJP, et al. The Mnn2 mannosyltransferase family modulates mannoprotein fibril length, immune recognition and virulence of Candida albicans. PLoS Pathog 2013; 9:e1003276; PMID:23633946; http://dx.doi.org/10.1371/journal.ppat.1003276
