Abstract
The interaction between the fungal pathogen Paracoccidioides brasiliensis and host cells is usually mediated by specific binding events between adhesins on the fungal surface and receptors on the host extracellular matrix or cell surface. One molecule implicated in the P. brasiliensis-host interaction is the 14-3-3 protein. The 14-3-3 protein belongs to a family of conserved regulatory molecules that are expressed in all eukaryotic cells and are involved in diverse cellular functions. Here, we investigated the relevance of the 14-3-3 protein to the virulence of P. brasiliensis. Using antisense RNA technology and Agrobacterium tumefaciens-mediated transformation, we generated a 14-3-3-silenced strain (expression reduced by ˜55%). This strain allowed us to investigate the interaction between 14-3-3 and the host and to correlate the functions of P. brasiliensis 14-3-3 with cellular features, such as morphological characteristics and virulence, that are important for pathogenesis.
Introduction
Paracoccidioidomycosis (PCM) is a systemic mycosis caused by fungi of the Paracoccidioides species complex (P. brasiliensis and P. lutzii). PCM is important due to the frequency and severity of the cases.Citation1 It is geographically confined to Latin America, with more than 10 million people possibly infected in this continent,Citation2 and the highest incidence occurs in Brazil (80%).Citation3,4
Fungi from the Paracoccidioides genus are thermal dimorphic fungi that grow both in a yeast form (at approximately 37°C) and as mycelia (below 25°C). Infection occurs mainly by inhalation of airborne propagules released by the mycelial form; once in the lungs and in contact with the host body temperature, it transforms to its yeast form, which is responsible for the symptoms and clinical manifestations during disease.Citation5,6
The development and the course of the microbial infection can be considered a multifactorial process that results from the interaction between the virulence factors of pathogen, leading to the pathogen's establishment in the host and causing disease, and the host's ability to prevent and resist microbial colonization or invasion.Citation7-10
The pathogen uses a large repertoire of surface molecules, specifically adhesins, that can bind to the extracellular matrix (ECM) of various cell types in the host, inducing endocytosis in normally non-phagocytic cells (epithelial and endothelial cells) during the invasion process.Citation11,12 This interaction with the ECM has been correlated with the processes of adhesion and invasion. The ECM is composed of components such as collagen, laminin, fibronectin, vitronectin and proteoglycans, which participate in the regulation of physiological processes including cell migration, signaling and the transport of solutes across cellular barriers. The respiratory tract, the primary focus of PCM, is composed of tissues rich in ECM, such as laminin and various types of collagen. Moreover, alveolar fibroblasts in the lungs secrete ECM components.Citation12-14
A required step for colonization and ultimately the development of disease is the ability of pathogens to adhere to host surfaces. The ability to adhere is a biological phenomenon that is widely distributed and shared by many pathogens, enabling the colonization of their respective habitats.Citation15,16 Some adhesins have been described for P. brasiliensis, such as gp43,Citation17,18 glyceraldehyde-3-phosphate dehydrogenase,Citation19 triose phosphate isomerase,Citation20 enolase,Citation21,22 a 32-Kda hydrolase,Citation23,24 malate synthase,Citation25 isocitrate lyase Citation26 and the 14-3-3 protein (30 KDa) Citation27,28; this last adhesin is the focus of the present study.
Strong evidence has resulted in the classification of the 14-3-3 protein of P. brasiliensis as an adhesin, including its ability to bind laminin and its association with virulent isolates of the fungus.Citation27 Additionally, immunocytochemical studies using both in vitro and in vivo models have demonstrated the ubiquitous distribution (cytosol and cell membrane) of the 14-3-3 protein in the yeast form of P. brasiliensis. A significant increase in the amount of 14-3-3 was observed in the fungal cell wall during interaction with pneumocytes and in an experimental acute infection in mice. Inhibition assays using the recombinant 14-3-3 protein and anti-14-3-3 antibody to study the interaction of P. brasiliensis with pneumocytes reported a variation in the rate of adherence at different stages during infection, suggesting an important role for 14-3-3 in fungus-pathogen interactions.Citation28
The aim of this study was to develop an isolate of P. brasiliensis with low expression of the 14-3-3 protein using antisense RNA technology. We used transformation mediated by Agrobacterium tumefaciens to elucidate the role of this protein in P. brasiliensis pathogenesis by investigating its biological function and involvement in virulence. These factors have not previously been described for this fungus.
Results
Knockdown of Pb14-3-3 expression
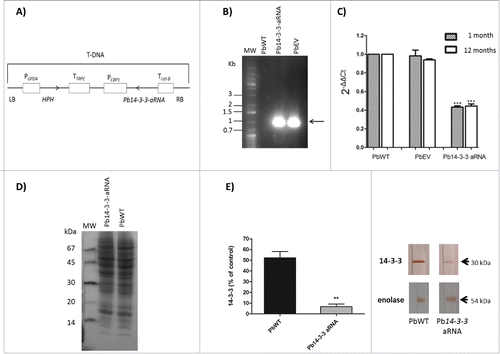
Using antisense (aRNA) technology and A. tumefaciens-mediated transformation (ATMT), we constructed an antisense RNA targeting the exon-coding sequence of the Pb14-3-3 gene to study the role of 14-3-3 in P. brasiliensis (). We confirmed the integration of the hygromycin cassette into the genomic DNA of one hygromycin-resistant transformant (). To pursue our main goals in this work, we selected a mitotically stable isolate with the highest decrease in Pb14-3-3 expression (˜55%) confirmed by RT-qPCR. The reduction in the transcript levels of Pb14-3-3 (band size of 30 kDa) was confirmed even after 12 months of successive subcultures of Pb14-3-3 aRNA (). Next, SDS-PAGE under reducing conditions and immunoblotting () was performed with monoclonal anti-14-3-3 antiserum and polyclonal anti-enolase (as an internal control, band of 54 kDa) (). The Pb14-3-3 aRNA strain expressed noticeably reduced levels of the 14-3-3 protein.
Figure 1. Inhibition of Pb14-3-3 expression using aRNA and A. tumefaciens-mediated transformation. (A) tDNA (T-DNA) inserted into P. brasiliensis by ATMT to silence Pb14-3-3. The anti-sense oligonucleotides were produced based on the Pb18 (PbWT) genomic sequence, as detailed in the Materials and Methods section. The sequence was cloned under the control of CBP-1 promoter (calcium-binding protein). The construct was sub-cloned into T-DNA region of the binary vector pUR5750 harboring hygromycin B phosphotransferase (HPH) driven by the glyceraldehyde-3-phosphate of Aspergillus nidulans (PGPDA) and with terminator (TTRPC) as described previously.Citation31 (B) PCR fragments amplified with an HPH-specific primer yielded a 1,000 bp internal fragment. Pb14-3-3 aRNA, PbWT and PbEV were used as the templates. MW, DNA molecular marker. C) Pb14-3-3 expression levels in PbWT, PbEV and Pb14-3-3 aRNA after 1 month and over 12 months of subculture. Gene expression levels obtained by RT-qPCR were normalized to the internal control TUB2; ***p < 0.0001 when compared with PbWT and PbEV; D) SDS-PAGE gel of total proteins of PbWT and Pb14-3-3 aRNA; E) Immunoblots of the total proteins tested with the anti-14-3-3 monoclonal antibody and anti-enolase (used as an internal control) and quantified using ImageJ software; p = 0.0021.
Pb14-3-3 silencing affects yeast cell morphology and morphological transition without affecting cell viability or fungal growth
The variability of budding of the yeast cells is shown in . PbWT and PbEV were not significantly different, with approximately 83% and 81% of yeast cells showing buds, respectively. The observed mean number of buds per yeast cell was 2.23 buds/yeast cell for PbWT and 1.82 buds/yeast cell for PbEV. In contrast, after knockdown with Pb14-3-3 aRNA, 67% of yeast cells possessed buds, with 0.95 buds/yeast cell, a significant reduction compared to the other isolates ().
Table 1. Percentage of budding cells and number of buds by cell exhibited by the isolates.
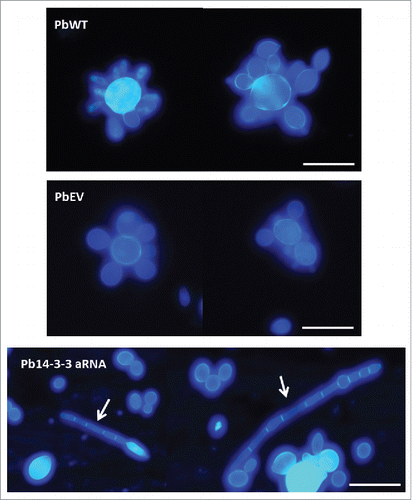
The estimated diameters are depicted in . Pb14-3-3 aRNA yeast cells were approximately 5 μm, significantly smaller than PbWT and PbEV, which averaged 9.19 μm and 8.67 μm in diameter, respectively. Microscopic observations indicated that some yeast cells (12%) of Pb14-3-3 aRNA were more elongated and filamentous than the wild-type cells and cells harboring the empty vector, but this was not the predominant phenotype ().
Figure 2. Knocking-down Pb14-3-3 in P. brasiliensis alters yeast cell size. Analysis of major diameter size of mother cells of PbWT, PbEV and Pb14-3-3 aRNA isolates. (***p < 0.0001); n = 100 mother cells.

Figure 3. Silencing of Pb14-3-3 leads to distinct P. brasiliensis yeast cell morphology. Microscopic evaluation of PbWT and PbEV yeast cells and yeast cells generated from the Pb14-3-3 aRNA strain using CalcoFluor; magnification 40X. White bars correspond to 20 μm. White arrows indicate yeast cells showing elongation and the presence of filamentation.
Coupled with the previous evidence, Pb14-3-3 silencing affected the fungal transition. To test the effect of Pb14-3-3 on fungal dimorphism, cells in each phase, yeast and mycelium, were incubated at 25°C and 37°C, respectively, until 100% conversion was observed.
At 25°C, after 24 h, PbWT, PbEV and Pb14-3-3 aRNA did not show differences, and we observed that approximately 15–20% of cells showed budding with neck elongation (transforming yeast). By 48 h, the percentage of transforming yeasts increased (to approximately 34.6–48.1%), and chlamydospore-like cells (differentiating hyphae) were present in all isolates PbWT (13.3%), PbEV (10%) and Pb14-3-3 aRNA (7.1%); however, at this time, hyphae were found only in PbWT (5%) and PbEV (2%). At 72 h, the majority of cells of PbWT (68%) and PbEV (58%) were hyphae, while those with Pb14-3-3 aRNA were yeast. Incubation for 120 h was sufficient to obtain 100% transition to mycelium for PbWT and PbEV. For the Pb14-3-3 aRNA isolate, this transition occurred gradually and slowly, requiring 10 days for completing the yeast-mycelium transition ().
Figure 4. Influence of Pb14-3-3 in the temperature morphological transition. Mycelia or yeast were induced to transform by changing the incubation temperature from 25°C to 37°C or from 37°C to 25°C. Morphological units were classified as described by Nunes et al., 2005Citation83: (A) Yeast (multiple budding), (B) Transforming yeast (yeast with neck elongation), (C) Differentiating hyphae (chlamydospore-like cells) and (D) hyphae. (E) Yeast-Mycelium transition at 25°C. (F) Mycelium-Yeast transition at 37°C. A total of 300 cells were counted at each time, and the results are shown as the mean.
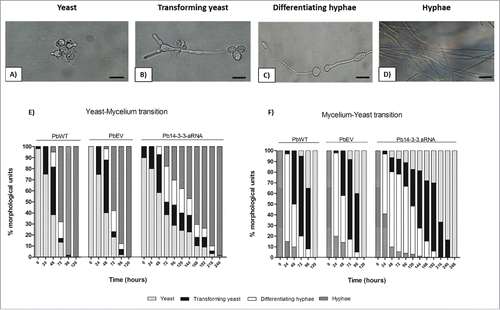
At 37°C, the same test was performed to verify the mycelium-yeast transition. After 24 h, most of the hyphae (58–82%) became enlarged, indicating the start of differentiation (differentiating hyphae) for all isolates. However, by 48 h of incubation, the Pb14-3-3 aRNA isolate showed impairment of the mycelium-yeast transition, as determined by the slight growth of transforming yeasts (13.3%) when compared to PbWT (50%) and PbEV (42%). While PbWT and PbEV advanced in the mycelium-yeast transition, achieving complete mycelium-yeast transition within 5 days, the silenced isolate with Pb14-3-3 aRNA required 11 days to complete the same process ().
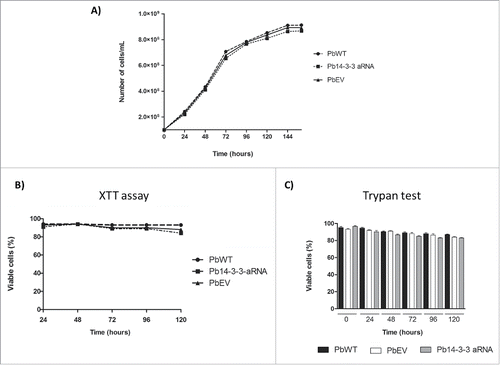
The decrease in expression of Pb14-3-3 did not alter the yeast cell vitality or viability. Moreover, no significant differences between the Pb14-3-3 aRNA strain and the control were detected during batch culture growth of yeast cells (). We also studied the viability of Pb14-3-3 aRNA, PbWT and PbEV yeast cells at different time points during batch culture using XTT and Trypan blue exclusion assays, calculating the percentage of live cells in each test strain based on the OD measured from the heat-killed P. brasiliensis, used as dead control cells, and no major differences were found in either viability test ().
Figure 5. Down-regulation of Pb14-3-3 does not affect cell viability or vitality of P. brasiliensis yeast cell. P. brasiliensis isolates PbWT, PbEV and Pb14-3-3 aRNA were evaluated by (A) cell counts in a Neubauer chamber, (B) a colorimetric XTT reduction assay and (C) Trypan blue exclusion test at different time points.
Differential interaction kinetics of Pb14-3-3 aRNA with pneumocytes
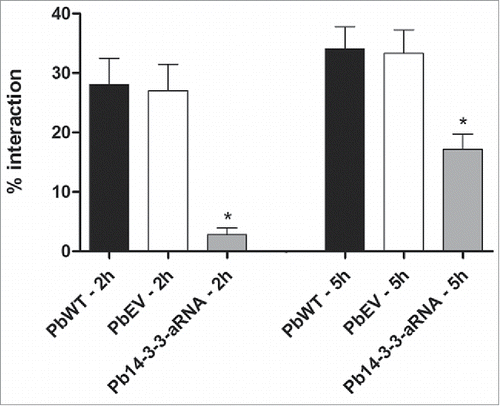
To elucidate the role of Pb14-3-3 in the interaction (adhesion/internalization) of P. brasiliensis with host cells, we infected the A549 epithelial human lung cell line with PbWT, PbEV and Pb14-3-3 aRNA strain yeast cells. After 2 h of interaction, the interaction capacity of Pb14-3-3 aRNA yeast cells was 10-fold decreased compared with PbWT. It was also possible to observe 2-fold decrease in the Pb14-3-3 aRNA interaction rate after 5 h of incubation when compared with PbWT, suggesting the involvement of Pb14-3-3 in interactions between P. brasiliensis and host cells. Yeast cells of the PbWT and PbEV strains showed no changes in interaction rates at the evaluated times ().
Figure 6. Interaction of Pb14-3-3 aRNA yeast cells with pneumocytes is affected during the early stage of infection. The interaction was assessed by indirect immunofluorescence and analyzed by flow cytometry at different time points. *p < 0.05 when comparing PbWT and PbEV with Pb14-3-3 aRNA at 2 and 5 hours after infection.
14-3-3 in P. brasiliensis is important for binding to ECM proteins
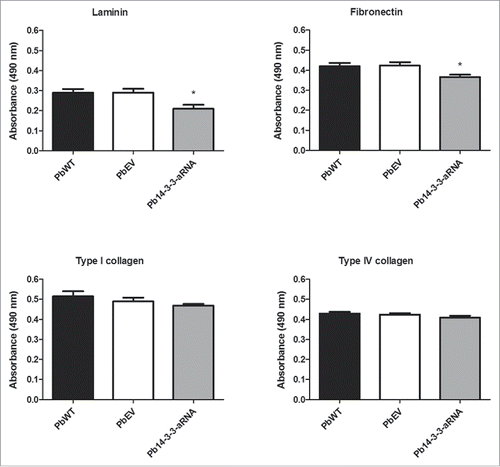
The influence of Pb14-3-3 on binding to ECM proteins was evaluated using an ELISA with an anti-cell free antibody that recognized the isolates equally (data not shown). Pb14-3-3 aRNA cells showed a significant reduction in binding to laminin (28%) and fibronectin (14%) when compared to PbWT. No differences were observed regarding the adhesion of the strains to type I and type IV collagen ().
Figure 7. Binding of the Pb14-3-3 mutant to ECM is reduced compared to the PbWT. Binding yeast cells of the strains PbWT, PbEV and Pb14-3-3 aRNA to the ECM components laminin, fibronectin, and type I and type IV collagen (10 µg mL−1). The interaction was determined by ELISA with peroxidase. *p < 0.05 when Pb14-3-3 aRNA was compared to PbWT and PbEV for laminin and fibronectin.
14-3-3 is an important virulence factor for P. brasiliensis
The relevance of Pb14-3-3 during the host-pathogen interaction was further evaluated using a G. mellonella infection model. To evaluate this hypothesis, the isolates PbWT, PbEV and Pb14-3-3 aRNA (5 × 106 cells/larva) were used to infect G. mellonella larvae that were maintained at 37°C, and the virulence of these fungi was evaluated for 7 days. Comparing the percentage of survival at the end of the experimental for all strains tested demonstrated that larvae infected with the Pb18 strain (PbWT) began to die at day 3, with no surviving larvae at the end of the experimental period. In contrast, larvae infected with the Pb14-3-3 aRNA strain survived longer: the larvae began to die at day 3, but the survival percentage of larvae infected with Pb14-3-3 aRNA was significantly (p < 0.0001) higher (35% survival) than for PbWT and PbEV at the end of the 7-day experiment (). No significant difference was observed between PbEV and PbWT, and no mortality was observed in the PBS-injected G. mellonella larvae (). Additionally, a control group inoculated with heat-killed fungal cells showed 100% survival, indicating that the larvae did not die from osmotic stress.
Figure 8. Silencing of Pb14-3-3 decreases the virulence of P. brasiliensis in a Galleria mellonella infection model. (A) Representative survival curves of an experimental infection performed in G. mellonella with PBS; 5×106 PbWT, PbEV, Pb14-3-3 aRNA and heat-killed yeast cells; p < 0.0001 when Pb14-3-3 aRNA were compared with PbWT and PbEV and B) Number of CFUs in larvae sacrificed and homogenized at 0, 24 and 48 h after infection (*p < 0.05 when compared Pb14-3-3 aRNA with PbWT and PbEV).
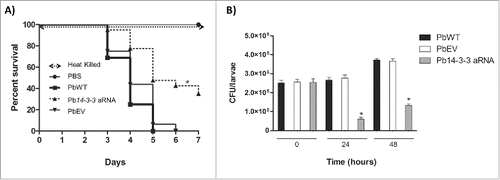
To compare the fungal replication kinetics over the course of the infection of wild-type (PbWT) and the silenced strain (Pb14-3-3 aRNA), the numbers of colony-forming units (CFUs) per larva were evaluated. At time zero, the isolates showed no differences based on CFU counting (). After 24 h of infection, PbWT and PbEV showed no differences in CFUs compared with the initial time, but for Pb14-3-3 aRNA, the number of CFUs was significantly lower than that for PbWT (˜4.4-fold lower) and PbEV (˜4.5-fold lower) (). By 48 h of infection, PbWT, PbEV and Pb14-3-3 aRNA underwent replication, evidenced by the increase in the fungal burden. Nevertheless the silenced isolate, Pb14-3-3 aRNA, was also observed to have significantly fewer CFUs than PbWT (˜2.8-fold lower) and PbEV (˜2.7-fold lower) despite showing an increase in CFUs compared to the 24 h time point (), supporting the results of the survival curve.
Discussion
Antisense RNA silencing has proven to be a strong molecular biological tool for studying the functions of cellular genes and identifying the genetic factors involved in the virulence and the dimorphic nature of P. brasiliensis.Citation29-31
Previous studies focused on elucidating the function of the 14-3-3 protein in the pathogenesis of P. brasiliensis.Citation27,28 Those studies were limited to the use of the recombinant 14-3-3 protein and the anti-14-3-3 antibody, and they therefore indirectly explored the role of 14-3-3 without manipulating gene expression. This is the first report to use a strain of P. brasiliensis with reduced transcript and protein levels of 14-3-3 to elucidate the role of this protein in host-pathogen interactions and in the biology and virulence of P. brasiliensis.
The Pb14-3-3 aRNA-silenced strain of P. brasiliensis was used in all of the experiments, with PbWT and PbEV serving as controls. The reductions in both gene and protein expression levels in the Pb14-3-3 aRNA strain were confirmed prior to its experimental use. The silenced strain demonstrated a yeast cell viability and growth rate similar to the controls. Based on these results, we can hypothesize that this protein is not important for fungal metabolism. In contrast, our results suggest a role in fungal adherence with an effect on virulence.
Interestingly, the decreased expression of the Pb14-3-3 gene affected the morphology of yeast cells maintained at 37°C, resulting in smaller yeast cells, fewer buds per yeast cell and the presence of a few yeast cells with a more elongated morphology, including the presence of filamentation. The report by Peraçoli et al. in 1999Citation32 demonstrated that the isolate Pb-T, recently isolated from armadillos and more virulent than Pb18, showed a significantly higher number of buds per mother cell than did Pb18. Likewise, the silenced isolate Pb14-3-3 aRNA, with fewer budding yeast cells, proved to be less virulent than Pb18 (PbWT). These findings support the previous hypothesis, that the most virulent isolate has more buds, but evaluating the contribution of morphological differences to disease progression requires further investigation of the polymorphisms present in P. brasiliensis.Citation33
Other evidence pointing to the possible involvement of the 14-3-3 protein in P. brasiliensis morphology includes the findings that both the yeast-to-mycelium transition and the reverse transition were hampered in the isolate with reduced expression of this protein. This suggests that Pb14-3-3 is important for pathogenicity that its closely associated with dimorphic transition: P. brasiliensis mycelium of strains that are unable to transform into yeast are not virulentCitation34,35 because the yeast form is more resistant to the killing mechanisms of various host cells, thereby contributing to the survival of the pathogen in this environment.Citation36
Previous studies with dimorphic fungi such as P. brasiliensis have demonstrated that these fungi require their morphological transitions for efficient adherence and invasion of the host to establish infection.Citation37 Morphological transitions in dimorphic fungi are highly complex events triggered by a wide range of environmental factors and consisting of a reversible change in the growth pattern, alternating between the unicellular yeast form and the filamentous form (hyphae and pseudohyphae).Citation38 Understanding the mechanisms that regulate these events is of extreme interest due to implications for pathogenesis and cell differentiation, as observed with the inhibition of the transition to the parasitic form by the female hormone estrogen.Citation39 The proteins Bmph1p and Bmph2p in Saccharomyces cerevisiae are homologous to Pb14-3-3 and regulate pseudohyphal growth through the MAPK (mitogen-activated protein kinase) signaling pathway. This interaction has been shown to regulate fungal morphogenesis.Citation40 Further, Kraus et al. Citation41 demonstrated that the ArtA protein of Aspergillus nidulans, which also has high homology to the Pb14-3-3 protein, is involved in preventing septum formation.
Additionally, 14-3-3 has been shown to be involved in the filamentation of other dimorphic fungi such as Candida albicans Citation42 and Yarrowia lipolytica. In these microorganisms, the filamentous growth is proportional to the level of expression of the 14-3-3 protein.Citation38
Studies have shown that the variability of cell size within a fungal cell population alters pathogenicity via altering phagocytosis.Citation43,44 Phagocytosis, along with the resultant killing by macrophages and monocytes in the lungs, is one of the first lines of defense by the host immune system,Citation45 thus demonstrating that in P. brasiliensis, a decrease in cell size leads to increased phagocytosis and decreased virulenceCitation44.
The evidence presented in this study demonstrated that reduced Pb14-3-3 expression induced morphological alterations, as change in size, which may affect the ability of host cells to phagocytose the fungus, suggesting that this protein plays an important role in the yeast form in P. brasiliensis, thereby contributing to its virulence. However, the specific mechanism by which this gene silencing was able to generate morphological changes was not elucidated in the present study and requires further investigation.
Many molecular interaction events and the subsequent establishment of infection that occurs between the pathogenic fungi and host are mediated by adhesion proteins on the fungal surface that are capable of binding to the host cell's ECM.Citation46 Mendes-Giannini et al.Citation47 observed a relationship between virulence and adherence to the ECM when comparing several P. brasiliensis isolates that showed different levels of virulence in vivo; the authors demonstrated that the isolate Pb18, which was recently isolated from an animal, had a greater ability to adhere to ECM components. Confirming the results of Andreotti et al., 2005,Citation27 we demonstrated that low expression of Pb14-3-3 resulted in a reduction of binding to laminin; this interaction was assessed by ELISA, which has greater sensitivity and specificity than the immunoblotting that was performed in the study cited above. Additionally, the Pb14-3-3 protein has been reported to interact with other ECM components, including fibronectin, suggesting that Pb14-3-3 is likely involved in the virulence of P. brasiliensis through interaction with ECM components. This interaction represents a critical step in the adherence and invasion of host tissues. For collagen type I and IV, we found no differences in the binding of isolates, suggesting that this component of the ECM is not relevant to the interaction between Pb14-3-3 and host cells.
P. brasiliensis uses a sequence of different mechanisms to establish successful infections, beginning with the first contact with host cells and extending through the later stages of disease.Citation48,49
The reduced initial adhesion observed in this study suggests that the Pb14-3-3 gene products strongly influence the initial adhesion, as reported by Andreotti et al., 2005,Citation27 Da Silva et al., 2013 Citation28 and De Oliveira et al., 2015,Citation50 but the requirement for these proteins was bypassed following extended contact with the host cell.
Together, these data suggest that the adhesion of P. brasiliensis to pneumocytes relies on a combination of factors, including the expression of different adhesins, and that the defects elicited by the silencing of a single gene, as shown herein for Pb14-3-3, are offset by prolonged contact with host cells.
The huge armamentarium of virulence factors is controlled by an extremely complicated regulatory network that coordinates and synchronizes all of the elements involved.Citation51 This regulation is important not only from the perspective of guaranteeing the expression of individual virulence elements but also to confer cross-talk between these determinants to ensure the appropriate response of the fungus in which all of the stages are activated following a temporal hierarchy.
To verify the importance of the Pb14-3-3 protein to the virulence of P. brasiliensis, we used the invertebrate infection model Galleria mellonella. Several studies have established G. mellonella as a useful model for evaluating fungal virulence,Citation52-56 including for P. lutzii, another species belonging to the Paracoccidioides species complex,Citation57 with the advantage that the innate immune response is highly similar between insects and mammals, Citation58,59 Citation60 including antimicrobial peptides and cells with phagocytic activity in hemolymph that function in a manner similar to human phagocytes.Citation61-63
Decreased expression of Pb14-3-3 resulted in a significant reduction of the virulence phenotype as reflected in the survival curve and CFU experiment with G. mellonella model. There was a reduction in the fungal burden in the larvae infected with the silenced strain, probably due to difficulty in the establishment in the host, leading to a lower resistance to killing by the host's immune system. Therefore, the silenced isolate was considered less virulent than Pb18 (PbWT), supporting the hypothesis that Pb14-3-3 is involved in the pathogenicity of P. brasiliensis.
A panel of mutants of the 14-3-3 protein (Bmh1p) of C. albicans was used to verify that specific mutations affect different pathways associated with virulence, including those involved with the formation of filaments and interactions with cells of the host immune system.Citation64 The involvement of the 14-3-3 protein has recently been reported to modulate the TLR (toll-like receptor) signaling pathway. It was demonstrated that 14-3-3 impairs TLR2, TLR3, TLR4, TLR7/8, and TLR9 ligand-induced IL-6, TNFα, and IFN-β production, thereby playing a major regulatory role in balancing the host inflammatory response to viral and bacterial infections by modulating the TLR signaling pathway.Citation65 Toll-like receptors are a group of pattern-recognition receptors that play a crucial role in the induction of innate immunity, the first line of defense against bacteria, viruses and fungi.Citation66 Roles for TLR2 and TLR4 in the recognition and internalization of P. brasiliensis have been reported in human monocytes and neutrophils.Citation67 Considering that the 14-3-3 protein of P. brasiliensis is more abundant at the cell surface when in contact with the host (da Silva et al., 2013), we suggest that decreased levels of this protein results in the reduced interaction of the fungus with host structures and lead to a decreased ability to evade host responses, possibly due to the modulation of the initial immune response, thereby resulting in the reduced virulence of Pb14-3-3 aRNA.
The characterization of virulence factors is essential for improving our understanding of fungal pathogenesis and of the interactions between fungal pathogens and the host. These studies may also serve as a baseline for exploring the development of prophylactics or new therapeutic strategies. This study established that Pb14-3-3 silencing differentially regulated important of the interaction with the host cells, such as morphology, the attachment index for ECM/pneumocytes and virulence. Therefore, 14-3-3 is an interesting therapeutic target for combating infection and modulating inflammatory conditions associated with PCM due to P. brasiliensis.
Materials and Methods
Microorganisms and culture media
For this study we used yeast cells of P. brasiliensis, strain Pb18 (referred to herein as PbWT, wild- type). The fungus was grown in BHI medium supplemented with 1% glucose (Beckton Dickinson and Company, Sparks, MD) at 37°C with aeration in a mechanical shaker and was collected during the early exponential phase (72–96 h). The isolate Pb18 was used in the genetic manipulation; this study is the first to address the transformation of the isolate Pb18.
The Agrobacterium tumefaciens strain LBA1100Citation68 was used to carry the binary vector used in this study; recombinant A. tumefaciens containing the binary vector pUR5750 (conferring kanamycin resistance in A. tumefaciens and Escherichia coli) was maintained at 28°C in LB medium supplemented with 100 µg mL−1 kanamycin.
E. coli DH5α grown at 37°C on LB medium supplemented with 0 µg mL−1 kanamycin was used as host for plasmid propagation and cloning.Citation69
Obtaining P. brasiliensis with 14-3-3 silencing
We followed the anti-sense strategy and A. tumefaciens-mediated transformation protocol described previously by Almeida et al. Citation70 and Ruiz et al. Citation71 to obtain P. brasiliensis mutants with silencing of Pb14-3-3. DNA from Pb18 was extracted from yeast cultures during exponential growth, and a high-fidelity proofreading DNA polymerase (Taq DNA Polymerase High-Fidelity, Invitrogen, USA) was employed to amplify an aRNA oligonucleotide sequence targeting the exon coding sequence of Pb14-3-3 (GenBank accession number AY462124); the oligonucleotide primers used are Pb14-3-3 Forward (5′- CCGCTCGAGCGGAAGGGGGACTACCACCGCT-3′) and Pb14-3-3 Reverse (5′-GGCGCGCCCTGAGCAACCTCAGTTGCAT-3′).
The Pb14-3-3 fragment was inserted into the pCR35 plasmid under the control of the promoter region of the calcium-binding protein gene (CBP-1) from Histoplasma capsulatum Citation72 and propagated in E. coli DH5α.Citation69 The CBP-1 promoter-AS cassette was subcloned into the pUR5750 plasmid (harboring a hygromycin B phosphotransferase gene), allowing the parental binary vector to harbor the transferred DNA (T-DNA) with the antisense molecule. Pb18 yeasts transformed with an empty vector (PbEV) were used as a Pb18 recombination control as previously described.Citation24,44 After transformation, the cells were spread onto selective BHI media supplemented with 200 µg mL−1 hygromycin B (Sigma-Aldrich). Prior to incubation, the plates were air-dried in a safety cabinet for 30 min. The selection plates were monitored for colony transformation and for colony-forming ability at 36°C for 15–20 days.Citation44 Randomly selected hygromycin-resistant transformants were tested for mitotic stability.
Molecular detection of the hygromycin resistance gene (HPH)
Genomic DNA from PbWT, PbEV and Pb14-3-3 aRNA yeast cells were isolated according to the glass beads protocol described by van Burik et al.,.Citation73 To confirm the presence of the hygromycin B resistance cassette, PCR (polymerase chain reaction) amplification was performed to detect the HPH 1000-bp amplification product using oligonucleotides primers hphF (5′-AACTCACCGCGACGTCTGTCGA-3′) and hphR (5′-CTACACAGCCATCGGTCCAGA-3′). The PCR cycling temperatures were as follows: 94°C for 10 min, 40 cycles of 15 s at 94°C, 1 min each at 56°C and 65°C and finally one cycle at 65°C for 10 min. The reaction products were analyzed using 1% agarose gels and visualized with GelRed under UV light.
Gene expression analysis
Total RNA was obtained from PbWT, PbEV and Pb14-3-3 aRNA yeast cells using the TRIzol® reagent (Invitrogen). cDNA synthesis was performed with RevertAid™ H Minus Reverse Transcriptase (Fermentas) according to the manufacturer's instructions. cDNAs were subjected to a qRT-PCR reaction to measure Pb14-3-3 expression levels; this reaction was performed using Maxima SYBR Green/ROX qPCR Master Mix 2X according to the manufacturer's instructions (Fermentas) in a 7500 Cycler Real-Time System (Applied Biosystems®). Pb14-3-3 expression was evaluated in PbWT, PbEV and Pb14-3-3 aRNA yeast cells; β-tubulin was selected as the endogenous control.Citation74 The sequences of the oligonucleotide primers used for the amplification of the target gene were Pb14-3-3RT-forward (5′- AATCTGCTTTCCGTTGCCTA-3′) and Pb14-3-3RT-reverse (5′ GTTTTGGCGGTACTCCTTGA-3′), the qRT-PCR product span any of the region targeted by antisense. For the β-tubulin gene, the primer sequences were β-tubulin RT-forward (5′- GTGGACCAGGTGATCGATGT-3′) and β-tubulin RT-reverse (5′- ACCCTGGAGGCAGTCACA-3′). Melting curve analysis was performed after the amplification phase to eliminate the possibility of nonspecific amplification or primer-dimer formation. Fold changes in mRNA expression were calculated using the 2−ΔΔCT method, where ΔΔCT is the difference between experimental conditions (silenced vs. non-silenced strains) after normalization to β-tubulin.Citation75 Each experiment was performed in triplicate, and the expression levels were measured 3 times.
Western blotting
To obtain the total protein extracts, log-phase cells of PbWT and Pb14-3-3 aRNA were concentrated and subsequently washed 3 times with ultrapure water via centrifugation 10,000 × g. The obtained pellets were resuspended in 10 mM Tris-HCl (pH 8.8) added with 1 mL PBS containing 100 mM phenylmethylsulfonyl fluoride and 1 mg mL−1 protease inhibitors (pepstatin, leupeptin, aprotinin, antipain, and chymostatin). The cells were frozen in liquid nitrogen and disrupted by maceration with glass beads, the mixture was vortexed twice 15 min and centrifuged at 5,000 x g for 15 min, and the supernatant was analyzed. The protein concentration was estimated by the Bradford method. SDS-PAGE was performed with equivalent amounts of proteins (0.025 mg) isolated from PbWT and Pb14-3-3 aRNA according to Laemmli.Citation76 For Western blotting experiments, the proteins were transferred onto nitrocellulose membranes (Hybond-C extra, Amersham Biosciences®) as described by Sambrook et al.Citation69. The membranes were blocked with a solution containing PBS, 5% skim milk and 1% bovine serum albumin (BSA) for 4 h at room temperature. After three washes with PBS, the membranes were incubated with the monoclonal anti-14-3-3 (Imuny®, diluted 1:500) antibody overnight, and this assay was also performed with polyclonal anti-enolaseCitation21 as an internal control. The anti-IgG secondary antibody conjugated with peroxidase (Sigma®) diluted 1:1000 was added after further washing, and the 14-3-3 and enolase proteins were visualized by adding 3,3′ diaminobenzidine and hydrogen peroxide as a substrate. Quantification of bands was performed using ImageJ software (NIH) for all blots. Values were calculated as the ratio of the protein of interest (14-3-3) to enolase.
Growth curve and viability assays
For the growth curve, the yeast cells of all isolates were grown in BHI broth at 37°C with shaking, and samples were collected to determine cell number using a standard Neubauer systemCitation77 at different times until the stationary phase was reached. All cultures were begun with an initial concentration of 1 × 108 cells mL−1.
Viability was assessed by measuring the metabolic activity of 2,3-bis(2-methoxy-4-nitro-5-sulfphenyl)-(2H)-tetrazolium-5-car-boxanilide (XTT) (Sigma) in a reduction assay as indicated by the manufacturer. Individual inoculums (200 μL) of PbWT, PbEV and Pb14-3-3 aRNA with 106 cells mL−1, 50 µL of XTT salt solution (1 mg mL−1 in PBS) and 4 µL of menadione solution (1 mM in acetone; Sigma) were added to develop the reaction. Microtiter plates were incubated at 37°C for 5 h.Citation78,79 Fungal mitochondrial dehydrogenase activity reduces XTT tetrazolium salt to XTT formazan, resulting in a colorimetric change that correlates with cell viability. The colorimetric change was measured using a microtiter plate reader (Kasvi®) at 492 nm. In all experiments, microtiter wells containing heat-killed P. brasiliensis and minimal medium alone were included as negative controls.
To confirm the viability results obtained from the XTT assay, a trypan blue dye-exclusion test was performed based on the principle that intact cellular membranes of live cells exclude the dye.Citation80,81 For counting yeast cells, the samples were collected at various times, and 10 μL of the cell solution was added to 190 μL trypan blue solution and diluted to a final volume of 1 mL as described by Do Carmo Silva et al., 2015.Citation82 Yeast cells were analyzed under a light microscope with a 40X lens.
Microscopy
To determine morphometric alterations of yeast cell populations, samples of PbWT, PbEV and Pb14-3-3 aRNA were evaluated. The yeast cells were grown under standard conditions to the exponential phase as described above. Then, they were collected and washed with PBS, followed by the preparation of suspensions with 106 cells mL−1 (in PBS). Then, 50 μL samples of these suspensions were stained using a 25 μM solution of Calcofluor White M2R® (Life Technologies) and observed under a Leica DMLB fluorescence microscope using the 40X objective.
A DFC350FX Leica camera was used for image capture. Under microscopic observation, we recorded the following parameters: the percentage of budding cells, the number of buds exhibited by each cell and the diameter (the major transverse or longitudinal axes) for 100 yeasts from each preparation. Pictures were processed using ImageJ (http://rsb.info.nih.gov/ij).
Additionally, the morphological temperature shift from 25°C to 37°C and vice versa was examined to investigate the influence of Pb14-3-3 in P. brasiliensis dimorphism. For this, fungal cells of PbWT, PbEV and Pb14-3-3 aRNA were cultivated in modified YPD liquid medium (0.5% yeast extract, 0.5% casein peptone, 1.5% glucose, pH 6.5) at either 37°C (yeast phase) or 25°C (mycelial phase) at 150 rpm for several days until the conversion was complete. Cultures were collected every 24 h and evaluated under brightfield microscopy, and a total of 300 cells from random fields were counted at 40X magnification. The transition was classified in 4 different morphological states as described by Nunes et al., 2005Citation83: hyphae, differentiating hyphae, transforming yeast and yeast.
Interaction of P. brasiliensis to pneumocytes
Pneumocytes monolayers (A549 epithelial lineage) were cultured for approximately 24 h in HAM-F12 medium (Cultilab) supplemented with 10% fetal bovine serum. Then, these monolayers were washed and infected with 105 cells mL−1 of PbWT, PbEV and Pb14-3-3 aRNA yeast cells previously stained for 30 min at 37°C with 10 μM of CFSE (5,6 – carboxyfluorescein diacetate N-succinimidyl ester) (BioChemika), which stains the fungal cell wall, washed and resuspended in HAM-F12 medium. The infected cells were incubated for 2 h and 5 h (early times post-inoculation) at 37°C, 5% CO2. Subsequently, after each incubation period, the cells were washed with PBS to remove non-adherent cells, detached from plastic with cold PBS and a rubber cell scraper on ice,Citation84 and then fixed in 4% paraformaldehyde.
The interaction rate was determined by flow cytometry using a FACsCanto™ (Becton Dickinson); uninfected cells and unlabeled yeast were used as internal controls to define the gates and for evaluating autofluorescence. The excitation wavelength was 488 nm, and emitted light was collected via the 530/30 nm band pass-filter. Data were processed and analyzed using FACsDiva software. The assays were performed in triplicate, and the mean results are expressed as the percentage of interaction (fluorescent cells) detected in the samples.
Ligand affinity of P. brasiliensis to ECM components
Next, 96-well polypropylene ELISA microtiter plates (Corning®) were precoated with ECM proteins (10 µg mL−1) in carbonate buffer (pH 9.6) for 1 h at room temperature. After blocking with 1% BSA (w/v) and 0.05% PBS-Tween, the plates were incubated individually with 5 × 106 cells mL−1 of PbWT, PbEV and Pb14-3-3 aRNA for 15 h at 37°C in triplicate wells. The cells were washed, and then anti-cell free antibody (1:100) (a polyclonal antibody against the P. brasiliensis cell wall-associated antigens) in 0.5% BSA and 0.05% PBS-Tween was added for 1 h at 37°C. Peroxidase-conjugated anti-rabbit IgG (1:2000; Sigma®) in 0.05% BSA and 0.05% PBS-Tween was added for 1 h at 37°C. The reaction was developed using citrate (pH 4.9) conjugated with o-phenylenediamine as a chromogenic substrate. Negative controls were performed by incubating the matrix with the primary and secondary antibodies alone and with 0.05% BSA. The absorbance was measured at 490 nm. The capacity of anti-cell free antibody to recognize the isolates equally was verified by Western blotting analysis (data not shown).
Evaluation of the virulence of P. brasiliensis using the Galleria mellonella model
Galleria mellonella larvae (number of larvae per group=16) weighing from 0.1 to 0.2 g were selected and used for the virulence assay. PbWT, PbEV and Pb14-3-3 aRNA yeast cells were harvested at the exponential phase of growth in BHI liquid medium at 37°C. Prior to the infection, the cells were washed thrice with PBS, passed through a syringe to eliminate cells clumps, and a total of 5 × 106 cells prepared in PBS were injected into each larva through the last left pro-leg using a 10 μL Hamilton syringe (Hamilton, USA) after cleaning the pro-leg with 70% ethanol. Each experiment was repeated 3 times independently. One additional group of larvae was inoculated with PBS to monitor the effects on survival due to physical injury, and another control group was inoculated with heat-killed fungal cells to test whether larvae were dying from osmotic stress because of the inoculum size. After the infection, the larvae were incubated in Petri dishes at 37°C for 7 days and checked daily for mortality (based on lack of movement after manipulation with forceps).Citation85
For evaluating the fungal burden of the different isolates, 5 larvae per group were infected as detailed above and sacrificed at each time point (0, 24 and 48 h). The larvae were homogenized in 1 mL sterile PBS, and the homogenates were plated on BHI agar containing chloramphenicol (30 mg mL−1) as described by Singulani et al. (unpublished data). Plates were incubated at 37°C, and colonies were counted daily until no increase in counts was observed.
Statistical analysis
One-way ANOVA with Tukey's coefficient was used to analyze the results obtained in this study. The results of the statistical analyses were considered significant when the p value was < 0.05. Prism 5 software (GraphPad Software Inc..) was used for the analyses and for graph construction. The Western blotting analyses were performed using Student´s t-test with the p-value set at p < 0.05.
For the survival curves, statistical analyses from the results of 3 independent experiments were performed using the Log-rank (Mantel-Cox) test in GraphPad Prism software with the p-value set at p < 0.05 and p < 0.0001.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by FAPESP (2011/18038-9 and 2015/03700-9), CNPq, CAPES and Programa de Apoio ao Desenvolvimento Científico da Faculdade de Ciências Farmacêuticas da UNESP (PADC/FCF). We would like to acknowledge Suélen Andreia Rossi and all members of Biología Celular y Molecular (Corporación para Investigaciones Biológicas – CIB, Medellín-Colômbia).
References
- Lacaz CS. Historical evolution of the knowledge on Paracoccidioides brasiliensis. 1994.
- Brummer E, Castaneda E, Restrepo A. Paracoccidioidomycosis: an update. Clin Microbiol Rev 1993; 6:89-117; PMID:8472249
- Restrepo A, McEwen JG, Castañeda E. The habitat of Paracoccidioides brasiliensis: how far from solving the riddle? Med Mycol 2001; 39:233-41; PMID:11446526; http://dx.doi.org/10.1080/714031028
- Colombo AL, Tobón A, Restrepo A, Queiroz-Telles F, Nucci M. Epidemiology of endemic systemic fungal infections in Latin America. Med Mycol 2011; 49:785-98; PMID:21539506
- Bocca AL, Amaral AC, Teixeira MM, Sato PK, Sato P, Shikanai-Yasuda MA, Soares Felipe MS. Paracoccidioidomycosis: eco-epidemiology, taxonomy and clinical and therapeutic issues. Future Microbiol 2013; 8:1177-91; PMID:24020744; http://dx.doi.org/10.2217/fmb.13.68
- San-Blas G, Niño-Vega G, Iturriaga T. Paracoccidioides brasiliensis and paracoccidioidomycosis: molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med Mycol 2002; 40:225-42; PMID:12146752; http://dx.doi.org/10.1080/mmy.40.3.225.242
- Hube B, Naglik J. Candida albicans proteinases: resolving the mystery of a gene family. Microbiology 2001; 147:1997-2005; PMID:11495978; http://dx.doi.org/10.1099/00221287-147-8-1997
- Franco M. Host-parasite relationships in paracoccidioidomycosis. J Med Vet Mycol 1987; 25:5-18; PMID:3553526; http://dx.doi.org/10.1080/02681-218780000021
- Kurokawa CS, Sugizaki MF, Peraçoli MT. Virulence factors in fungi of systemic mycoses. Rev Inst Med Trop Sao Paulo 1998; 40:125-35; PMID:9830725; http://dx.doi.org/10.1590/S0036-46651998000300001
- VAN DE, Wouw AP, Howlett BJ. Fungal pathogenicity genes in the age of ‘omics’;. Mol Plant Pathol 2011; 12:507-14; PMID:21535355; http://dx.doi.org/10.1111/j.1364-3703.2010.00680.x
- Filler SG, Sheppard DC. Fungal invasion of normally non-phagocytic host cells. PLoS Pathog 2006; 2:e129; PMID:17196036; http://dx.doi.org/10.1371/journal.ppat.0020129
- Amenta PS, Gil J, Martinez-Hernandez A. Connective tissue of rat lung. II: Ultrastructural localization of collagen types III, IV, and VI. J Histochem Cytochem 1988; 36:1167-73; PMID:3403967; http://dx.doi.org/10.1177/36.9.3403967
- Schwartz MA. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb Perspect Biol 2010; 2:a005066; PMID:21084386; http://dx.doi.org/10.1101/cshperspect.a005066
- Geiger B, Yamada KM. Molecular architecture and function of matrix adhesions. Cold Spring Harb Perspect Biol 2011; 3:pii: a005033; PMID:21441590; http://dx.doi.org/10.1101/cshperspect.a005033
- Sohn K, Senyürek I, Fertey J, Königsdorfer A, Joffroy C, Hauser N, Zelt G, Brunner H, Rupp S. An in vitro assay to study the transcriptional response during adherence of Candida albicans to different human epithelia. FEMS Yeast Res 2006; 6:1085-93; PMID:17042758; http://dx.doi.org/10.1111/j.1567-1364.2006.00130.x
- Bhavsar AP, Auweter SD, Finlay BB. Proteomics as a probe of microbial pathogenesis and its molecular boundaries. Future Microbiol 2010; 5:253-65; PMID:20143948; http://dx.doi.org/10.2217/fmb.09.114
- Vicentini AP, Gesztesi JL, Franco MF, de Souza W, de Moraes JZ, Travassos LR, Lopes JD. Binding of Paracoccidioides brasiliensis to laminin through surface glycoprotein gp43 leads to enhancement of fungal pathogenesis. Infect Immun 1994; 62:1465-9; PMID:8132354
- Hanna SA, Monteiro da Silva JL, Giannini MJ. Adherence and intracellular parasitism of Paracoccidioides brasiliensis in Vero cells. Microbes Infect 2000; 2:877-84; PMID:10962270; http://dx.doi.org/10.1016/S1286-4579(00)00390-7
- Barbosa MS, Báo SN, Andreotti PF, de Faria FP, Felipe MS, dos Santos Feitosa L, Mendes-Giannini MJ, Soares CM. Glyceraldehyde-3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect Immun 2006; 74:382-9; PMID:16368993; http://dx.doi.org/10.1128/IAI.74.1.382-389.2006
- Pereira LA, Báo SN, Barbosa MS, da Silva JL, Felipe MS, de Santana JM, Mendes-Giannini MJ, de Almeida Soares CM. Analysis of the Paracoccidioides brasiliensis triosephosphate isomerase suggests the potential for adhesin function. FEMS Yeast Res 2007; 7:1381-8; PMID:17714474; http://dx.doi.org/10.1111/j.1567-1364.2007.00292.x
- Donofrio FC, Calil AC, Miranda ET, Almeida AM, Benard G, Soares CP, Veloso SN, Soares CM, Mendes Giannini MJ. Enolase from Paracoccidioides brasiliensis: isolation and identification as a fibronectin-binding protein. J Med Microbiol 2009; 58:706-13; PMID:19429745; http://dx.doi.org/10.1099/jmm.0.003830-0
- Marcos CM, de Fátima da Silva J, de Oliveira HC, Moraes da Silva RA, Mendes-Giannini MJ, Fusco-Almeida AM. Surface-expressed enolase contributes to the adhesion of Paracoccidioides brasiliensis to host cells. FEMS Yeast Res 2012; 12:557-70; PMID:22443156; http://dx.doi.org/10.1111/j.1567-1364.2012.00806.x
- González A, Gómez BL, Diez S, Hernández O, Restrepo A, Hamilton AJ, Cano LE. Purification and partial characterization of a Paracoccidioides brasiliensis protein with capacity to bind to extracellular matrix proteins. Infect Immun 2005; 73:2486-95; http://dx.doi.org/10.1128/IAI.73.4.2486-2495.2005
- Hernández O, Almeida AJ, Gonzalez A, Garcia AM, Tamayo D, Cano LE, Restrepo A, McEwen JG. A 32-kilodalton hydrolase plays an important role in Paracoccidioides brasiliensis adherence to host cells and influences pathogenicity. Infect Immun 2010; 78:5280-6; http://dx.doi.org/10.1128/IAI.00692-10
- da Silva Neto BR, de Fátima da Silva J, Mendes-Giannini MJ, Lenzi HL, de Almeida Soares CM, Pereira M. The malate synthase of Paracoccidioides brasiliensis is a linked surface protein that behaves as an anchorless adhesin. BMC Microbiol 2009; 9:272; PMID:20034376; http://dx.doi.org/10.1186/1471-2180-9-272
- Zambuzzi-Carvalho PF, Cruz AH, Santos-Silva LK, Goes AM, Soares CM, Pereira M. The malate synthase of Paracoccidioides brasiliensis Pb01 is required in the glyoxylate cycle and in the allantoin degradation pathway. Med Mycol 2009; 47:734-44; PMID:19888806; http://dx.doi.org/10.3109/13693780802609620
- Andreotti PF, Monteiro da Silva JL, Bailão AM, Soares CM, Benard G, Soares CP, Mendes-Giannini MJ. Isolation and partial characterization of a 30 kDa adhesin from Paracoccidioides brasiliensis. Microbes Infect 2005; 7:875-81; PMID:15862780; http://dx.doi.org/10.1016/j.micinf.2005.02.005
- da Silva JEF, de Oliveira HC, Marcos CM, da Silva RA, da Costa TA, Calich VL, Almeida AM, Mendes-Giannini MJ. Paracoccidoides brasiliensis 30 kDa adhesin: identification as a 14-3-3 protein, cloning and subcellular localization in infection models. PLoS One 2013; 8:e62533; PMID:23638109; http://dx.doi.org/10.1371/journal.pone.0062533
- Menino JF, Almeida AJ, Rodrigues F. Gene knockdown in Paracoccidioides brasiliensis using antisense RNA. Methods Mol Biol 2012; 845:187-98; PMID:22328375; http://dx.doi.org/10.1007/978-1-61779-539-8_12
- Hernández O, Almeida AJ, Tamayo D, Torres I, Garcia AM, López A, Restrepo A, McEwen JG. The hydrolase PbHAD32 participates in the adherence of Paracoccidioides brasiliensis conidia to epithelial lung cells. Med Mycol 2012; 50:533-7; http://dx.doi.org/10.3109/13693786.2011.619583
- Tamayo D, Muñoz JF, Torres I, Almeida AJ, Restrepo A, McEwen JG, Hernández O. Involvement of the 90 kDa heat shock protein during adaptation of Paracoccidioides brasiliensis to different environmental conditions. Fungal Genet Biol 2013; 51:34-41; PMID:23207691; http://dx.doi.org/10.1016/j.fgb.2012.11.005
- Peraçoli MT, Sugizaki MF, Mendes RP, Naiff R, Montenegro MR. Paracoccidioides brasiliensis isolated from armadillos is virulent to Syrian hamsters. Mycopathologia 1999; 148:123-30; http://dx.doi.org/10.1023/A:1007119403572
- Svidzinski TI, Miranda Neto MH, Santana RG, Fischman O, Colombo AL. Paracoccidioides brasilienses isolates obtained from patients with acute and chronic disease exhibit morphological differences after animal passage. Rev Inst Med Trop Sao Paulo 1999; 41:279-83; PMID:10602541; http://dx.doi.org/10.1590/S0036-46651999000500003
- Borges-Walmsley MI, Chen D, Shu X, Walmsley AR. The pathobiology of Paracoccidioides brasiliensis. Trends Microbiol 2002; 10:80-7; PMID:11827809; http://dx.doi.org/10.1016/S0966-842X(01)02292-2
- San-Blas G, Niño-Vega G. Paracoccidioides brasiliensis: virulence and host response. In Cihlar, RL and Calderone, RA (ed.). Fungal pathogenesis: principles and clinical applications. pp. 205–226. New York, NY. 2001.
- Rooney PJ, Klein BS. Linking fungal morphogenesis with virulence. Cell Microbiol 2002; 4:127-37; PMID:11906450; http://dx.doi.org/10.1046/j.1462-5822.2002.00179.x
- Böhmer M, Colby T, Böhmer C, Bräutigam A, Schmidt J, Bölker M. Proteomic analysis of dimorphic transition in the phytopathogenic fungus Ustilago maydis. Proteomics 2007; 7:675-85; PMID:17340586; http://dx.doi.org/10.1002/pmic.200600900
- Hurtado CA, Rachubinski RA. YlBMH1 encodes a 14-3-3 protein that promotes filamentous growth in the dimorphic yeast Yarrowia lipolytica. Microbiology 2002; 148:3725-35; PMID:12427962; http://dx.doi.org/10.1099/00221287-148-11-3725
- Aristizabal BH, Clemons KV, Stevens DA, Restrepo A. Morphological transition of Paracoccidioides brasiliensis conidia to yeast cells: in vivo inhibition in females. Infect Immun 1998; 66:5587-91; PMID:9784579
- Roberts RL, Mösch HU, Fink GR. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell 1997; 89:1055-65; PMID:9215628; http://dx.doi.org/10.1016/S0092-8674(00)80293-7
- Kraus PR, Hofmann AF, Harris SD. Characterization of the Aspergillus nidulans 14-3-3 homologue, ArtA. FEMS Microbiol Lett 2002; 210:61-6; PMID:12023078; http://dx.doi.org/10.1111/j.1574-6968.2002.tb11160.x
- Palmer GE, Johnson KJ, Ghosh S, Sturtevant J. Mutant alleles of the essential 14-3-3 gene in Candida albicans distinguish between growth and filamentation. Microbiology 2004; 150:1911-24; PMID:15184577; http://dx.doi.org/10.1099/mic.0.26910-0
- Almeida AJ, Cunha C, Carmona JA, Sampaio-Marques B, Carvalho A, Malavazi I, Steensma HY, Johnson DI, Leão C, Logarinho E, et al. Cdc42p controls yeast-cell shape and virulence of Paracoccidioides brasiliensis. Fungal Genet Biol 2009; 46:919-26; PMID:19686860; http://dx.doi.org/10.1016/j.fgb.2009.08.004
- Almeida AJ, Cunha C, Carmona JA, Sampaio-Marques B, Carvalho A, Malavazi I, Steensma HY, Johnson DI, Leão C, Logarinho E, et al. Cdc42p controls yeast-cell shape and virulence of Paracoccidioides brasiliensis. Fungal Genet Biol 2009; 46:919-26; PMID:19686860; http://dx.doi.org/10.1016/j.fgb.2009.08.004
- Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chrétien F, Heitman J, Dromer F, Nielsen K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog 2010; 6:e1000953; PMID:20585559; http://dx.doi.org/10.1371/journal.ppat.1000953
- Lengeler KB, Davidson RC, D'souza C, Harashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J. Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev 2000; 64:746-85; PMID:11104818; http://dx.doi.org/10.1128/MMBR.64.4.746-785.2000
- Mendes-Giannini MJ, Andreotti PF, Vincenzi LR, da Silva JL, Lenzi HL, Benard G, Zancopé-Oliveira R, de Matos Guedes HL, Soares CP. Binding of extracellular matrix proteins to Paracoccidioides brasiliensis. Microbes Infect 2006; 8:1550-9; PMID:16698299; http://dx.doi.org/10.1016/j.micinf.2006.01.012
- André DC, Lopes JD, Franco MF, Vaz CA, Calich VL. Binding of laminin to Paracoccidioides brasiliensis induces a less severe pulmonary paracoccidioidomycosis caused by virulent and low-virulence isolates. Microbes Infect 2004; 6:549-58; http://dx.doi.org/10.1016/j.micinf.2004.02.010
- Kurokawa CS, Lopes CR, Sugizaki MF, Kuramae EE, Franco MF, Peraçoli MT. Virulence profile of ten Paracoccidioides brasiliensis isolates: association with morphologic and genetic patterns. Rev Inst Med Trop Sao Paulo 2005; 47:257-62; PMID:16302108; http://dx.doi.org/10.1590/S0036-46652005000500004
- de Oliveira HC, da Silva JeF, Scorzoni L, Marcos CM, Rossi SA, de Paula E, Silva AC, Assato PA, da Silva RA, Fusco-Almeida AM, Mendes-Giannini MJ. Importance of adhesins in virulence of Paracoccidioides spp. Front Microbiol 2015; 6:303; PMID:25914695
- Fàbrega A, Vila J. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev 2013; 26:308-41; http://dx.doi.org/10.1128/CMR.00066-12
- Fuchs BB, Eby J, Nobile CJ, El Khoury JB, Mitchell AP, Mylonakis E. Role of filamentation in Galleria mellonella killing by Candida albicans. Microbes Infect 2010; 12:488-96; PMID:20223293; http://dx.doi.org/10.1016/j.micinf.2010.03.001
- Lev S, Desmarini D, Li C, Chayakulkeeree M, Traven A, Sorrell TC, Djordjevic JT. Phospholipase C of Cryptococcus neoformans regulates homeostasis and virulence by providing inositol trisphosphate as a substrate for Arg1 kinase. Infect Immun 2013; 81:1245-55; PMID:23381992; http://dx.doi.org/10.1128/IAI.01421-12
- Mesa-Arango AC, Forastiero A, Bernal-Martínez L, Cuenca-Estrella M, Mellado E, Zaragoza O. The non-mammalian host Galleria mellonella can be used to study the virulence of the fungal pathogen Candida tropicalis and the efficacy of antifungal drugs during infection by this pathogenic yeast. Med Mycol 2013; 51:461-72; PMID:23170962; http://dx.doi.org/10.3109/13693786.2012.737031
- Jacobsen ID. Galleria mellonella as a model host to study virulence of Candida. Virulence 2014; 5:237-9; PMID:24384470; http://dx.doi.org/10.4161/viru.27434
- Firacative C, Duan S, Meyer W. Galleria mellonella model identifies highly virulent strains among all major molecular types of Cryptococcus gattii. PLoS One 2014; 9:e105076; PMID:25133687; http://dx.doi.org/10.1371/journal.pone.0105076
- Thomaz L, García-Rodas R, Guimarães AJ, Taborda CP, Zaragoza O, Nosanchuk JD. Galleria mellonella as a model host to study Paracoccidioides lutzii and Histoplasma capsulatum. Virulence 2013; 4:139-46; PMID:23302787; http://dx.doi.org/10.4161/viru.23047
- Cotter G, Doyle S, Kavanagh K. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol Med Microbiol 2000; 27:163-9; PMID:10640612; http://dx.doi.org/10.1111/j.1574-695X.2000.tb01427.x
- Salamitou S, Ramisse F, Brehélin M, Bourguet D, Gilois N, Gominet M, Hernandez E, Lereclus D. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology 2000; 146 (Pt 11):2825-32; PMID:11065361; http://dx.doi.org/10.1099/00221287-146-11-2825
- Trevijano-Contador N, Herrero-Fernández I, García-Barbazán I, Scorzoni L, Rueda C, Rossi SA, García-Rodas R, Zaragoza O. Cryptococcus neoformans induces antimicrobial responses and behaves as a facultative intracellular pathogen in the non mammalian model Galleria mellonella. Virulence 2015; 6:66-74; PMID:25531532; http://dx.doi.org/10.4161/21505594.2014.986412
- Tojo S, Naganuma F, Arakawa1 K, Yokoo1 S. Involvement of both granular cells and plasmatocytes in phagocytic reactions in the greater wax moth, Galleria mellonella. J Insect Physiol 2000; 46:1129-35; PMID:10817839; http://dx.doi.org/10.1016/S0022-1910(99)00223-1
- Mowlds P, Barron A, Kavanagh K. Physical stress primes the immune response of Galleria mellonella larvae to infection by Candida albicans. Microbes Infect 2008; 10:628-34; PMID:18457977; http://dx.doi.org/10.1016/j.micinf.2008.02.011
- Vogel H, Altincicek B, Glöckner G, Vilcinskas A. A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genomics 2011; 12:308; PMID:21663692; http://dx.doi.org/10.1186/1471-2164-12-308
- Kelly MN, Johnston DA, Peel BA, Morgan TW, Palmer GE, Sturtevant JE. Bmh1p (14-3-3) mediates pathways associated with virulence in Candida albicans. Microbiology 2009; 155:1536-46; PMID:19372164; http://dx.doi.org/10.1099/mic.0.027532-0
- Butt AQ, Ahmed S, Maratha A, Miggin SM. 14-3-3ε and 14-3-3σ inhibit Toll-like receptor (TLR)-mediated proinflammatory cytokine induction. J Biol Chem 2012; 287:38665-79; PMID:22984265; http://dx.doi.org/10.1074/jbc.M112.367490
- Qi H, Denning TL, Soong L. Differential induction of interleukin-10 and interleukin-12 in dendritic cells by microbial toll-like receptor activators and skewing of T-cell cytokine profiles. Infect Immun 2003; 71:3337-42; PMID:12761116; http://dx.doi.org/10.1128/IAI.71.6.3337-3342.2003
- Bonfim CV, Mamoni RL, Blotta MH. TLR-2, TLR-4 and dectin-1 expression in human monocytes and neutrophils stimulated by Paracoccidioides brasiliensis. Med Mycol 2009; 47:722-33; PMID:19888805; http://dx.doi.org/10.3109/13693780802641425
- Beijersbergen A, Dulk-Ras AD, Schilperoort RA, Hooykaas PJ. Conjugative Transfer by the Virulence System of Agrobacterium tumefaciens. Science 1992; 256:1324-7; PMID:17736763; http://dx.doi.org/10.1126/science.256.5061.1324
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press. 1,626 pp. New York, NY. 1989.
- Almeida AJ, Carmona JA, Cunha C, Carvalho A, Rappleye CA, Goldman WE, Hooykaas PJ, Leão C, Ludovico P, Rodrigues F. Towards a molecular genetic system for the pathogenic fungus Paracoccidioides brasiliensis. Fungal Genet Biol 2007; 44:1387-98; PMID:17512227; http://dx.doi.org/10.1016/j.fgb.2007.04.004
- Ruiz OH, Gonzalez A, Almeida AJ, Tamayo D, Garcia AM, Restrepo A, McEwen JG. Alternative oxidase mediates pathogen resistance in Paracoccidioides brasiliensis infection. PLoS Negl Trop Dis 2011; 5:e1353; PMID:22039556; http://dx.doi.org/10.1371/journal.pntd.0001353
- Rappleye CA, Engle JT, Goldman WE. RNA interference in Histoplasma capsulatum demonstrates a role for alpha-(1,3)-glucan in virulence. Mol Microbiol 2004; 53:153-65; PMID:15225311; http://dx.doi.org/10.1111/j.1365-2958.2004.04131.x
- van Burik JA, Schreckhise RW, White TC, Bowden RA, Myerson D. Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Med Mycol 1998; 36:299-303; PMID:10075499; http://dx.doi.org/10.1080/02681219880000471
- Goldman GH, dos Reis Marques E, Duarte Ribeiro DC, de Souza Bernardes LA, Quiapin AC, Vitorelli PM, Savoldi M, Semighini CP, de Oliveira RC, Nunes LR, et al. Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot Cell 2003; 2:34-48; PMID:12582121; http://dx.doi.org/10.1128/EC.2.1.34-48.2003
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402-8; http://dx.doi.org/10.1006/meth.2001.1262
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227:680-5; PMID:5432063; http://dx.doi.org/10.1038/227680a0
- Menino JF, Saraiva M, Gomes-Rezende J, Sturme M, Pedrosa J, Castro AG, Ludovico P, Goldman GH, Rodrigues F. P. brasiliensis virulence is affected by SconC, the negative regulator of inorganic sulfur assimilation. PLoS One 2013; 8:e74725; PMID:24066151; http://dx.doi.org/10.1371/journal.pone.0074725
- Meshulam T, Levitz SM, Christin L, Diamond RD. A simplified new assay for assessment of fungal cell damage with the tetrazolium dye, (2,3)-bis-(2-methoxy-4-nitro-5-sulphenyl)-(2H)-tetrazolium-5-carboxanil ide (XTT). J Infect Dis 1995; 172:1153-6; PMID:7561202; http://dx.doi.org/10.1093/infdis/172.4.1153
- Chandra J, Mukherjee PK, Leidich SD, Faddoul FF, Hoyer LL, Douglas LJ, Ghannoum MA. Antifungal resistance of candidal biofilms formed on denture acrylic in vitro. J Dent Res 2001; 80:903-8; PMID:11379893; http://dx.doi.org/10.1177/00220345010800031101
- Strober, W. 2001. Trypan Blue Exclusion Test of Cell Viability. Current Protocols in Immunology. 21:3B:A.3B.1–A.3B.2
- Prado RS, Bailão AM, Silva LC, de Oliveira CM, Marques MF, Silva LP, Silveira-Lacerda EP, Lima AP, Soares CM, Pereira M. Proteomic profile response of Paracoccidioides lutzii to the antifungal argentilactone. Front Microbiol 2015; 6:616; PMID:26150808
- do Carmo Silva L, Tamayo Ossa DP, Castro SV, Bringel Pires L, Alves de Oliveira CM, Conceição da Silva C, Coelho NP, Bailão AM, Parente-Rocha JA, Soares CM, et al. Transcriptome Profile of the Response of Paracoccidioides spp. to a Camphene Thiosemicarbazide Derivative. PLoS One 2015; 10:e0130703; PMID:26114868; http://dx.doi.org/10.1371/journal.pone.0130703
- Nunes LR, Costa de Oliveira R, Leite DB, da Silva VS, dos Reis Marques E, da Silva Ferreira ME, Ribeiro DC, de Souza Bernardes LA, Goldman MH, Puccia R, et al. Transcriptome analysis of Paracoccidioides brasiliensis cells undergoing mycelium-to-yeast transition. Eukaryot Cell 2005; 4:2115-28; PMID:16339729; http://dx.doi.org/10.1128/EC.4.12.2115-2128.2005
- Bernardino S, Pina A, Felonato M, Costa TA, Frank de Araújo E, Feriotti C, Bazan SB, Keller AC, Leite KR, Calich VL. TNF-α and CD8+ T cells mediate the beneficial effects of nitric oxide synthase-2 deficiency in pulmonary paracoccidioidomycosis. PLoS Negl Trop Dis 2013; 7:e2325; http://dx.doi.org/10.1371/journal.pntd.0002325
- Scorzoni L, de Lucas MP, Mesa-Arango AC, Fusco-Almeida AM, Lozano E, Cuenca-Estrella M, Mendes-Giannini MJ, Zaragoza O. Antifungal efficacy during Candida krusei infection in non-conventional models correlates with the yeast in vitro susceptibility profile. PLoS One 2013; 8:e60047; PMID:23555877; http://dx.doi.org/10.1371/journal.pone.0060047
