Cryptosporidium is a parasite that causes the majority of waterborne protozoan outbreaks of gastrointestinal disease worldwide.Citation1 Cryptosporidiosis usually presents as self-limiting, although often prolonged, diarrhea with abdominal pain, nausea, vomiting and low-grade fever. Infection can be life threatening in some severely immunocompromised groups.Citation2 Post infection sequelae have also been reported.Citation3 On a global level, Cryptosporidium hominis and Cryptosporidium parvum cause the majority of human infections and drinking waterborne outbreaks. Recently, Cryptosporidium cuniculus has also caused a sizeable waterborne outbreak.Citation4 Molecular detection of human infective Cryptosporidium species is usually based on conserved housekeeping genes, as is the case for other microorganisms.Citation5 However, novel detection targets have been investigated for Cryptosporidium, but previous investigations aiming to identify C. hominis and C. parvum specific genes had limited success because the majority of putative species-specific genes were subsequently found to be present in the other species.Citation6,7 This was attributed to the limited numbers of genome sequences available and their low quality that hindered the accurate prediction of genes' presence. Nevertheless, evidence was found for one C. hominis specific gene (Chos-1), which was further validated independently using comparative genomics of newly sequenced clinical isolates.Citation8 Chos-1 has a telomeric location and the predicted secreted protein of 50 kDa has interesting features, (highly glycosylated, serine rich and several internal repeats), suggestive of a role in host pathogen interaction.Citation6 It is plausible that other Cryptosporidium species-specific genes and virulence factors are also located at the telomeres. Subtelomeres are hotspots for genetic recombination in other parasitic protozoa, resulting in significant chromosome length and sequence composition polymorphisms and an over-representation of highly-diverged loci which have been utilized for discriminatory diagnostic purposes.Citation9-12
Thanks to decreased costs and technological advances, an increasing number of full genome sequences are becoming available for Cryptosporidium spp. and are contributing to improved comparative genomics analyses. While many investigators used these genomes to identify genes and proteins based on their presence in other parasites and bacteria, we have continued to look for species-specific genes that are likely to be involved in host parasite interaction and virulence. In addition, these genes provide a source of highly specific and stable detection markers. In this study, we used an increased number of genome sequences to identify species-specific genes, focusing on genes close to the chromosome ends. Subsequently, primers and probes targeting these loci were designed and evaluated as novel real-time PCR assays for the specific detection of the major human infective waterborne parasites C. hominis and C. parvum.
Previous investigation of putative species-specific genes led to the characterization of the telomeric Chos-1 gene.Citation6 This finding suggested that further genes are likely to be analogously positioned and warranted further investigation of subtelomeric regions of C. hominis and C. parvum genome sequences. In the first instance, the reference genome sequences of C. hominis TU502 (gp60 subtype IaA25R3) Citation13 and C. parvum Iowa (gp60 subtype IIaA15G2R1) Citation14 were retrieved from CryptoDB (http://cryptodb.org) and used for analysis. Subsequently, genomic data of newly sequenced genomes from C. hominis and C. parvum clinical isolates (UKH4 gp60 subtype IaA14R3 and UKP6 gp60 IIaA15G2R1, respectively) were used to verify our findings.Citation15 Comparative analysis of sequence data was achieved by aligning subtelomeric regions spanning 30000 bp from the 5′- and 3′-ends of the 8 C. hominis and C. parvum chromosomes using the Artemis Comparison Tool (ACT) Citation16 http://www.webact.org/WebACT/home. The subtelomeric regions were scanned in order to identify species-specific coding sequences. The specificity of these sequences was further validated against other Cryptosporidium isolates with different gp60 subtypes: 2 C. hominis (UKH5 gp60 IbA10G2, UKH3 gp60 IbA10G2) and 4 C. parvum (UKP2 gp60 IIaA19G1R2, UKP3 gp60 IIaA18G2R1, UKP7 gp60 IIaA17G1R1, UKP8 gp60 IIdA22G1) using a standalone nucleotide blast test generated using BioEdit (version 7.2.5) and against all non-Cryptosporidium isolates using blastn software tool provided by NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch).
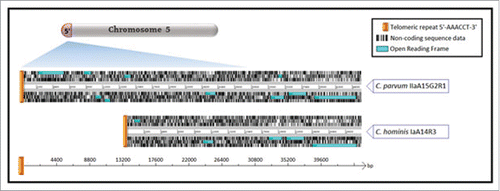
The analysis showed that a 14000 bp stretch of genomic sequence was putatively specific to C. parvum and missing from C. hominis along the subtelomeric arm extending from the 5′ telomeric repeat (5′-AAACCT-3′) on chromosome 5 (). In C. parvum, a 419 bp stretch of genomic sequence was missing at the 3′ telomeric end of chromosome 5 and therefore putatively C. hominis specific. Four open reading frames (ORFs) were encoded within the C. parvum specific sequence and the C. hominis specific sequence partially spanned a single ORF. Two of the C. parvum specific ORFs are the result of a C. parvum specific gene duplication event of gene cgd5_4580, which is located at the subtelomeric 3′-end of chromosome 6 in C. parvum, and which contains an ortholog in C. hominis called Chro.50507. The third open reading frame, identified as gene cgd6_5500, also contains an ortholog in C. hominis, which has non syntenic localization compared to its C. parvum counterpart as it is located on a different chromosome. The fourth coding sequence, Cgd6_5510 (Cops-2) encoding an insulinase-like peptidase, was wholly specific to C. parvum, and hence selected for inclusion in this study. The genomic sequence identified in C. hominis, Chro.00007(Chos-2), was not entirely species-specific, with an orthologous stretch of 100 bp sequence identified at the 3′ end of chromosome 7 in C. parvum. However, the degree of divergence for this orthologous segment (88% nucleotide similarity) and the specificity of the remaining 319 bp stretch warranted inclusion of this marker for discriminatory purposes. Chos-1, Chos-2 and Cops-2 were the 3 species-specific loci used to develop and evaluate real-time PCR assays for the specific detection of C. hominis and C. parvum.
Figure 1. Subtelomeric sequence data alignment at the 5′-end of chromosome 5 reveals significant sequence incongruence between C. hominis and C. parvum. Subtelomeric sequence data extending <30,000bp inwards from the 5′-telomeric end of chromosome 5 was extracted from newly-sequenced whole genome sequences for C. parvum subtype IIaA15G2R1 (UKP6) and C. hominis subtype IaA14R3 (UKH4). Sequences were aligned and compared using the Artemis Comparison Tool (ACT).Citation16 Orthologous coding sequences were independently validated through The European Molecular Biology Open Software Suite (EMBOSS) Stretcher pairwise sequence alignment of nucleotide ORFs.Citation18
Sequence data for each locus (Chos-1, Chos-2 and Cops-2) from all available C. hominis and C. parvum isolates (3 C. hominis UKH3, UKH4, UKH5 and 5 C. parvum UKP2, UKP3, UKP6, UKP7, UKP8) were retrieved and aligned separately (for each species) using BioEdit version 7.0.9.0 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Primer sequences were selected to amplify fragments of interest and appropriate length (70–150 bp), while avoiding undesirable molecular interactions such as cross reactivity or self-annealing. Primer length, annealing temperature, %GC content and the potential for undesirable molecular interactions were assessed using the Primer Express software program (Applied Biosystems). The predicted PCR products were analyzed in silico and no orthologs were found in other Cryptosporidium spp. MGB hydrolysis probes were designed according to the optimum criteria: probe having an annealing temperature 10°C higher than the primers and avoiding runs of identical nucleotides and nucleotides likely to quench the reporter fluorescence. Primers and probes sequences and characteristics are presented in . For Chos-1, 2 different reverse primers with a common forward primer and probe were tested. For Chos-2, 2 different forward primers with a common reverse primer and probe were tested and for Cops-2, a single forward and reverse primer and probe were used. Primer and probe sequences were checked for cross-reactions with non-target sequences on the GenBank database using the Basic Local Alignment Search Tool (BLAST; http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). No complete (both primers and probe) cross reactions with non-Cryptosporidium sequences were detected. Primers and probes (all MGB-non-fluorescent quencher, NFQ probes) were ordered from Life Technologies.
Table 1. Sequences of forward (F) and reverse (R) primers and probes (T) designed to investigate the potential of Chos-1 and Chos-2 for the specific detection of C. hominis and Cops-2 for the specific detection of C. parvum.
The primer and probe sets were tested in a singleplex real-time PCR format. PCR conditions were TaqMan Environmental Master Mix 2.0 (ABI, Life Technologies), 600 nM of each primer and 100 nM probe. Cycling conditions were 95°C for 10 min, 95°C for 15 seconds and 60°C for 1 min, 50 cycles. PCRs were run on a Rotorgene thermocycler (Qiagen). Additionally, 2 primer and probe sets were evaluated in a duplex format for the detection of single and dual C. parvum and C. hominis infections.
Each real time assay was evaluated using target and non-target genomic DNA from Cryptosporidium species and genotypes isolated from human and animal infections, which included human infective species (such as C. cuniculus and C. viatorum) as well as non-human infective species (such as C. andersoni, C. baileyi and C. bovis). All samples were from anonymised clinical human and animal sources and provided by the Cryptosporidium Reference Unit ().
Table 2. Real time PCR results using a comprehensive Cryptosporidium DNA panel from clinical and animal sources. Results are presented as Ct values.
While optimising PCR conditions, a small number of samples were tested using the primer sets in a conventional PCR format. This resulted in DNA amplification from species other than C. hominis and C. parvum (data not shown). The amplified bands were very faint and did not allow sequence analysis of PCR products. However, the use of probe-based real-time PCR format substantially increased the specificity of the assay. Real-time PCR results presented as Ct values are summarised in . Ct values ranged from 22.30 for TU502 isolate detection using Chos-1 assay to 38.44 for UKH15 detection using Chos-2 assay. Overall, only C. hominis and C. cuniculus were detected using Chos-1 and Chos-2 primer-probe sets. This result is not surprising because C. cuniculus is closely related to C. hominis and has been documented to amplify using C. hominis assays.Citation17 For Cops-2, only C. parvum DNA was amplified. However, one of the 7 C. parvum isolates tested (UKP12) did not amplify using the Cops-2 assay. Our results support the species specificity of these detection assays within the genus Cryptosporidium, demonstrated by the testing of a comprehensive panel of human and non-human infective Cryptosporidium species and genotypes. This specificity is conferred by the use of primers and probes in a real time PCR format.
The performance of a duplex assay for the simultaneous detection of both C. hominis and C. parvum was assessed using Chos2-F2 and Cops-2 primers and probes. This is particularly important for clinical settings as the duplex PCR would allow detection of both human pathogens in a single test at a decreased cost and time compared to individual singleplex PCRs. Individual C. hominis and C. parvum DNA as well as an artificially mixed DNA sample were tested using singleplex and duplex formats. The samples were correctly identified by both assay formats, but the duplex format was associated with some increase in Ct values compared with those from the simplex PCR (). This is a well-known trade off of the multiplex PCR. However, for the Cops-2 assay the Ct difference between singleplex and duplex formats was negligible, showing that no competition was occurring when a duplex format is used.
Table 3. Real time PCR results (Ct values) comparing single and duplex Chos-2F2 and Cops-2 assays.
Comparative genomics are an important aspect of the post-genomic era. Analysis of genome sequences is continuously improving our understanding of pathogen biology, pathogenicity, evolution and host adaptation. Comparative genomics also allow identification of conserved and specific genes. We exploited the increasing numbers of published Cryptosporidium genome sequences to identify putative unique genes that are likely to be involved in virulence and host adaptation. Subsequently, we developed assays based on these genes as specific detection tools. Our previous attempt had a limited success as only one C. hominis specific gene was found.Citation6 This was mainly attributed to the low quality of the TU502 genome sequence available at the time. Technological advances have enabled the generation of an ever increasing number of genome sequences of improved quality.Citation8,15 These improvements are likely to contribute to more accurately predicted species-specific genes. Indeed, assays targeting these genes were confirmed experimentally as species-specific as shown by our results. The use of probe-based real-time PCR assay dramatically increased the specificity of the assays.
These novel real-time PCR assays for the detection of C. hominis and C. parvum were evaluated using a comprehensive panel of human and non-human infective Cryptosporidium species and genotypes. Only DNA from C. parvum for real time PCR Cops-2 assay and C. hominis and C. cuniculus for Chos-1 and Chos-2 real time PCR assays were amplified. However, further specificity assessment needs to be performed using DNA from other human-infective parasites likely to be present in human stool samples such as Giardia duodenalis, Cyclospora cayetanensis, Entamoeba histolytica/dispar, Toxoplasma gondii, Blastocystis hominis and Dientamoeba fragilis. Further validation work needs to be undertaken, including assessment of analytical and diagnostic sensitivity and specificity, repeatability and reproducibility, as well as consideration of clinical and epidemiological application prior to use these assays in a clinical setting.
For the Cops-2 assay, 1 out the 7 C. parvum isolates tested (UKP12) did not amplify. The reason for this is unknown. It could be due to sequence variation, DNA quality or quantity or presence of inhibitors, although the latter is unlikely as the sample amplified using other detection targets. Therefore, it would be useful to include an internal positive control in any diagnostic PCR based assay. The positive control could target a Cryptosporidium conserved gene and the performance of the modified assay will need to be assessed.
While extensively characterized genes continue to be the targets of choice for the detection of human infective Cryptosporidium species, identification and validation of species-specific genes are sometimes preferable, especially when investigating host adaptation and evolution. Additionally, the use of these loci is likely to add robustness to the assay especially in comparison to detection and typing assays relying on small sequence variation and even on a single nucleotide polymorphism for successful amplification and sequence analysis. More genes continue to be discovered in the post genomic era and are likely to contribute to major breakthroughs in the fight against infectious diseases.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
The research leading to these results has received funding from the European Union Seventh Framework Program [FP7/2007–2013] under Grant agreement no: 311846.
References
- Baldursson S, Karanis P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks - An update 2004–2010. Water Res 2011; 45:6603-14; PMID:22048017; http://dx.doi.org/10.1016/j.watres.2011.10.013
- Bouzid M, Hunter PR, Chalmers RM, Tyler KM. Cryptosporidium Pathogenicity and Virulence. Clin Microbiol Rev 2013; 26:115-34; PMID:23297262; http://dx.doi.org/10.1128/CMR.00076-12
- Hunter PR, Hughes S, Woodhouse S, Raj N, Syed Q, Chalmers RM, Verlander NQ, Goodacre J. Health sequelae of human cryptosporidiosis in immunocompetent patients. Clin Infect Dis 2004; 39:504-10; PMID:15356813; http://dx.doi.org/10.1086/422649
- Puleston RL, Mallaghan CM, Modha DE, Hunter PR, Nguyen-Van-Tam JS, Regan CM, Nichols GL, Chalmers RM. The first recorded outbreak of cryptosporidiosis due to Cryptosporidium cuniculus (formerly rabbit genotype), following a water quality incident. J Water Health 2014; 12:41-50; PMID:24642431; http://dx.doi.org/10.2166/wh.2013.097
- Bouzid M, Steverding D, Tyler KM. Detection and surveillance of waterborne protozoan parasites. Curr Opin Biotech 2008; 19:302-6; PMID:18524569; http://dx.doi.org/10.1016/j.copbio.2008.05.002
- Bouzid M, Hunter PR, McDonald V, Elwin K, Chalmers RM, Tyler KM. A new heterogeneous family of telomerically encoded Cryptosporidium proteins. Evol Appl 2013; 6:207-17; PMID:23467513; http://dx.doi.org/10.1111/j.1752-4571.2012.00277.x
- Bouzid M, Tyler KM, Christen R, Chalmers RM, Elwin K, Hunter PR. Multi-locus analysis of human infective Cryptosporidium species and subtypes using ten novel genetic loci. Bmc Microbiol 2010; 10; PMID:20696051
- Guo YQ, Tang K, Rowe LA, Li N, Roellig DM, Knipe K, Frace M, Yang CF, Feng YY, Xiao LH. Comparative genomic analysis reveals occurrence of genetic recombination in virulent Cryptosporidium hominis subtypes and telomeric gene duplications in Cryptosporidium parvum. Bmc Genomics 2015; 16; http://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-015-1517-1
- Barros RRM, Marini MM, Antonio CR, Cortez DR, Miyake AM, Lima FM, Ruiz JC, Bartholomeu DC, Chiurillo MA, Ramirez JL, et al. Anatomy and evolution of telomeric and subtelomeric regions in the human protozoan parasite Trypanosoma cruzi. Bmc Genomics 2012; 13; PMID:22681854
- Callejas S, Leech V, Reitter C, Melville S. Hemizygous subtelomeres of an African trypanosome chromosome may account for over 75% of chromosome length. Genome Res 2006; 16:1109-18; PMID:16899654; http://dx.doi.org/10.1101/gr.5147406
- Cornejo OE, Fisher D, Escalante AA. Genome-wide patterns of genetic polymorphism and signatures of selection in Plasmodium vivax. Gen Biol Evolut 2015; 7:106-19; http://dx.doi.org/10.1093/gbe/evu267
- Jiang H, Li N, Gopalan V, Zilversmit MM, Varma S, Nagarajan V, Li J, Mu J, Hayton K, Henschen B, et al. High recombination rates and hotspots in a Plasmodium falciparum genetic cross. Gen Biol 2011; 12:R33; PMID:21463505; http://dx.doi.org/10.1186/gb-2011-12-4-r33
- Xu P, Widmer G, Wang YP, Ozaki LS, Alves JM, Serrano MG, Puiu D, Manque P, Akiyoshi D, Mackey AJ, et al. The genome of Cryptosporidium hominis. Nature 2004; 431:1107-12; PMID:15510150; http://dx.doi.org/10.1038/nature02977
- Abrahamsen MS. Cryptosporidium parvum genome project. Compar Funct Genom 2001; 2:19-21; http://dx.doi.org/10.1002/cfg.67.
- Hadfield SJ, Pachebat JA, Swain MT, Robinson G, Cameron SJ, Alexander J, Hegarty MJ, Elwin K, Chalmers RM. Generation of whole genome sequences of new Cryptosporidium hominis and Cryptosporidium parvum isolates directly from stool samples. Bmc Genomics 2015; 16:650; PMID:26318339; http://dx.doi.org/10.1186/s12864-015-1805-9
- Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: the Artemis comparison tool. Bioinformatics 2005; 21:3422-3; PMID:15976072; http://dx.doi.org/10.1093/bioinformatics/bti553
- Robinson G, Wright S, Elwin K, Hadfield SJ, Katzer F, Bartley PM, Hunter PR, Nath M, Innes EA, Chalmers RM. Re-description of Cryptosporidium cuniculus Inman and Takeuchi, 1979 (Apicomplexa: Cryptosporidiidae): morphology, biology and phylogeny. Int J Parasitol 2010; 40:1539-48; PMID:20600069; http://dx.doi.org/10.1016/j.ijpara.2010.05.010
- Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends Genet 2000; 16:276-7; PMID:10827456; http://dx.doi.org/10.1016/S0168-9525(00)02024-2
