abstract
Highly pathogenic (HP) avian influenza viruses (AIV) evolve from low pathogenic (LP) precursors after circulation in poultry by reassortment and/or single mutations in different gene segments including that encoding NS1. The carboxyl terminal end (CTE) of NS1 exhibits deletions between amino acid 202 and 230 with still unknown impact on virulence of AIV in chickens. In this study, NS1 protein sequences of all AIV subtypes in birds from 1902 to 2015 were analyzed to study the prevalence and distribution of CTE truncation (ΔCTE). Thirteen different ΔCTE forms were observed in NS1 proteins from 11 HA and 8 NA subtypes with high prevalences in H9, H7, H6 and H10 and N9, N2, N6 and N1 subtypes particularly in chickens and minor poultry species. With 88% NS217 lacking amino acids 218–230 was the most common ΔCTE form followed by NS224 (3.6%). NS217 was found in 10 and 8 different HA and NA subtypes, respectively, whereas NS224 was detected exclusively in the Italian HPAIV H7N1 suggesting relevance for virulence. To test this assumption, 3 recombinant HPAIV H7N1 were constructed carrying wild-type HP NS1 (Hp-NS224), NS1 with extended CTE (Hp-NS230) or NS1 from LPAIV H7N1 (Hp-NSLp), and tested in-vitro and in-vivo. Extension of CTE in Hp NS1 significantly decreased virus replication in chicken embryo kidney cells. Truncation in the NS1 decreased the tropism of Hp-NS224 to the endothelium, central nervous system and respiratory tract epithelium without significant difference in virulence in chickens. This study described the variable forms of ΔCTE in NS1 and indicated that CTE is not an essential virulence determinant particularly for the Italian HPAIV H7N1 but may be a host-adaptation marker required for efficient virus replication.
Introduction
Avian influenza virus (AIV) belongs to the family Orthomyxoviridae with a genome consisting of 8 segments of single-stranded RNA with negative polarity. Variations in the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA) of AIV result in 16 HA and 9 NA different subtypes present in birds. All H1-H16 subtypes were isolated from birds without overt disease but few viruses of the H5 and H7 subtypes can cause mass lethality in few days.Citation1 These highly pathogenic (HP) AIV strains evolve from low pathogenic (LP) ancestors by reassortment and/or point mutations. Although virulence of HPAIV is a complex multigenic trait in nature,Citation2,3 in some H5N1 and H7N9 viruses mutations in the gene encoding the non-structural protein 1 (NS1) contribute to virus virulence in chickens.Citation4-6 NS1 is a multifunctional protein encoded by the smallest segment 8 of AIV encompassing typically 890 nucleotides. In birds, NS1 can appear in 2 distinct alleles, where allele A is more common than allele B.Citation7-9 NS1 contains 2 functional domains, an RNA binding domain (RBD) and an effector domain (ED). The RBD (amino acids 1–73) binds different RNA species (e.g. viral RNA, viral mRNAs, poly(A) RNA and double stranded RNA) preventing the activation of host cellular sensors. The ED (amino acids 74–230) interacts with host factors via several motifs.Citation10 In addition to its well-defined role as an antiviral antagonist, variations in the Postsynaptic density protein 95, Drosophila disc large tumor suppressor, and Zonula occludens 1 protein domain (PDZ) at the carboxyl terminal end (CTE) of NS1 resulted in expansion of the host range of some influenza viruses in different cell cultures and modulated virulence in mice and poultry.Citation11-13 Size variation of NS1 has been described, mainly due to a stop codon in the ED which results in truncation of the C-terminal end (ΔCTE).Citation9,14 Nonetheless, prevalence and distribution of NS1 ΔCTE among AIV subtypes and their biological impact in poultry have not been well studied.
There are several epizootics caused by HPAIV in poultry which evolved from LPAIV previously circulating in the poultry population.Citation15 The largest of these epidemics in a western country was caused by H7N1 in poultry in Italy in 1999: the LPAIV emerged in March and the HPAIV emerged in December.Citation16 Genetic changes accompanying shift of the Italian LPAIV to HPAIV H7N1 have been well-defined.Citation17,18 Among other mutations, transformation of LPAIV to HPAIV has been also accompanied with alterations in NS1, where the contribution of these mutations in virulence remained indeterminate.Citation14 Truncation of 6 amino acids (aa) at the C-terminal end (225 RVESEV230) of NS1 in the Italian HPAIV H7N1 vs. LPAIV is of special interest. Hence, in-vitro, ΔCTE increased the lethality of an Italian HPAIV H7N1 for chicken embryos Citation19 but did not affect LPAIV replication or virulence in chickens.Citation20 To date, it is not known whether this ΔCTE contributes to virulence of HPAIV H7N1 in chickens. In this study, prevalence and distribution of NS1 variants were studied by analysis of all full-length NS1 protein sequences of H1-H16/N1-N9 AIV in birds from 1902 to 2015 deposited in the GenBank database. Moreover, using reverse genetics the impact of NS1 variation on virus replication in-vitro and on virulence in-vivo was further assessed in specific-pathogen-free (SPF) chickens.
Results
Variable size of ΔCTE of NS1 among AIV
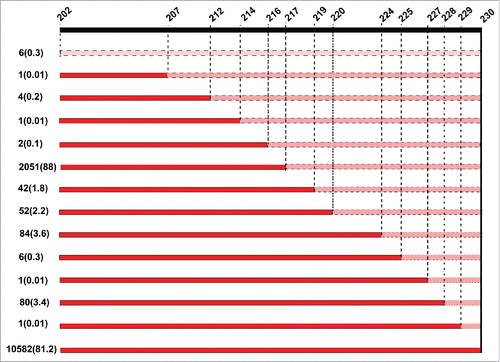
A total of 13026 sequences from AIV in different avian species from 1902 to 2015 were analyzed. NS1 in 10582 out of 13026 (81.2%) sequences encompassed 230 aa. Deletion within the CTE of the NS1 was exhibited by 13 variants in 2331 sequences (17.8%) (Table S1) denoted by the number of the last aa in the NS1 protein (). These were NS202, 207, 212, 214, 216, 217, 219, 220, 224, 225, 227, 228 and 229 (). The most prevalent form was NS217 present in 2051 out of 2331 (88%) sequences, followed by NS224 (n=84/2331; 3.6%), NS228 (n=80/2331, 3.4%), NS220 (n=52/2331, 2.2%) and NS219 (n= 42/2331, 1.8%). All other variants combined represented only 1%. Furthermore, a low prevalence of an extension in CTE to 237 (n= 112/13026, 0.9%) or 238 (n=1/13026, 0.01%) amino acids was also observed ().
Figure 1. Patterns of truncation of the C-terminal end of the NS1 protein in avian influenza viruses from 1902 to 2015. Shown are the truncations of the C-terminal end (ΔCTE) of NS1 among aa 202 to 230. Numbers on the left are total sequences pertaining the specific truncation form and between parentheses are the percentages of total ΔCTE divided by total 13026 analyzed AIV sequences. Truncated sequences are shown in pink while truncated sequences of CTE are in red. The naïve form of NS1, seen in 10582 out of 13026 sequences (81.2%) is 230 aa of length. Meanwhile, several truncations were observed in NS1 since 1954. The most common form is NS217 followed by NS224.
Distribution of NS1 ΔCTE in different HA and/or NA subtypes
The analyzed sequences represented all avian HA and NA subtypes. No NS1 truncation was observed in viruses specifying subtypes H8 and H12-H16 or N4-type neuraminidase (). About 88% of all ΔCTE, including the majority of NS217, are observed in H6, H7 and H9 types () and about 80% are in N2-, N6- and N9-type neuraminidase (). Truncation of NS1 was observed in 36 HxNx subtypes out of 123 HxNx combinations in the GenBank. shows that 70% of all ΔCTE are indeed represented by H6N2, H9N2 and H7N9 viruses. NS224 was only observed in the Italian HPAIV H7N1 viruses in 1999–2000 and compared to their LPAI H7N1 progenitors they also carried V136I and D139N in the NS1.
Table 1. Prevalence of NS1 truncation in the C-terminal end in 16 hemagglutinin subtypes of avian influenza viruses from 1902 to 2015.
Table 2. Prevalence of NS1 truncation in the C-terminal end in 9 neuraminidase subtypes of avian influenza viruses from 1902 to 2015.
Table 3. Prevalence of NS1 truncation in the C-terminal end among different HA-NA combination of avian influenza viruses from 1902 to 2015.
Distribution of NS1 ΔCTE in different years and countries
Years and origins of emergence of each NS1 ΔCTE form are summarized in . In viruses isolated from 1902 to 1955, NS1 ΔCTE was not detected and all viruses exhibited NS230. From 1956 to 2015, the most common truncation was NS217. In 1956–1990 there were 23/32 (71.8%) of NS217 mostly due to frequent isolation of H5N2 viruses particularly in the Pennsylvania outbreak in 1983–1984 in chickens, turkeys, goose and Guinea fowl as well as in H5N2 from chickens, ducks and little grebe in Taiwan, China and Vietnam in 2006–2014. In 1991–2000, there were 105 out of 225 (46.7%) analyzed sequences with NS217 due to circulation of H9N2, H6N2 (in China and Hong Kong) and H7N3 (in Pakistan). Also, in this period NS224 and NS220 were observed in 84/225 (37.3%) and 22/225 (9.8%) sequences, respectively. In 2000–2010, NS217 was common (n= 974/1111; 87.7%), due to circulation of H9N2, H6 (particularly in China) and H7N3 (in Pakistan). Since 2011, due to widespread of H9N2, H10N6 and H7N9 (which share similar NS segment likely from H9N2) resulted in 950/963 (98.7%) sequences pertaining NS217. Only two forms of truncation are currently circulating, NS217 and NS219.
Table 4. Years and countries of origin of each form of NS1 C-terminal end truncation among avian influenza viruses from 1902–2015.
Distribution of NS1 ΔCTE in different poultry species
Interestingly, only NS207 and NS219 were first observed in wild birds; in H10N8 in long-tailed duck and in H4N6 in budgerigar (). Other forms emerged in chickens, ducks, geese, and turkeys. Importantly, most of the avian viruses with NS1-ΔCTE were isolated in chickens, turkeys, quails and other non-aquatic bird species. In addition, in each of these species, NS1-ΔCTE viruses represented a large fraction of the total number of isolates in that species, ranging from ˜30% (turkeys) to 45% (chickens) to >70% (partridges) (). It is not clear from this study whether viruses in partridge, pheasants, pigeons and ostriches acquired this deletion upon or after transmission from other hosts (e.g., chickens or wild birds). Moreover, less prevalence was observed in Guinea fowl and some wild birds (e.g. sparrows, sandpiper, turnstone, moorhen).
Figure 2. Prevalence of truncations in the C-terminus of NS1 of AIV among different bird species Blue columns represent the total sequences exhibiting truncation in the C-terminus of NS1 (NS1 ΔCTE) in AIV of a given species to the total AIV sequences with truncation (2331 analyzed in this study). Red columns refer to the total NS1 ΔCTE sequences to total sequences collected from the respective species. Results are shown in percentages (vertical axes).
Phylogenetic analysis
Phylogenetic analysis of the NS1 gene indicated presence of 2 distinct alleles, A and B, which further subdivided into Eurasian and American lineages. The Italian H7N1 clustered in allele B, the less common allele of NS1, in the Eurasian lineage (b), whereas H9N2 and common reassortants (e.g., H7N9, H5N1, H5N6 and H6N2) as well as the Pennsylvanian H5N2/1983–1984 viruses with NS217 belonged to allele A. Allele B is also carried by viruses from H5N2 and H7N2 outbreaks in Mexico and USA in 1994/1995 and 1990s-2000s, respectively and by the earlier H5N1 viruses in Hong Kong 1996/1997. Most of the NS1 sequences of AIV in allele B are of full-length (230aa).
Generation of recombinant viruses and mutants
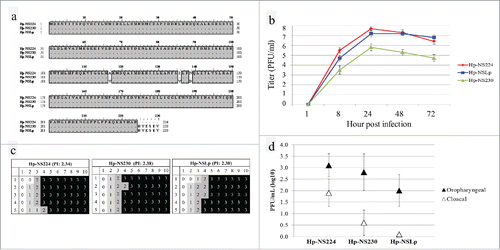
In an attempt to analyze whether NS1 truncation could impact the viral phenotype, we genetically engineered 3 recombinant viruses harbouring distinct variants of NS1. All gene segments of the Italian H7N1 Hp and the NS gene segment of Lp were successfully cloned and recombinant viruses were rescued in 293T/MDCK cell cultures. Three recombinant viruses were generated containing 7 segments (segments 1 – 7) from Hp and variable NS1: Hp-NS224 (contains NS from Hp), Hp-NS230 (NS from Hp with extended CTE) and Hp-NSLp (contains NS from Lp containing 230 aa) (). Of note, both Hp-NS224 and Hp-NS230 also possessed 3 substitutions relative to Hp-NSLp (NS230 of the LP virus): V117A, V136I and D139I. All viruses were propagated once in SPF ECE and full genome sequences of rescued viruses revealed no undesired mutations.
Figure 3. In-vitro and in vivo characterization of recombinant HPAI H7N1 viruses generated in this study (a) Sequence of the NS1 protein of viruses generated in this study. (b) Replication kinetics in CEK cells after 2 independent trials after 1, 8, 24, 48 and 72 hours post infection. (c) Pathogenicity index after clinical examination of oculonasally infected 6-week-old specific-pathogen-free white leghorn chickens with 104.5 PFU/ml of the indicated viruses. Healthy chickens were scored “0.” Chickens showing one clinical sign (depression, ruffled feathers, diarrhea, sneezing, coughing, conjunctivitis, discharges, or cyanosis of comb, wattle or shanks) were scored “1” and were defined as ill. Severely ill chickens showing 2 or more clinical signs were scored “2,” whereas dead chickens were scored “3.” The pathogenicity index (PI) was calculated as the mean sum of the daily arithmetic mean values divided by 10; the number of observation days. (d) Virus excretion in oropharyngeal and cloacal swabs at 4 dpi was done by plaque assay and the mean titer ± standard deviation of positive birds was expressed in plaque forming unit per ml (PFU/ml).
Replication kinetics and plaque morphology
Viruses were grown in CEK at MOI of 0.001 for 1, 8, 24, 48 and 72 hours. All viruses reached the maximum rate of replication after 24 hours post infection. However, Hp-NS230 replicated to significantly lower levels at each time point as indicated by plaque assay. Meanwhile, Hp-NS224 and Hp-NSLp had a comparable higher titer (). On MDCK cells, Hp-NS230 formed significantly smaller plaques than Hp-NS224 and Hp-NSLp.
Animal experiment
After infection of 6-week-old SPF chickens via oculonasal route, most of chickens developed moderate clinical signs at day 2 post-infection (pi) mainly depression, cyanosis of unfeathered skin, comb, wattles, conjunctivitis, and/or facial edema. At day 3 pi, 4/5, 5/5 and 4/5 of birds infected with Hp-NS224, Hp-NS230 and Hp-NSLp, respectively, developed severe clinical signs. At 4 dpi, 4/5 chickens infected with Hp-NS224 or Hp-NS230 and 2/5 of Hp-NSLp died, whereas remaining birds developed severe clinical signs and were humanely euthanized then scored 3 at day 5 pi. No significant difference in pathogenicity for all viruses was observed with average clinical scores of 2.34, 2.38 and 2.30, respectively ().
Virus excretion
Quantification of viral excretion in oropharyngeal and cloacal swabs at 4 dpi was done by plaque assay on MDCK cells without addition of exogenous trypsin. Quantity of Hp-NS224 virus excreted in oropharyngeal swabs was higher than in cloacal swabs. All oropharyngeal swabs were positive, and 5/5, 4/5 and 1/5 of birds infected with Hp-NS224, Hp-NS230 and Hp-NSLp excreted virus in cloacal swabs, respectively. Compared to the Hp-NS224 infected birds, Hp-NSLp infected animals excreted significantly lower titers in oropharyngeal and cloacal swabs ().
Pathology
At autopsy, regardless of the group, chickens exhibited tenacious mucus in the pharynx, hemorrhagic conjunctival lymphatic follicles, and severe atrophy of the bursa and the thymus. Besides pericardial petechiation, hemorrhagic-necrotizing proventriculitis and gastritis of variable degree was found. Almost all animals showed necrosis of the pancreas parenchyma, with a severe degree in Hp-NS224, and variable degree in Hp-NSLp, or Hp-NS230 infected chickens. A subcutaneous edema of the head was evident after Hp-NSLp and Hp-NS224 infection.
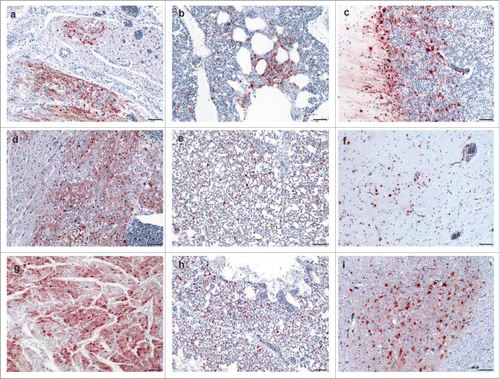
Generally, microscopic examination identified a widespread tropism to blood vessels in all chickens with antigen detection in the endothelial cells of almost all organs investigated (, for details see supplementary Table S2). Based on semi-quantification the NS from Hp-NSLp or extension of CTE of the NS1 (Hp-NS230) resulted in less frequent signals within endothelial cells compared to Hp-NS224, especially in the trachea, brain, and heart. In the lungs, influenza virus antigen was detected in pneumocytes to a high extent in most of the birds. However, the parabronchial epithelium was variably affected (3/5 Hp-NS224; 2/5 Hp-NS230; 0/5 Hp-NSLp) just as the bronchial epithelium (4/5 Hp-NS224; 0/5 Hp-NS230; 2/5 Hp-NSLp). In cardiac myocytes multifocal to diffuse signals were observed in all birds (), similar results were found in the liver (hepatocytes) and pancreas (acinar cells). Variation in immunoreactivity was observed in the brain of infected birds: whereas viral antigen was constantly found in neurons and glial cells of all animals with a marked, diffuse distribution after Hp-NS224 infection, meningeal cells were infrequently affected (5/5 Hp-NS224; 4/5 Hp-NS230; 1/5 Hp-NSLp).Viral antigen was rarely present in neurons of the peripheral nervous system in the intestine, heart and pancreas. Remarkably, the number of positive birds and extent of virus antigen in mesenchymal and endothelial cells of the spleen and endothelial cells in the duodenum after Hp-NS230 infection was lower than in other birds. The reason for this different tropism of Hp-NS230 to spleen and endothelium is unknown.
Figure 4. Detection of avian influenza virus nucleoprotein in infected chickens using immunohistochemistry Detection of influenza- nucleoprotein by immunohistochemistry (ABC method, intranuclear and intracytoplasmis bright red antigen signal by 3-amino-9-ethyl-carbazol chromogen and haematoxylin (blue) counterstain): heart (a, d, g), lung (b, e, h) and brain (c, f, i) samples obtained from birds infected with Hp-NS224 (a-c), HP- NS230 (d-f) and HP-NSLp (g-i) at 4 d post infection; for detailed data on the full tropism to different organs examined in this study readers are refered to Table S2. Bar = 50 µm.
Discussion
NS is a multifunctional protein. In addition to the well-known role as antiviral antagonist, it can affect virus replication, host-range, virulence and tropism mainly shown in mammal models or in-vitro, whereas only few studies have been performed in poultry. Citation3,12,13,21 Although truncation of the CTE of NS1 has been previously reported using a limited number of sequences,Citation9,14,22 little is known about the prevalence and distribution of NS1 ΔCTE in avian influenza viruses of all subtypes. Moreover, the impact of NS1 truncation on virulence particularly the HPAIV H7 viruses in poultry has not been adequately studied.
In this study, we analyzed all available NS1 protein sequences from H1-H16 AIV isolated between 1902 and 2015 from avian species deposited in the GenBank database. 13 different variants of NS1 ΔCTE were observed. Truncation was first reported in AIV, both LPAIV and HPAIV, from domestic poultry and was not common in the wild bird reservoir. Thus, ΔCTE is probably advantageous for the adaptation of AIV to domestic poultry. Generally, it is notable that (1) some forms of truncation emerged for a limited time and disappeared thereafter. This may be due to elimination of infected birds, adverse effect of ΔCTE on virus replication, reversion of the premature stop codon to a sense codon or other unknown reasons. (2) NS1 truncation occurred mostly in countries with a history of AIV endemicity in poultry (e.g. China, Vietnam, USA, Italy); mainly in the South-eastern Asia and North-eastern part of the USA (Fig. S1).Thus, truncation of NS1 appeared to be the result of extended circulation of AIV in poultry. Among other ΔCTE, NS217 and NS224 were the most prevalent forms. NS217 was reported frequently in LPAIV of non-H5/H7 subtypes particularly H9N2, H10 and H6 viruses which are known to be widespread and mostly (semi)endemic in land-based poultry. Thus, it seems to be less likely a virulence marker for AIV. The second most prevalent form was NS224 which is unique to H7N1 viruses isolated in Italy in 1999/2000. Since it was seen only in HPAIV it may play a role in virulence of the HPAIV in chickens. Additional reasons for the assumption that NS224 in H7N1 viruses may be a virulence marker are (1) the prevalence of NS1 truncation in allele B viruses is low (e.g., NS1 of Mexican H5N2/1994 and American H7N2/1990s-2000s are 230 aa in length) and (2) the H7N1 virus was isolated from a wide range of hosts including pheasants, quails and ostriches.Citation23
To test for influence of the NS1 variations on virulence, 3 isogenic HPAI viruses were reverse-engineered to carry NS from Hp (224 aa in length), NS from HP with extended C-terminal domain (230 aa), or Lp (230 aa). HPAIV replication was impaired in avian cell culture after extension of the CTE which is in accordance with reduced virus replication in chicken fibroblast monolayers due to extension of NS1 CTE of an Italian HPAIV H7N1 of ostrich origin.Citation19 Although, the Lp NS contain also a full NS, it did not affect virus replication which may be due to synergism of other mutations in NS1 (e.g. V136I, and D139N). On the other hand, an Italian LPAIV H7N1 with deletion of226ESEV230 replicated as well as the LPAIV with naïve NS in chicken embryo fibroblasts. Citation20 Another study showed that extension of NS1 of an HPAIV H5N1 did not attenuate viral replication on chicken fibroblasts.Citation13 Accordingly, the effect of NS1 ΔCTE on virus replication in-vitro is most likely to be cell- and strain-dependent.
Contribution of the NS1 to the virulence of an H5N1 virus Citation4 as well as to the virulence of a historic HPAIV H7N1 with NS gene segment from HPAIV H5N1 Citation24 has been described. Our findings indicate that NS1 variation did not play a significant role in virulence of the Italian H7N1 viruses in chickens and major virulence markers reside outside the NS1 gene. This is supported by our recent findings that virulence of this HPAIV H7N1 is modulated by amino acids in the HA2 domain in addition to the multibasic cleavage site. Citation25 Likewise, truncation of NS1 from an Italian LPAIV H7N1 did not increase virulence of the virus in chickens Citation20 and extension of the C-terminal end of NS1 of an HPAIV H5N1 did not decrease virulence in chickens.Citation13
In our study, compared to the Hp virus, the NS1 mutants exhibited impaired virus excretion from infected birds at 4dpi particularly via the cloacal route. A reduced amount of virus excretion due to NS mutations may reduce bird-to-bird transmission Citation26 which, however, was not studied in our experiment. Moreover, spread of influenza viruses to the brain usually occurs by the hematogenic route and also through the olfactory nerve.Citation27 Decreased replication in the brain in birds infected with Hp-NS230 or Hp-NSLp may be due to decreased tropism to endothelial cells; which is an important feature of HPAIV in domestic poultry.Citation28 These results indicated that NS1 can (1) modulate tropism to different tissues limiting systemic spread of the virus and (2) truncation is involved in this restriction but also other mutations in the NS1 play an additional role. Previously, it has been shown that the NS1 gene facilitates spread of HPAIV H5N1 from the lung to the brain of experimentally infected mice.Citation29
Taken together; our comprehensive analysis of the NS1 gene segments and animal experiments support that truncation of the C-terminal end of NS1 protein is not an essential virulence determinant of AIV, particularly for the Italian HPAIV H7N1, in chickens, however, it may be a host adaptation marker as indicated by efficient replication in-vitro and in-vivo.
Materials and Methods
Sequence comparison and molecular analysis
All available full-length NS1 sequences from avian species were retrieved from the Influenza Virus Resource database and analyzed for the prevalence and distribution of NS1ΔCTE. A total of 13026 NS1 protein sequences from H1-H16/N1-N9 AIV from birds excluding laboratory strains were analyzed. For the Italian H7N1, 167 NS1 protein sequences of H7N1 isolated between 1999 to 2002 from poultry in Italy were retrieved from GenBank (n= 159) and Global Initiative on Sharing All Influenza Data (GISAID) (n=8). Budt, M.; Schweiger, B.; Wolff, T. from the Robert Koch Institut and anonymous submitter from Istituto Zooprofilattico Sperimentale Delle Venezie are acknowledged for depositing the NS1 protein sequences in GISAID. All sequences were aligned by MAFFT,Citation30 visualized by BioEdit 7.1.7Citation31 and further edited manually. Amplification of 8 gene segments of an HPAIV H7N1 and the NS segment of an LPAIV H7N1 (see below) used in this study for cloning was done after plaque purification.Citation32 Target segments were successfully amplified using a one-step RT-PCR Citation32 and amplicons were purified from agarose gel slices using QIAquick Gel Extraction Kit (Qiagen, Cat. 28704). Sequences were generated using an ABI BigDye Terminator v.1.1 Cycle Sequencing Kit (Applied Biosystems, Cat. 4337451). Gene sequences generated in this study are available on the GISAID database under accession numbers: EPI651417 to EPI651424 for HPAIV and EPI624440 for Lp NS gene. Phylogenetic analysis of available NS1 genes from AIV in the GenBank was generated after removal of identical sequences using Maximum Likelihood algorithm with 1000 bootstrap replications as implemented in IQ-TREE Citation33 and was further visualized and edited using FigTree (http://tree.bio.ed.ac.uk/software/figtree/) and Inkscape 0.91 (www.inkscape.org).
Viruses and cells
Two H7N1 viruses, LPAIV A/chicken/Italy/473/1999 (referred to here as Lp) and HPAIV A/chicken/Italy/445/1999 (referred to as Hp), were obtained from the repository of the Friedrich-Loeffler-Institut (FLI). Madin-Darby canine kidney cells (MDCK) were used for titration and purification by plaques assay. A mixture of 293T cells and MDCK cells (1:5) was used for rescue of viruses by reverse genetics. Both cell lines were obtained from the Cell Culture Collection in Veterinary Medicine of the FLI. Monolayers of primary chicken embryo kidney (CEK) cells were prepared from the kidneys of 18-day-old SPF chicken embryos and used for replication kinetics.
Virus isolation and propagation
All viruses except Lp were handled in biosafety level 3 (BSL3) facilities at the FLI. They were propagated in the allantoic cavity of 9–11 d old SPF embryonated chickens eggs (ECE) (Lohmann Tierzucht, Cuxhaven, Germany) as described.Citation34 Inoculated eggs were incubated at 37°C for 5 d and assessed daily using light source to determine embryo viability. Eggs with dead embryos or those harvested at the end of the incubation period were kept at 2 –8°C for 24 hours. The allantoic fluid was then harvested. The hemagglutinating activity was determined using the standard hemagglutination test after mixing with 1% chicken erythrocytes and phosphate buffer saline (PBS) in U-bottom shaped 96-well plates (Greiner, Cat. 650101).Citation34 Virus containing allantoic fluids with HA titer over 16 (24) were streaked on Columbia agar with 5% sheep blood (VWR International) and incubated at 37°C for 48–72 hours. Bacteria-free allantoic fluid was pooled, aliquoted, labeled and kept at −80°C until further use.
Generation of recombinant viruses and mutants
Viral RNA was extracted using QIAamp® Viral RNA Mini Kit according to the manufacturer recommendation (Qiagen, Cat. 52904). All eight gene segments of Hp and NS of Lp were amplified as previously described Citation32,35 using Omniscript RT Kit (Qiagen, Cat. 205111) and Phusion High-Fidelity DNA Polymerase (New England BioLabs, Cat. M0530S) following the manufacturers' recommendations. Amplicons were purified in 1.5% agarose gel (wt/vol) and extracted using QIAquick Gel Extraction Kit (Qiagen, Cat. 28704). Each gene segment was cloned into pHWSccdB plasmid according to the published protocol.Citation36 Plasmids containing the insert were used for transfection of competent E.coli strains SURE2® (Agilent Technologies, Cat. 200152) or One Shot® TOP10 (Invitrogen, C4040-10) as per the manufacturers' protocols. Transfected bacteria were plated on Luria-Bertani (LB) agar supplemented with 100 µg/ml ampicillin, and plates were incubated for 16 hours at 37°C. Single colonies were picked and propagated overnight in 2 – 4 ml LB broth with ampicillin overnight at 37°C. Ultrapure transfection-grade plasmids were isolated from these cultures using QIAGEN Plasmid Mini and Midi Kits (Qiagen, Cats. 12125 and 12143) following the producer's manuals. Insertion of AIV gene segments was confirmed after digestion of each plasmid with NheI (New England BioLabs, Cat. R0131) and sequence analysis. Concentration of each plasmid was determined in Nanodrop ND-1000 spectrophotometer (Peqlab GmbH, Germany). Recombinant viruses were rescued by transfection of 293T/MDCK cells using 1 µg of each of the 8 plasmids in presence of Lipofectamine™ 2000 Reagent and Opti-MEM® (Invitrogen, Cats. 11668–030 and 31985–070).Citation37 Introduction of the selected mutations into the NS1 coding region of Hp was done using site-directed mutagenesis according to the QuikChange™ protocol (Agilent Technologies, Cat. 200518). All viruses and/or mutants were rescued and propagated in SPF ECE.Citation37 Rescued viruses were sequenced to confirm presence of the desired mutations and to exclude any other genetic alterations.Citation32
Replication kinetics
Replication efficiency of recombinant viruses was assessed by infecting CEK at a multiplicity of infection (MOI) of 0.001 for one hour. Extracellular virions were inactivated by addition of citrate buffer pH 3.0 for 2 minutes. Cells were then washed twice with PBS. The cells were overlaid with MEM supplemented with 0.2% bovine serum albumin (BSA) (Sigma) and kept at 37°C/5% CO2 for 1, 8, 24, 48 and 72 hours. The cells and supernatant were harvested at given time points and stored at −80°C until titration by plaque assay as described below. Results are shown as mean ± standard deviation of 2 independent trials for each time point.
Plaque assay
Plaque assay was used for purification of wild type Lp and Hp viruses, titration of rescued viruses expressed as plaque forming units per mL (PFU/mL) and determination of plaque phenotypes. Ten-fold serial dilutions of each virus were incubated with MDCK cells in 6-well plates for one hour at 37°C. Infected cells were washed with PBS twice then overlaid by 1.8% agar (BD, Cat. 214010) in DMEM supplemented with 4% BSA. For the growth of Lp virus 2 μg/ml of N-tosyl-L-phenylalanine chloromethyl ketone (TPCK)-treated trypsin (Sigma, Cat. T1426) was added. All plates were incubated at 37°C/5% CO2 for 3 d and then fixed with formaldehyde (10%) containing crystal violet (0.1%).Citation38 Plaques were counted and plaque sizes were measured by using Nikon NIS-Elements software.
Animal experiments
Animal experiments were conducted in the BSL3 animal facilities of the FLI following the German Regulations for Animal Welfare after approval by the authorized ethics committee of the State Office of Agriculture, Food Safety, and Fishery in Mecklenburg – Western Pomerania (LALLF M-V) under the registration number TSD/7221.3–1.1–018/07. Fifteen 6-week old white leghorn SPF chickens were purchased from Lohmann Tierzucht (Cuxhaven, Germany). Each recombinant virus was used to infect 5 birds per group with 104.5 PFU/bird oculonasally. All birds were monitored daily for clinical signs (depression, respiratory disorders, diarrhea, cyanosis of the comb, wattles or shanks, facial edema and nervous signs) over a 10 d observation period. Clinical scoring was done following the standard protocolCitation34 where healthy birds were scored (0), sick birds showing one of one of the mentioned clinical signs were scored (1), severely sick birds showing 2 or more signs were scored (2) and dead birds were scored (3). Moribund birds that were not able to eat or drink were humanely euthanized using isoflurane (CP-Pharma, Germany) inhalation and scored as dead on the next observation day. The pathogenicity index (PI) was calculated as the sum of the daily arithmetic mean values of all infected birds divided by 10 (the number of observation days) with a final range from 0 (avirulent) to 3 (highly virulent).
Virus excretion
Cloacal and oropharyngeal swabs were collected from all birds at 4 dpi. Swabs were immersed in 1.5 ml DMEM containing antibiotic in sterile safe-lock Eppendorf, mixed by vigorous vortexing for 30 seconds and kept at −80°C until use. The amount of virus excretion was determined by titration of the swabs samples using plaque assay as described above.
Histopathology and immunohistochemistry
Samples from trachea, lungs, heart, proventriculus, gizzard, duodenum, cecum, pancreas, liver, kidney, spleen, bursa of Fabricius, thymus and brain from dead birds were used to study the viral tropism and lesions after infection by different recombinant viruses. Samples were fixed in 10% neutral buffered formalin and paraffin-embedded sections were used for immunohistochemistry with a primary antibody directed against the influenza-nucleoprotein (NP, 1:750) and biotinylated goat anti-rabbit IgG1 (Vector, Burlingame, BA-1000) as secondary antibody (1:200) as previously published.Citation39 The extent of nucleoprotein antigen detection was semi-quantified by scoring (blinded study) on a 0 to 4+ severity scale for tissues: 0 = (−) negative; 1+ single cells, 2+ scattered foci, 3+ numerous foci, 4+ coalescing foci or diffuse and on a scale of 0 to 3+ for endothelium: 0 = (−) negative; 1+ single blood vessel, 2+ multiple blood vessels, 3+ diffuse.
Statistics
Virus titers in CEK cell culture as well as the amount of influenza virus nucleoprotein in processed tissues of infected birds using IHC were compared using an ANOVA with post hoc Tukey tests. Moreover, Kruskal-Wallis and Wilcoxon tests with Bonferroni correction were used to assess significance differences in plaque morphology and virus titers in swabs collected 4 dpi. Comparison of the severity of clinical symptoms between groups was assessed by comparing the mean clinical score per bird during a 10 d observation period. Exact Fisher tests were used to compare number of birds with positive swabs as well as frequency of ΔCTE per HA-subtypes, NA-subtypes or species. Results were considered significant at a p-value < 0.05. All analysis was done using R version 2.14.0 from the R Foundation for Statistical Computing (http://www.r-project.org).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary files
Download Zip (687.7 KB)Acknowledgments
The authors would like to thank Frank Klipp, Thorsten Arnold and Doreen Fiedler for their support in the animal experiment, Silvia Schuparis for excellent technical assistance, Heinz-Günther Strebelow for sequencing and Timm C. Harder from the FLI and Ilaria Capua, Istituto Zooprofilattico Sperimentale delle Venezie, Padova, Italy for providing the viruses.
References
- Alexander DJ. A review of avian influenza in different bird species. Vet Microbiol 2000; 74:3-13; PMID:10799774; http://dx.doi.org/10.1016/S0378-1135(00)00160-7
- Khaliq Z, Leijon M, Belak S, Komorowski J. A complete map of potential pathogenicity markers of avian influenza virus subtype H5 predicted from 11 expressed proteins. BMC Microbiol 2015; 15:128; PMID:26112351; http://dx.doi.org/10.1186/s12866-015-0465-x
- Imai H, Shinya K, Takano R, Kiso M, Muramoto Y, Sakabe S, Murakami S, Ito M, Yamada S, Le MT, et al. The HA and NS genes of human H5N1 influenza A virus contribute to high virulence in ferrets. PLoS Pathogens 2010; 6:e1001106; PMID:20862325; http://dx.doi.org/10.1371/journal.ppat.1001106
- Li Z, Jiang Y, Jiao P, Wang A, Zhao F, Tian G, Wang X, Yu K, Bu Z, Chen H. The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J Virol 2006; 80:11115-23; PMID:16971424; PMID:16971424; http://dx.doi.org/10.1128/JVI.00993-06
- Ayllon J, Domingues P, Rajsbaum R, Miorin L, Schmolke M, Hale BG, Garcia-Sastre A. A single amino acid substitution in the novel H7N9 influenza A virus NS1 protein increases CPSF30 binding and virulence. J Virol 2014; 88:12146-51; PMID:25078692; http://dx.doi.org/10.1128/JVI.01567-14
- Long JX, Peng DX, Liu YL, Wu YT, Liu XF. Virulence of H5N1 avian influenza virus enhanced by a 15-nucleotide deletion in the viral nonstructural gene. Virus Genes 2008; 36:471-8; PMID:18317917; http://dx.doi.org/10.1007/s11262-007-0187-8
- Lin YP, Shu LL, Wright S, Bean WJ, Sharp GB, Shortridge KF, Webster RG. Analysis of the influenza virus gene pool of avian species from southern China. Virology 1994; 198:557-66; PMID:8291238; http://dx.doi.org/10.1006/viro.1994.1067
- Kawaoka Y, Gorman OT, Ito T, Wells K, Donis RO, Castrucci MR, Donatelli I, Webster RG. Influence of host species on the evolution of the nonstructural (NS) gene of influenza A viruses. Virus Res 1998; 55:143-56; PMID:9725667; http://dx.doi.org/10.1016/S0168-1702(98)00038-0
- Suarez DL, Perdue ML. Multiple alignment comparison of the non-structural genes of influenza A viruses. Virus Res 1998; 54:59-69; PMID:9660072; http://dx.doi.org/10.1016/S0168-1702(98)00011-2
- Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol 2008; 89:2359-76; PMID:18796704; http://dx.doi.org/10.1099/vir.0.2008/004606-0
- Ma W, Brenner D, Wang Z, Dauber B, Ehrhardt C, Hogner K, Herold S, Ludwig S, Wolff T, Yu K, et al. The NS segment of an H5N1 highly pathogenic avian influenza virus (HPAIV) is sufficient to alter replication efficiency, cell tropism, and host range of an H7N1 HPAIV. J Virol 2010; 84:2122-33; PMID:20007264; http://dx.doi.org/10.1128/JVI.01668-09
- Soubies SM, Volmer C, Croville G, Loupias J, Peralta B, Costes P, Lacroux C, Guerin JL, Volmer R. Species-specific contribution of the four C-terminal amino acids of influenza A virus NS1 protein to virulence. J Virol 2010; 84:6733-47; PMID:20410267; http://dx.doi.org/10.1128/JVI.02427-09
- Zielecki F, Semmler I, Kalthoff D, Voss D, Mauel S, Gruber AD, Beer M, Wolff T. Virulence determinants of avian H5N1 influenza A virus in mammalian and avian hosts: role of the C-terminal ESEV motif in the viral NS1 protein. J Virol 2010; 84:10708-18; PMID:20686040; http://dx.doi.org/10.1128/JVI.00610-10
- Dundon WG, Milani A, Cattoli G, Capua I. Progressive truncation of the Non-Structural 1 gene of H7N1 avian influenza viruses following extensive circulation in poultry. Virus Res 2006; 119:171-6; PMID:16464514; http://dx.doi.org/10.1016/j.virusres.2006.01.005
- Abdelwhab el SM, Veits J, Mettenleiter TC. Genetic changes that accompanied shifts of low pathogenic avian influenza viruses toward higher pathogenicity in poultry. Virulence 2013; 4:441-52; PMID:23863606; http://dx.doi.org/10.4161/viru.25710
- Capua I, Mutinelli F, Marangon S, Alexander DJ. H7N1 avian influenza in Italy (1999 to 2000) in intensively reared chickens and turkeys. Avian Pathol 2000; 29:537-43; PMID:19184849; http://dx.doi.org/10.1080/03079450020016779
- Monne I, Fusaro A, Nelson MI, Bonfanti L, Mulatti P, Hughes J, Murcia PR, Schivo A, Valastro V, Moreno A, et al. Emergence of a highly pathogenic avian influenza virus from a low-pathogenic progenitor. J Virol 2014; 88:4375-88; PMID:24501401; http://dx.doi.org/10.1128/JVI.03181-13
- Fusaro A, Tassoni L, Hughes J, Milani A, Salviato A, Schivo A, Murcia PR, Bonfanti L, Cattoli G, Monne I. Evolutionary trajectories of two distinct avian influenza epidemics: Parallelisms and divergences. Infect, Genet Evol : J Mol Epidemiol Evolution Genet Infect Dis 2015; 34:457-66; PMID:26003682; http://dx.doi.org/10.1016/j.meegid.2015.05.020
- Keiner B, Maenz B, Wagner R, Cattoli G, Capua I, Klenk HD. Intracellular distribution of NS1 correlates with the infectivity and interferon antagonism of an avian influenza virus (H7N1). J Virol 2010; 84:11858-65; PMID:20844052; http://dx.doi.org/10.1128/JVI.01011-10
- Soubies SM, Hoffmann TW, Croville G, Larcher T, Ledevin M, Soubieux D, Quere P, Guerin JL, Marc D, Volmer R. Deletion of the C-terminal ESEV domain of NS1 does not affect the replication of a low-pathogenic avian influenza virus H7N1 in ducks and chickens. J Gen Virol 2013; 94:50-8; PMID:23052391; http://dx.doi.org/10.1099/vir.0.045153-0
- Pica N, Langlois RA, Krammer F, Margine I, Palese P. NS1-truncated live attenuated virus vaccine provides robust protection to aged mice from viral challenge. J Virol 2012; 86:10293-301; PMID:22787224; http://dx.doi.org/10.1128/JVI.01131-12
- Dundon WG, Capua I. A closer look at the NS1 of influenza virus. Viruses 2009; 1:1057-72; PMID:21994582; http://dx.doi.org/10.3390/v1031057
- Mutinelli F, Capua I, Terregino C, Cattoli G. Clinical, gross, and microscopic findings in different avian species naturally infected during the H7N1 low- and high-pathogenicity avian influenza epidemics in Italy during 1999 and 2000. Avian Dis 2003; 47:844-8; PMID:14575075; http://dx.doi.org/10.1637/0005-2086-47.s3.844
- Vergara-Alert J, Busquets N, Ballester M, Chaves AJ, Rivas R, Dolz R, Wang Z, Pleschka S, Majo N, Rodriguez F, et al. The NS segment of H5N1 avian influenza viruses (AIV) enhances the virulence of an H7N1 AIV in chickens. Vet Res 2014; 45:7; PMID:24460592; http://dx.doi.org/10.1186/1297-9716-45-7
- Abdelwhab EM, Veits J, Tauscher K, Ziller M, Teifke J, Stech J, Mettenleiter TC. A unique multibasic proteolytic cleavage site and three mutations in the HA2 domain confer high virulence of H7N1 avian influenza virus in chickens. J Virol 2016; 90:400-11; PMID:26491158; http://dx.doi.org/10.1128/JVI.02082-15
- Kong W, Liu L, Wang Y, He Q, Wu S, Qin Z, Wang J, Sun H, Sun Y, Zhang R, et al. C-terminal elongation of NS1 of H9N2 influenza virus induces a high level of inflammatory cytokines and increases transmission. J Gen Virol 2015; 96:259-68; PMID:25326314; http://dx.doi.org/10.1099/vir.0.071001-0
- van Riel D, Verdijk R, Kuiken T. The olfactory nerve: a shortcut for influenza and other viral diseases into the central nervous system. J Pathol 2015; 235:277-87; PMID:25294743; http://dx.doi.org/10.1002/path.4461
- Short KR, Veldhuis Kroeze EJ, Reperant LA, Richard M, Kuiken T. Influenza virus and endothelial cells: a species specific relationship. Front Microbiol 2014; 5:653; PMID:25520707; http://dx.doi.org/10.3389/fmicb.2014.00653
- Spesock A, Malur M, Hossain MJ, Chen LM, Njaa BL, Davis CT, Lipatov AS, York IA, Krug RM, Donis RO. The virulence of 1997 H5N1 influenza viruses in the mouse model is increased by correcting a defect in their NS1 proteins. J Virol 2011; 85:7048-58; PMID:21593152; http://dx.doi.org/10.1128/JVI.00417-11
- Katoh K, Standley DM. MAFFT: iterative refinement and additional methods. Methods Mol Biol 2014; 1079:131-46; PMID:24170399; http://dx.doi.org/10.1007/978-1-62703-646-7_8
- Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999; 41:95-8; PMID:1726757
- Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 2001; 146:2275-89; PMID:11811679; http://dx.doi.org/10.1007/s007050170002
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015; 32:268-74; PMID:25371430; http://dx.doi.org/10.1093/molbev/msu300
- Alexander, D. Avian Influenza: OIE Terrestrial Manual. Chapter 2.3.4. Available online at: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.04_AI.pdf 2015 (Last accessed 04-04-2016).
- Kreibich A, Stech J, Mettenleiter TC, Stech O. Simultaneous one-tube full-length amplification of the NA, NP, M, and NS genes of influenza A viruses for reverse genetics. J Virolog Methods 2009; 159:308-10; PMID:19406155; http://dx.doi.org/10.1016/j.jviromet.2009.04.020
- Stech J, Stech O, Herwig A, Altmeppen H, Hundt J, Gohrbandt S, Kreibich A, Weber S, Klenk HD, Mettenleiter TC. Rapid and reliable universal cloning of influenza A virus genes by target-primed plasmid amplification. Nucleic Acids Res 2008; 36:e139; PMID:18832366; http://dx.doi.org/10.1093/nar/gkn646
- Stech O, Veits J, Weber S, Deckers D, Schroer D, Vahlenkamp TW, Breithaupt A, Teifke J, Mettenleiter TC, Stech J. Acquisition of a polybasic hemagglutinin cleavage site by a low-pathogenic avian influenza virus is not sufficient for immediate transformation into a highly pathogenic strain. J Virol 2009; 83:5864-8; PMID:19297482; http://dx.doi.org/10.1128/JVI.02649-08
- Gohrbandt S, Veits J, Hundt J, Bogs J, Breithaupt A, Teifke JP, Weber S, Mettenleiter TC, Stech J. Amino acids adjacent to the haemagglutinin cleavage site are relevant for virulence of avian influenza viruses of subtype H5. J Gen Virol 2011; 92:51-9; PMID:20881092; http://dx.doi.org/10.1099/vir.0.023887-0
- Breithaupt A, Kalthoff D, Dale J, Bairlein F, Beer M, Teifke JP. Neurotropism in blackcaps (Sylvia atricapilla) and red-billed queleas (Quelea quelea) after highly pathogenic avian influenza virus H5N1 infection. Vet Pathol 2011; 48:924-32; PMID:20974871; http://dx.doi.org/10.1177/0300985810386467
