ABSTRACT
The Cek1 MAP kinase (MAPK) mediates vegetative growth and cell wall biogenesis in the fungal pathogen Candida albicans. Alterations in the fungal cell wall caused by a defective Cek1‑mediated signaling pathway leads to increased β‑1,3‑glucan exposure influencing dectin‑1 fungal recognition by immune cells. We show here that cek1 cells also display an increased exposure of α‑1,2 and β‑1,2‑mannosides (α‑M and β‑M), a phenotype shared by strains defective in the activating MAPKK Hst7, suggesting a general defect in cell wall assembly. cek1 cells display walls with loosely bound material as revealed by transmission electron microscopy and are sensitive to tunicamycin, an inhibitor of N‑glycosylation. Transcriptomal analysis of tunicamycin treated cells revealed a differential pattern between cek1 and wild type cells which involved mainly cell wall and stress related genes. Mapping α‑M and β‑M epitopes in the mannoproteins of different cell wall fractions (CWMP) revealed an important shift in the molecular weight of the mannan derived from mutants defective in this MAPK pathway. We have also assessed the role of galectin‑3, a member of a β‑galactoside‑binding protein family shown to bind to and kill C. albicans through β‑M recognition, in the infection caused by cek1 mutants. Increased binding of cek1 to murine macrophages was shown to be partially blocked by lactose. Galectin-3−/− mice showed increased resistance to fungal infection, although galectin-3 did not account for the reduced virulence of cek1 mutants in a mouse model of systemic infection. All these data support a role for the Cek1‑mediated pathway in fungal cell wall maintenance, virulence and antifungal discovery.
Introduction
Candida albicans is an opportunistic fungus that forms part of the human microbiota. Upon alteration of the host defenses, this microbe may disseminate within the human body and gain access to internal organs causing severe infections. Although several virulence factors have been identified in the last years,Citation1 cell wall components are considered to be most relevant: their external location makes them essential in processes such as adhesion, colonization and immune recognition, playing therefore a major role during infection.Citation2
The C. albicans cell wall is a complex dynamic structure based on a core assembly of β‑1,3‑glucan (covalently linked to β‑1,6‑glucan) and chitin and an outer layer of mannose‑glycosylated proteins.Citation3,4 Chitin, a polymer of β‑1,4‑linked N‑acetylglucosamine, is bound to a network of glucan which is usually masked by the cell wall outer layer and is only exposed due to certain cell wall alterations.Citation5 The outer layer of the fungal cell wall is mainly composed of mannoproteins and phosphopeptidomannan (PPM), which is a polymer of O‑linked mannoses (α‑1,2 type or α‑1,2/α‑1,3 type depending on the fungus) and N‑linked mannoses (an inner core elongated by α‑1,6 linear chain with branches of α‑1,2 and α‑1,3 mannose). C. albicans PPM and mannoproteins carry α‑ and β‑1,2‑mannosides; however, while C. albicans and S. cerevisiae α‑linked mannosides are rather similar, the β‑1,2‑type of linkage is specific to the pathogenic yeast, contributing to its virulence and immunomodulatory responses.Citation6-8 β‑1,2‑mannosides (β‑M) are also a main component of the glycosphingolipid phospholipomannan (PLM) which is thought to be distributed through the cell wall, both in the inner and outer layer.Citation9 Since the fungal cell wall is the most external structure of the cell, it is at the interface between the host and the infective microbe, constituting the main source of pathogen-associated molecular patterns (PAMPs). These structures are recognized by the pattern recognition receptors (PRRs) from immune cells, mediating microbial uptake and killing, and modulating the immune response.Citation10 Toll‑like receptors (TLRs) have been identified as a major class of PRRs involved in the recognition of microbial structures, being TLR2 and TLR4, which recognize PLMs and O‑linked mannans respectively, the main TLRs involved in the signaling induced by C. albicans.Citation11,12 Fungal mannans are recognized by additional receptors: α‑mannans and hypha-specific high mannose containing structures are recognized by dectin‑2,Citation13,14 while N‑linked mannans are recognized by macrophage mannose receptors (MR)Citation15 and by DC‑SIGN (dendritic cell‑specific intercellular adhesion molecule 3‑grabbing non‑integrin).Citation16 The soluble (not membrane‑bounded) mannose‑binding lectin (MBL), also recognizes mannans and mediates Candida opsonisation and uptake.Citation17 Finally, galectin‑3, a member of a β‑galactoside‑binding protein family, contributes to the recognition of C. albicans by macrophages in cooperation with TLR2 and Dectin‑1,Citation7,18 a C‑type lectin that binds exposed β‑1,3‑glucan.Citation19 Galectin‑3 is highly produced and secreted by macrophages and is also expressed in dendritic cells (DCs), activated lymphocytes and epithelial cells.Citation6,20 It participates in a variety of cellular processes through either intracellular or extracellular mechanisms.Citation21 Intracellular galectin‑3 regulates cell survival, pre‑mRNA splicing and phagocytosisCitation22 while extracellular galectin‑3 modulates cell adhesion, activation and migrationCitation23 and it has been shown to possess a direct fungicidal activity against C. albicans through recognition of β‑M.Citation24
Adaptation to a changing environment and coping with host defenses is vital for pathogen survival. MAPK‑mediated signal transduction pathways are mechanisms that organisms employ for this purpose. In C. albicans, three main MAPKs pathways have been identified and their importance on fungal virulence and pathogenesis established.Citation25 The cell integrity pathway (PKC pathway) is mediated by the Mkc1 MAPK and plays an important role in cell wall biogenesis, stress response, morphogenesis, biofilm formation and virulence.Citation26-30 The HOG pathway is mainly involved in the response and adaptation to stress but is also implicated in morphogenesis, virulence and cell wall biogenesis,Citation31-34 possibly through repression of the SVG pathway.Citation35 The SVG pathway comprises the MAPK Cek1 which is activated through the MAPKKK Ste11 and MAPKK Hst7 module.Citation36,37 Signaling in this pathway is mediated through, at least, three upstream proteins: Sho1, Msb2 and Opy2.Citation38-40 This pathway is involved in morphogenesis and hyphal formation,Citation36 but it seems to be especially relevant for the adaptation to cell wall damage and glycosylation sensing.Citation41 Cek1 is phosphorylated in response to cell wall inhibitors and its activation is also regulated by quorum sensing and growth.Citation38,42 Due to its implication in cell wall biogenesis, strains defective in elements of the Cek1‑mediated pathway display a higher exposure of β‑(1,3)‑glucan at the cell surface with a consequent increase of the fungal cells binding to the immune receptor dectin‑1, increased phagocytosis and macrophage activation.Citation43 The cek1 mutant is also associated to a reduced virulence in the mouse model for systemic candidiasis although it doesn´t show hypersensitivity to macrophages or neutrophil-mediated killing.Citation44,45
In this study we have characterized the cell wall of cek1 mutants and their response to drugs that interfere with cell wall assembly. We demonstrate here that cell wall α‑1,2 and β‑1,2‑mannosides are differentially exposed in the surface of cek1 mutants (as well as in hst7 mutants) and that this altered cell wall structure influences cek1 recognition by host immune cells. We also address the role of galectin‑3 in mediating the recognition of C. albicans and its possible implication during the infection by cek1 cells.
Results
Transcriptional analysis reveals differentially expressed cell wall-related genes in cek1 mutants
Previous results from our group have uncovered a role for the Cek1‑mediated pathway in cell wall construction as inferred by the increased susceptibility to certain cell wall inhibitors (such as Congo red, calcofluor white or tunicamycin) of mutants altered in this pathway.Citation35,38,42,46 In order to obtain evidence on how this MAPK influences fungal cell wall composition and identify specific targets of the pathway, we compared the transcriptome of CAF2 and cek1 strains under standard exponential growth conditions (log‑phase cells at O.D. = 1 growing in YEPD at 37ºC). Genes whose transcript levels were altered compared to wild type CAF2 cells (>2 or <0.5) were analyzed (). Among the genes that are differently expressed, 27 were down‑regulated in cek1 mutants (ratio cek1/CAF2 < 0.5) while 12 were up‑regulated (ratio cek1/CAF2 > 2). Repressed genes in the cek1 mutant mainly encode proteins involved in the response to stress, including chemicals and drugs (HSP21, DDR48, KAR2, GLX3, CDR1, AHP1 among others) with some hits related to cell wall biogenesis (PGA13, IHD1). Genes that were induced in mutants lacking CEK1 encode proteins related to cell wall biogenesis (i.e. CHT2, PGA45) and the stress response (GDH3, FMA1) among others. Therefore an important number of stress response and cell wall related genes are differently expressed in cek1 mutants, supporting the role of Cek1 in the biogenesis of the cell wall and the suggested morphological alterations caused by the absence of this kinase.Citation36,43
Figure 1. Analysis of the effect of tunicamycin treatment on C. albicans transcriptional response and on cek1 cell wall. (A) Heat map showing differentially expressed genes in cek1 versus CAF2 cells under basal (exponentially growing, not treated with tunicamycin) conditions. (B) Venn diagrams showing up‑ and down‑regulated genes (left and right panel, respectively) upon 2 hours treatment of wild type (CAF2) and cek1 mutant with tunicamycin (TUN). Specific genes to the wild type strain or cek1 mutant are shown in green or red respectively, while commonly regulated genes are represented in yellow. (C) Cells from a stationary culture were diluted in fresh pre‑warmed YEPD or YEPD supplemented with tunicamycin (5 μg/mL), and grown at 37ºC for 2 hours before being collected and processed for TEM analysis. Magnifications of wt and cek1 untreated cells are shown to the right. (D) A representative histogram of cells´ β‑glucan exposure at the end of the experiment (2h). (E) Fold induction of β‑glucan exposure after tunicamycin treatment vs no treatment (induction TUN / no TUN). * indicates p < 0.05 (p = 0.028) as determined using an independent t-Student's test (n = 3).
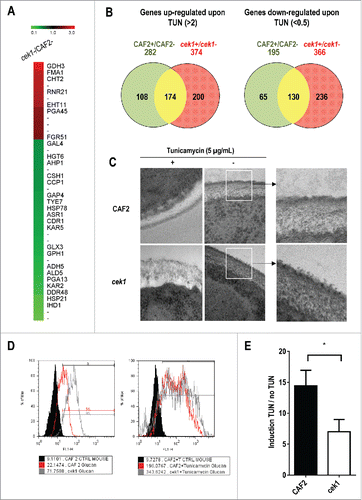
Activating stimuli of the Cek1 pathway include the resumption of growth from stationary phaseCitation38 and tunicamycin.Citation39 We therefore analyzed the different transcriptional response upon tunicamycin (an N‑glycosylation inhibitor) treatment in cek1 mutants compared to the wild type CAF2 in exponentially growing cells, with the aim of identifying putative targets of the route. We found 174 (up) and 130 (down) commonly regulated genes upon treatment in both strains (). As expected, CEK1 was found among upregulated genes in CAF2 (5.37x, 2 replicas only). Upregulated genes involved previously reported UPR targets genes,Citation47 such as ERO1 and others related to protein folding (JEM1, HAC1), genes encoding proteins involved in glycosylation and protein modification (DPM1, PMI1, MNN12), cell wall biogenesis (CRH11, ECM331, CHS7, PHR3), stress response (DDR48, GLX3, HAC1, GTT1) or cell cycle (PCL2). However, some genes from different categories were found to be specifically up‑regulated in CAF2 (PMT4, BMT3, MYO2, CDR1, SOD1) or cek1 (OPY2, PGA6, SOL1, PCL1, TSA1), described in Table S1. Downregulated genes (listed in Table S2) include genes encoding proteins involved in cell wall biogenesis (PGA38, MUC1, ALS2, ECM33), stress response (KIC1, PHR1, HSP60, NRG1, SSA2,) and others (SSB1, PHO84). cek1 mutants showed an important specific down‑regulation of genes involved in cell wall biogenesis (CHT2, SUN41, XOG1, MNT2, MSB2, RBT5, ALS3, EFG1), stress response (DCK1, CTA3, SOD4) and cell cycle (CLB2). Interestingly, we found that KAR2 was repressed in cek1 mutants while being upregulated in CAF2 cells. In summary, we observed an important differential behavior between CAF2 and cek1 in response to the N‑glycosylation inhibitor tunicamycin which mainly involved cell wall and stress related genes, in line with the previously reported cek1 susceptibility to cell wall inhibitors and glyco‑stressors.Citation39,48
cek1 cell wall alterations are augmented upon treatment with tunicamycin
Transmission electron microscopy (TEM) was used to analyze the appearance of the cell wall structure of cek1 mutants during exponential phase of growth. Contrasting to the morphology presented by wild type and cek1 strains in stationary phase of growth,Citation43 cek1 mutants showed, during an active phase of growth, a cell wall presenting a central layer with higher density than the wild type CAF2. Most interestingly, cek1 mutants showed an amorphous dispersed material that was apparently loosely bound to the surface. This suggests the existence of crosslinking defects and enhanced release of cell wall mannoproteins (CWMPs) to the external medium (). In order to reveal these defects, we used tunicamycin, which has been shown to mediate Cek1 activation,Citation49 in two different approaches. First, cek1 mutants were treated with this drug and its cell wall was analyzed by TEM. Although the addition of tunicamycin to CAF2 cells affected somehow the cell wall structure, discernible by a lower electro-dense central layer when compared to the wild type cells devoid of treatment, tunicamycin had a profound effect on the structural organization of the cek1 cell wall, which became enlarged and showed a reduced consistency with signs of mechanical disturbance (). Differences were also appreciated in the greater transparency to electrons of this layer in cek1 vs. wt cells. Second, we measured the effect of tunicamycin treatment on the exposure of glucan to detect an altered glucan masking.Citation50 As previously described, cek1 mutants showed an increased glucan exposure compared to wild type cells under standard growth conditions.Citation43 Flow cytometry analysis revealed that inhibition of mannosylation due to tunicamycin led to an almost 15‑fold increase in the exposure of β‑glucan in wild type cells (), while in cek1 mutants, this increase was only 5‑fold. Tunicamycin effect on β‑1,3‑glucan exposure is therefore reduced in cells with a compromised Cek1‑mediated pathway, probably as a consequence of the already increased basal levels of glucan exposure in these mutants. Collectively, these results indicate that cek1 mutants have an altered cell wall structure which is more evident upon treatment with the N‑glycosylation inhibitor tunicamycin.
The absence of a functional SVG pathway leads to increased levels of α‑1,2 and β‑1,2‑mannosides detected by specific antibodies at the surface of the cell wall
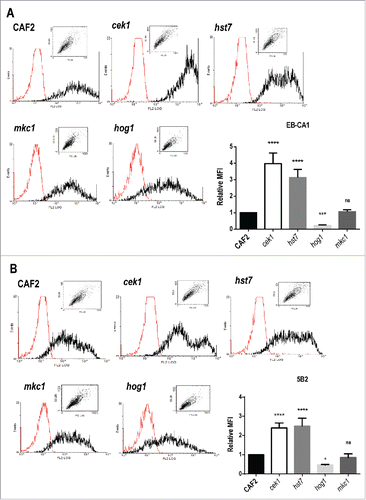
Although cellular β‑1,3‑glucan is able to trigger efficient and protective immune responses against C. albicans infectionsCitation51,52 the bulk of this polymer is usually masked by the outer layer of cell wall mannoproteins (CWMPs). We therefore wondered whether alterations could be also detected in the external outer layer of cek1 cells, where mannose units are α‑1,2 and β‑1,2 linked. These molecules are abundant in C. albicans and contribute to its virulence due to their immunomodulatory and adhesion properties.Citation53,54 We made use of flow cytometry analysis and specific antibodies to determine the levels of α‑1,2 and β‑1,2 linked mannosides (α‑M and β‑M) at the surface of cek1 mutants under standard conditions (YEPD, 37ºC). The absence of Cek1 led to an almost 4‑fold increase on the levels of α‑M in cells at stationary phase when compared to the wild type strain, as inferred using the EB‑CA1 specific monoclonal antibody (MAb)Citation55: 2600 ± 183.95 versus 665.8 ± 124.25 (). Similarly, β‑M levels were assessed using the mAb 5B2.Citation56 We again observed an increase in the levels of this signal in cek1 mutants, more than 2‑fold increase compared to wild type cells (402 ± 51.22 vs. 170.82 ± 37.86, ). Both linked mannosides were also increased in mutants lacking the MAPKK Hst7, the canonical Cek1 MAPKK, similarly to cek1 mutants (: 2037 ± 84.14 and 415 ± 29.93 for α‑M and β‑M, respectively), indicating that this is not a defect specific to the MAPK but to the pathway. On the contrary, hog1 mutants showed an almost 4‑fold reduction in α‑M (147.6 ± 28, ) and 2‑fold reduction in β‑M (78.65 ± 17.34, ). This effect was also observed in mutants defective in the upstream regulator of the HOG pathway, SSK1 (data not shown), which shows, in accordance with this, enhanced Cek1 activation.Citation38,57 No significant differences were, however, found between mkc1 mutants and wild type cells (). The alterations observed within cek1 and hst7 cell wall are not exclusive to α‑ and β‑M as the levels of detected chitin were also increased when compared to the wild type strain (Fig. S1). Therefore, α‑M and β‑M are more easily detected in mutants altered in the CEK1‑mediated pathway while they are less detected in hog1 mutants.
Figure 2. Levels of α‑ and β‑mannosides at the cell wall surface of strains defective in elements from the MAPK signaling network. Cells from stationary cultures of CAF2, cek1, hst7, mkc1 and hog1 mutants were analyzed by flow cytometry to detect the levels of α‑1,2‑ and β‑1,2‑mannosides (α‑M and β‑M), recognized by the specific antibodies EB‑CA1 (A) and 5B2 (B) respectively. Representative histograms (the red histogram refers to samples treated only with the secondary antibody) and mean fluorescence intensities (MFI) from three independent experiments with the corresponding standard deviation are shown. One way ANOVA followed by Dunnet's correction for multiple comparisons was applied to evaluate differences between mutants and wild type strain (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). MFI absolute values for CAF2 cells were 665.8 (A) and 170.8 (B).
cek1 mutants display an altered pattern of mannosylation on different cell wall mannoproteins
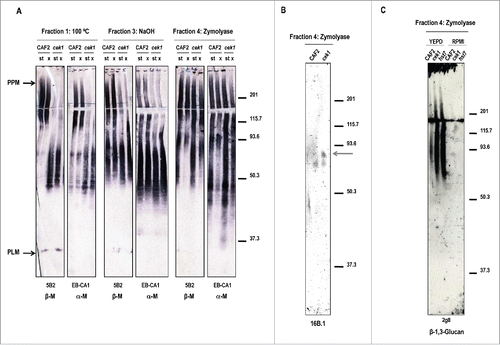
As our data indicated that this protein participates in the construction of the cell wall, we further characterized the structural alterations of the cek1 cell wall. For this purpose, we focused on the analysis of α‑M and β‑M present in specific cell wall fractions generated upon a sequential extraction procedure (described in Citation58). Different families of mannoproteins were gradually released from the cell wall: the non‑covalently bound CWMPs (such as PPM and PLM) were released by heat (fraction 1) and the mannoproteins covalently bound to β‑glucans were obtained either by mild alkali treatment (fraction 3) or zymolyase treatment (fraction 4). These cell wall fractions were then mapped for the presence of α‑M and β‑M by western blotting using the EB‑CA1 and 5B2 (respectively) specific antibodies. As the surface expression pattern of β‑M is differently modulated according to the growth conditions,Citation59 we analyzed cells from either stationary or exponential cultures. While no alterations were detected in whole cell extracts (Fig. S2), the analysis of cell wall fractions revealed differences between CAF2 and cek1 cell wall observed as a shift in the size of the PPM signal in the mutant compared to the wild type strain (, fraction 1). This different pattern of the mannan epitope was also observed in fraction 3 corresponding to PIR CWMPs. However, no significant differences were observed between cell extracts from stationary and exponentially growing cultures. The analysis of mannosylation in GPI‑anchored cell wall proteins (released by zymolyase treatment – fraction 4) revealed a different β‑ and α‑mannosylation of the CWMPs depending on the phase of growth; however these differences were similar in both strains. Similar results were observed in hst7 mutants defective in the upstream activating kinase (Fig. S3) suggesting that these defects may be associated to Cek1 activation. β‑M are not only associated to the non‑covalently bound molecules phosphopeptidomannan (PPM) and phospholipomannan (PLM), but also to other CWMPs either through O‑ or N‑glycosylation.Citation58,60,61 The hyphal specific GPI‑anchored protein Hwp1 contains β‑M that are associated through O‑glycosylation.Citation58 This protein is highly expressed under filamentous growth inducing conditionsCitation62 and it was analyzed in this study. Exponentially growing cells of CAF2 and cek1 strains were grown in YEPD and RPMI medium to O.D. = 1 at 37ºC to induce hyphal formation and were collected and processed for the extraction of the different CWMPs (). Zymolyase released extracts from the RPMI cultures were separated in SDS PAGE electrophoresis and Hwp1 was detected with the MAb 16B.1 which recognizes the recombinant N‑terminal Hwp1 fragment.Citation58 As shown, an additional band (gray arrow) was observed in cek1, suggesting that this protein may be differently glycosylated in this mutant (). We could also confirm in this fraction the enhanced presence of β‑1,3‑glucan in SVG mutants (cek1 and hst7) compared to the wild type strain, which is in agreement with previously described data (). No differences were observed between strains growing with RPMI medium, probably because of the induction of the hyphal formation in all strains and their similar morphology. Overall, these data demonstrate a clear alteration on the structural organization of both the cek1 and hst7 cell wall compared to wild type cells.
Figure 3. Mapping glycan epitopes in cek1 mutants. (A) Different cell wall fractions from stationary (st) and exponential (x) phase cultures of CAF2 and cek1 strains were separated by SDS-PAGE before blotting with 5B2 (β‑M) and EB-CA1 (α‑M) antibodies. CWMP extracts were released by heat treatment (Fraction 1), NaOH (Fraction 3) or zymolyase treatment (Fraction 4). (B) Zymolyase released CWMP extracts (fraction 4) from CAF2 and cek1 cells grown at exponential phase in RPMI media were separated by SDS-PAGE and analyzed by western blotting with the MAb 16B.1 for the detection of Hwp1. The predicted glycosylated form of Hwp1 is highlighted with an arrow. (C) Zymolyase (Fraction 4) extracts from the indicated strains grown at exponential phase in YEPD or RPMI media were analyzed with the mAb 2g8 for the detection of β‑1,3‑glucan.
The interaction of cek1 mutants with murine macrophages lead to increased expression and/or secretion of galectin‑3
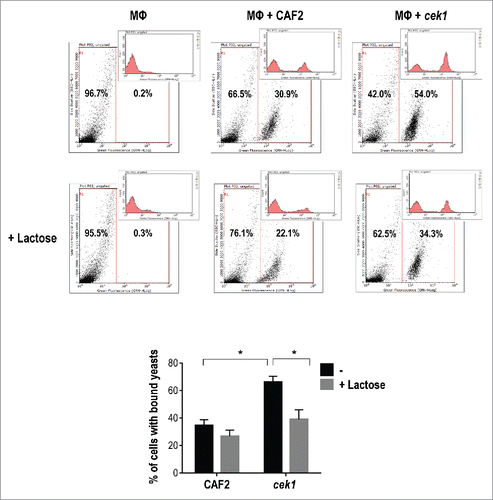
There is evidence that β‑M present Candida species may be the target of galectin-3.Citation24 Given the increased detection of β‑M in the cell surface of cek1 mutants, we analyzed whether galectin-3 could be relevant in this interaction. We first assessed the effect of lactose on C. albicans binding to murine macrophages, given its role as inhibitor of galectins carbohydrate binding site.Citation63 As shown in , cek1 mutants bind more efficiently to murine macrophages than the wild type but this increased binding is partially impaired by the presence of lactose. For these assays, C. albicans cells were labeled with FITC and added to a confluent monolayer of Raw 264.7 macrophages, allowing the interaction to occur at 4ºC for 30 min. Assessment of yeasts bound to macrophages was performed by flow cytometry. The binding behavior showed by cek1 cells in the absence and presence of lactose is dependent on the signaling pathway as hst7 mutants and a strain expressing a non‑activable allele of CDC42 (CDC42D118A) also display increased binding to murine macrophages which is reduced to the wild type levels by the addition of lactose (data not shown). These results suggest a role for galectins in cek1 recognition (and/or binding) by immune cells.
Figure 4. Lactose blockage of C. albicans binding to murine macrophages. Representative flow cytometry analysis of CAF2 and cek1 (marked with FITC) binding to murine macrophages (MΦ – Raw 264.7 cell line) at MOI 10:1 (yeast: MΦ) and 30 min. interaction time on ice. Blockage with lactose (100 mM) was made 30 min. prior infection. The graphic shows the percentage of bound yeasts from independent experiments (n = 4). * p < 0.05.
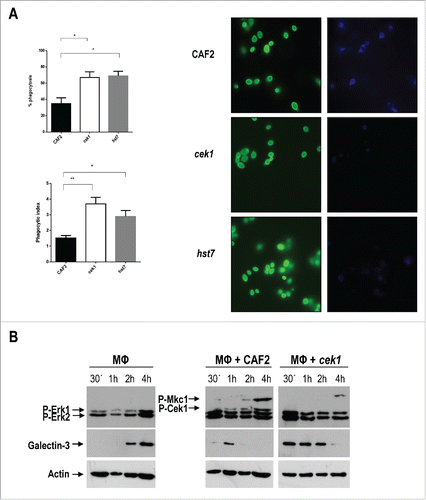
Previous work from our group has demonstrated that cek1 mutants display not only an increased binding to immune cells, but also an augmented phagocytosis by human macrophages and dendritic cells which is, in part, due to an increased exposure of β‑1,3‑glucan via dectin‑1.Citation43 We have here analyzed cek1 and hst7 phagocytosis by murine macrophages from the Raw 264.7 cell line through a double staining method. Blastoconidia from the indicated strains were pre‑stained with Oregon green and incubated with murine macrophages for 3 hours at MOI 1:1. After fixing, non‑phagocytosed yeasts were stained by calcofluor white which cannot penetrate inside the macrophages under our conditions. Cells lacking either CEK1 or HST7 were more quickly phagocytosed by Raw 264.7 macrophages (, compare 35 ± 5% for wt to 69 ± 4 for hst7 and 67 ± 5 for cek1 mutant). This was also reflected in a different phagocytic index, 1.52 ± 0.16 for CAF2, 3.7 ± 0.4 for cek1 and 2.9 ± 0.38 for hst7.
Figure 5. Analysis of galectin‑3 expression by murine macrophages upon interaction with C. albicans cells. (A) Phagocytosis by murine macrophages of the cek1 and hst7 mutants vs. the CAF2 wild type strain. Percentage of phagocytosed fungal cells (Upper subfigure) or Phagocytic index (Lower subfigure) assessed by counting number of internalized yeast per 100 phagocytes (at least 10 fields on each immunofluorescence carried out in duplicate). Data from phagocytic assays were collected in at least 3 independent experiments. (*p < 0.05, ** p < 0.01). Fluorescent microscopy images showing extracellular yeast (blue) and both extra and intracellular yeast (green) are shown. (B) Immunodetection analysis of cells recovered from interaction studies between C. albicans and murine macrophages. Wild type (CAF2) or cek1 mutant cells were added to a confluent monolayer of macrophages (Raw 264.7 – MΦ) at MOI 10:1 (yeast: MΦ) and let to interact at 37ºC. Cells were collected at the indicated time-points and treated for immunoblot detection of MAPKs and ERKs activation pattern (anti-phospho-p44/42) as well as for detection of galectin‑3 expression (anti‑galectin‑3). Anti-actin was used as loading control. (C) Immunofluorescence microscopy analysis of C. albicans cells (CAF2 and cek1) interacting with murine macrophages (MOI: 1:1, yeast: MΦ). Yeast cells are constitutively expressing RFP (visible in red) while galectin‑3 is visible in green. Right pictures are fluorescence merged images of green and red channels, while left pictures include the phase contrast image for clarity. Control macrophages without Candida cells are shown as -.
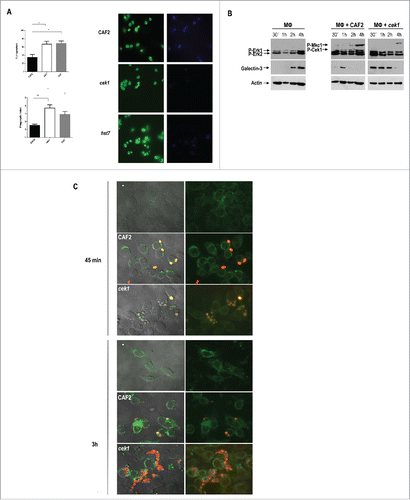
Although lactose is not a specific inhibitor of galectin‑3, we focused on this receptor and analyzed its expression and secretion pattern by macrophages upon interaction with CAF2 or cek1 via protein gel blotting and immunofluorescence assays. First, we added pre‑washed yeast cells from overnight grown cultures to confluent monolayers of macrophages and recovered the co‑culture at different time points. A MOI of 10:1 (yeast:MΦ) was used for the western blotting assays in order to ensure the detection of C. albicans MAPKs. As controls, we also recovered macrophages incubated without C. albicans as well as yeast cells incubated without macrophages. Galectin‑3 expression, as well as the MAPK and ERK activation patterns of yeasts and macrophages (respectively), was analyzed using anti‑galectin‑3 and anti‑phospho‑p44/42 antibodies. Upon interaction with fungal cells, Erk1 and Erk2 became phosphorylated reaching a maximum level at 15‑30 min post‑infection () and no substantial differences were seen regarding the level and activation pattern of Erk1 proteins between CAF2 and cek1 strains. Cek1 and Mkc1 followed a similar phosphorylation pattern as the one observed in the sample control without macrophages (data not shown). Interestingly, cek1 cells induced earlier the expression of galectin‑3 by macrophages (30 min. compared to 1h in CAF2 cells) and this was more sustained in time, being detected at later time points compared to the wild type ().
In a different approach, galectin‑3 expression and localization within the cell was analyzed by immunofluorescence staining (). For this, we marked both CAF2 and cek1 with a red fluorescent protein (RFP) and used an anti‑rat antibody conjugated to FITC to detect anti‑galectin‑3 binding. Samples were collected after 45 min. and 3h interaction at MOI 1:1 (yeast: MΦ). Analysis of the fluorescence microscopy images revealed an increased phagocytosis of cek1 mutants by immune cells compared to wild type cells, especially evident at 45 min, which is in accordance with previous results ( and Citation43). At this time point (45 min), galectin-3 seems to surround cek1 yeast cells while localizing at the surface of macrophages upon interaction with CAF2. At 3h cek1 mutants seem to stimulate expression of galectin-3 to a higher extent compared to wild type cells. Moreover, galectin‑3 appeared to be more diffuse in the extracellular medium, providing blurrier images when cells were incubated with cek1, suggesting once more an altered galectin‑3 mediated immune regulation promoted by the absence of the Cek1 MAP kinase.
Ex vivo and in vivo analyses show a non‑dependency on galectin‑3 for cek1‑mediated infection
Galectin‑3 knock‑out mice were used to specifically assess the role of galectin‑3 in C. albicans infection, and particularly, to analyze the immune response displayed by this receptor upon interaction with cek1. For these experiments we used thioglycollate‑induced peritoneal macrophages as well as bone marrow derived macrophages (BMMs) and dendritic cells (BMDCs). MAPKs defective mutants were incubated with immune cells from wild type (galectin-3+/+) or knockout (galectin-3−/−) mice for viability studies. Strains defective in the Cek1 MAPK have been show to display an increased survival in the presence of human polymorphonuclear cells (PMNs) or the promyelocytic leukemia cell line HL-60.Citation45 Although we assessed CAF2 and cek1 survival to BMDCs, BMMs and peritoneal macrophages, we were not able to observe any substantial difference between both strains. Moreover, no significant changes in C. albicans mortality were observed between galectin-3+/+ and galectin-3−/− cells (Fig. S4A). Regarding cek1, the minor differences observed were similar to the ones showed by CAF2 suggesting that this immune receptor does not interfere with cek1 killing by immune cells. We also analyzed the cytokine pattern response of BMDCs challenged with C. albicans CAF2 and cek1 strains by immunoenzymatic assays (ELISA). The cytokine profile showed by BMDCs upon interaction with C. albicans cells is not significantly affected by the absence of CEK1 and this similarity between CAF2 and cek1 mutants does not lead to an evident alteration attributable to the absence of the immune receptor galectin‑3 (Fig. S4B). Interaction with any strains, led however to a higher release of Th17‑related cytokines (IL‑6 and IL‑23) which is in accordance to published results.Citation64
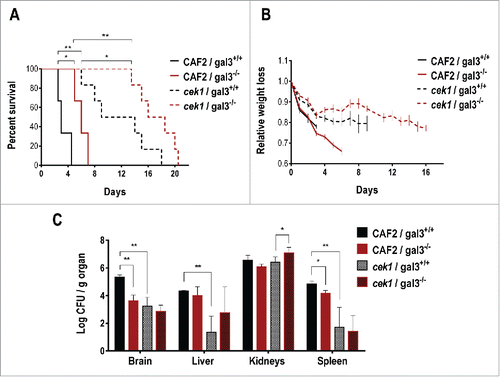
Strains defective in the Cek1 MAPK have been shown to display a reduced virulence in a mouse model for systemic infection.Citation36 The cell wall alterations caused by a defective SVG pathway leads to increased recognition and phagocytosis of C. albicans cells by the immune system. In order to determine the role of galectin‑3 in this process, we performed a virulence assay using knockout mice for this lectin. CAF2 or cek1 cells were inoculated via the tail vein into mice deficient (or not) in galectin‑3 (galectin-3−/− and galectin-3+/+). The infected mice were weighed regularly and, after death, the target organs (kidneys, spleen, brain and liver) were processed for fungal burden assessment. As described before,Citation36 cek1 mutants displayed a reduced virulence in this infection model and showed less fungal loads in every target organs analyzed except kidneys, where levels of fungal burden were generally high (and similar) for both C. albicans CAF2 and cek1 strains (). Interestingly, galectin‑3 influenced virulence in this model, and mice defective in this lectin were found to be more resistant to fungal infection compared to galectin-3+/+ mice, showing a less pronounced weight loss and in general, less fungal burden on target organs. The mean survival time (MST) for CAF2 was 3 days for galectin-3+/+ mice and 6 days for galectin-3−/−, similarly, these values were 9 and 16 for cek1 cells. Thus the minor virulence of cek1 is not associated to the galectin‑3 mediated response.
Figure 6. Role of galectin‑3 in a mouse model of systemic infection. 0.6 × 106 cells of C. albicans were infected via the tail vein in galectin-3+/+ (wild type) and galectin-3−/− (knockout) C57BL/6J mice (CAF2: n = 3; cek1: n = 6). (A) Mice survival after C. albicans systemic infection. Statistical analysis was measured by log rank (Mantel-Cox) test, *p < 0.05, **p < 0.01. (B) Weight loss of infected mice during the survival assay. Data is shown as the weight mean of all mice until 50% of the infected group remained alive, related to initial individual weight prior to the infection. (C) Post-mortem analysis of fungal burden in target organs. *p < 0.05, **p < 0.01.
Discussion
In this work we have analyzed the contribution of the Cek1‑mediated MAP kinase pathway in the composition and organization of the C. albicans cell wall and its relevance during the interaction with immune cells. It is well accepted that C. albicans recognition by the immune system is a multifactorial process in which several PRRs may interact in order to trigger an efficient immune response.Citation65 Alterations in the cell wall architecture may lead therefore to a better recognition by the host and a more efficient fungal clearance, therefore influencing the outcome of the disease. Protection or infection, depend on both the accessibility of fungal components and the differential expression of different receptors on immune system cells. The specific interaction favors a Th response (Th1/Th2/Th17) according to the cytokine production profile.Citation65,66 cek1 mutants are less virulent in a mouse model of systemicCitation36 and localizedCitation44 infection; they present defects in hyphal formation although in vivo they are perfectly able to undergo the dimorphic transition (unpublished observations). Furthermore, no relevant differences regarding the induction of specific oxidative stress genes are observed in cek1 mutants.Citation45 The reduced virulence of this mutant could be therefore explained by the altered cell wall described here. This is evident in TEM micrographs from stationaryCitation43 and exponentially growing cells: the structured cell wall layers appear, in this mutant, disorganized and loosely bound when compared to the wild-type. The lack of a functional Cek1 pathway leads also to higher levels of exposed cell wall components (β‑1,3‑glucans, α‑1,2‑mannans, β‑1,2‑mannans and chitin) in the fungal surface and the increased β‑1,3‑glucan exposure (typically hidden beneath the mannoprotein outer layer) has been already reported to contribute to the enhanced immune cells recognition and phagocytosis observed in both cek1 and hst7 mutants.Citation43 The observations described in this work regarding the increased levels of α‑1,2‑ and β‑1,2‑linked mannosides (α‑M and β‑M) in cek1 mutants, could also account for the attenuated virulence due to a more efficient immune response. Another possibility could involve antibodies against mannan, that have been described as protective, increasing the opsonisation of yeast cells, favoring phagocytosis and killing in vivo.Citation67 In fact, cek1 mutant induces higher levels of IL‑10 and TNF‑α during murine macrophage interaction compared to the wild type strain, which is in accordance to previously reported data.Citation43 The data presented here demonstrating the increased levels of α‑M and β‑M in cek1 mutants, together with the already published results describing an alteration of the structural integrity of cell wall mannans in these strains,Citation68 show that the absence of the Cek1 MAPK has a strong impact on the fungal cell wall architecture and consequent glycans exposure/expression.
The observed alterations on the cell wall of cek1 mutants could rely on their transcriptional profile. When compared to the wild type strain grown in exponential phase, important differences arise. cek1 mutants exhibit an up-regulation of genes that encode different GPI‑anchored proteins such as PGA45, FGR41 (>2 fold), YWP1, PGA18 and KRE1 (>1.5 fold). Among genes down‑regulated in cek1 we also found different GPI‑anchored proteins like PGA13 and IHD1, a putative β‑1,4‑glucan branching enzyme GLC3 and different stress response genes (DDR48, HSP21). A different regulation of genes encoding protein mannosyltranferases was also observed: cek1 mutants display lower levels of PMT6 and MNN12 expression while the gene coding for BMT2 is up‑regulated. Interestingly, we also found altered expression of genes whose induction/repression is regulated by farnesol like MRF1, GRE3, DDR48, IFD6 and GPM2, which is in accordance with the role of the SVG pathway in quorum sensing and the regulation of Cek1 activation in response to farnesol.Citation42
The transcriptomic differences between CAF2 and cek1 were also evident in the presence of the N‑glycosylation inhibitor tunicamycin. Treatment with this drug leads to Cek1 phosphorylationCitation49 and drastically affects the already altered cell wall of the cek1 mutant, indicated by the TEM micrographs. Tunicamycin up-regulates genes involved in protein glycosylation (for instance the ones that encode protein mannosyltransferases: MNN21, MNN15, MNN12, PMT4, BMT3) and cell wall biogenesis (such as ECM331, KRE1, CHS3, PHR3, PIR1). This drug is also an inducer of endoplasmic reticulum (ER) stress, leading to the activation of the unfolded protein response (UPR).Citation69 In agreement with previously reported data, treatment with tunicamycin upregulated the bZIP transcription factor HAC1 which is required for ER stress resistance in C. albicansCitation70 and S. cerevisiae.Citation71 Hac1 further influences the synthesis of cell wall components and the transcription of genes involved in secretory and vesicle trafficking processes, at least under stress conditions.Citation70 The connection between tunicamycin and the Cek1 pathway is evidenced by the different regulation of KAR2 (encoding a ER stress chaperoneCitation72,73 of the UPR pathway) between CAF2 (up-regulated) and cek1 (downregulated) in response to tunicamycin and the recent finding that Msb2, a signaling mucin essential for the activation of Cek1,Citation39 also regulates UPR-targetsCitation74 under heat stress.
Mutations in pathways involved in O‑ and N‑glycosylation reduce the presence of β‑M expression.Citation58 och1 mutants, defective in the outer branched N‑linked glycans, have reduced levels of α‑M and β‑M epitopes compared to the wild type strain. The pmt1 mutant, defective in the first steps of the O‑glycosylation and partially responsible for Hwp1 glycosylation,Citation75,76 shows also a decrease in β‑M epitopes observed in fraction 1 (100ºC), 3 (NaOH) and 4 (zymolyase) and a shift in mannans´ molecular weight. Interestingly, cells defective in either OCH1 or PMT1 constitutively display an increased Cek1 activation,Citation49,77 supporting therefore the idea that the Cek1‑mediated pathway is essential for cell wall remodeling in C. albicans. A shift in both α‑M and β‑M (lower molecular weight) is also observed in cek1 and hst7 mutants when compared to the wild type strain, suggesting that crosslinking of cell wall components may be affected in these mutants. However, analysis by protein gel blotting does not reveal a drastic decrease of α‑M and β‑M amounts, which could be due to either shorter α‑1,6 chains in cek1 mutants or to a lesser crosslinked mannan. We support the idea that these mutants expose normally hidden cell wall molecules like glucans and inner PIR and GPI mannoproteins that carry α‑M and β‑M favoring recognition (but not necessarily the amount) of these components by antibodies. The repressive role that the HOG pathway exerts on the SVG pathwayCitation35 may also account for the cell wall alterations observed in cek1 and hst7 mutants: strains defective in the MAPK Hog1 display significant lower levels of both α‑ and β‑M exposure than the wild-type.
The overexposed β‑M at the cell wall of cek1 and hst7 mutants led us to hypothesize the implication of galectin‑3 in cek1-mediated infection. In fact, galectin‑3 is involved in the macrophage recognition of C. albicans by specifically binding β‑1,2‑oligomannosides, an uncommon linkage associated to mannan which is present in this fungal pathogen but absent from the non‑pathogenic yeast S. cerevisiae.Citation6,7 This immune receptor has been shown to associate with dectin‑1, that specifically recognizes fungal β‑1,3‑glucan, to mediate fungal uptake, although galectin‑3 is not essential for this process, working together in the proinflammatory response of macrophages.Citation18 Interestingly, dectin‑1 mediates the increased binding and phagocytosis of cek1 mutants due to its enhanced cell wall glucan exposure, and the activation of intracellular signaling pathways of human dendritic cells and macrophages.Citation43 The observation of galectin-3 surrounding cek1 yeast cells during their interaction with macrophages, especially at early time points, suggest a role for both dectin‑1 and galectin‑3 in the increased recognition and phagocytosis of this mutant. Galectin‑3 is moreover a pleiotropic protein participating in a variety of cellular processes and being expressed in, and secreted by, several cells.Citation22,23 The secretion of galectin‑3 by human gingival epithelial cells has been shown to be significantly and rapidly upregulated by C. albicans and C. parapsilosis.Citation78 Furthermore, its secretion by neutrophils seems to act as a proinflammatory autocrine/paracrine signal for phagocytosis: exogenous galectin‑3 increases yeast phagocytosis while the presence of a galectin-3 blocking antibody reduced phagocytosis.Citation79 In addition to the indicated roles of this lectin in promoting phagocytic uptake and prevention of microbial dissemination, galectin‑3 has a direct fungicidal activity upon binding to the specific β‑M present in the C. albicans cell wall.Citation24 In this work, we show that the increased binding of cek1 mutants is partially impaired by lactose (a blocking agent of galectins binding site) suggesting a role for this receptor in cek1 recognition. Through western blotting and immunofluorescence analysis of C. albicans interaction with murine macrophages, we uncovered an increased stimulation of galectin‑3 secretion caused by cek1 infection, when compared to the wild type strain, and possible increased expression at the macrophage membrane and cytoplasm. This could be explained by the increased levels of cell wall mannans exposed in cek1 mutants, as galectin‑3 secretion by neutrophils has been reported to be induced by treatment with mannan isolated from C. albicans.Citation79
Host cell galectins can recognize glycan ligands not only from fungi, but also from viruses, bacteria and parasites, and its role is not always associated to microbial clearance and prevention of infection. Although they are able to stimulate host defense, by directly or indirectly triggering leucocyte activation, phagocytosis, complement fixation and cytokine production, they can act as a bridge to target cells, promoting microbial infection (reviewed in Citation80). In this study we show no significant differences regarding the dendritic cells and macrophage‑mediated killing of cek1 or wild type strains between immune cells derived from either galectin-3+/+ or galectin-3−/- mice. This is somehow in agreement with the fact that galectin‑3 by itself is not essential for fungal uptake and that recognition of β‑M does not seem to be crucial for fungal eradication.Citation7 However, galectin‑3 knockout mice displayed an enhanced resistance to C. albicans infection, associated to lower fungal burden in target organs, especially in brain and spleen. Interestingly, such differences were minimal or absent in kidneys, suggesting that the ultimate cause of death was kidney failure in both strains and/or the mechanisms controlling proliferation in this microenvironment were different. These results are unexpected as it was reported that mice defective in galectin‑3 are more sensitive to C. albicans infection.Citation81,82 While we do not have an explanation for this, differences could exist between both set of experiments regarding the infectious dose used and the origin of the used galectin-3+/+ and galectin-3−/− mice (our strain is backcrossed for over 9 generations to C57BL/7 mice while this number is lower in commercial (Jackson Laboratories) mice) that could be maybe responsible for the observed discrepancies. Nevertheless, an increased resistance of galectin‑3 null mice has been also observed in infections with Trypanosoma brucei,Citation83 SalmonellaCitation84 and Rhodococcus equi,Citation85 and mice galectin-3−/− demonstrated reduced levels of bacteremia, compared to wild type mice, after challenge with live N. meningitides.Citation86 The pattern of released cytokines from bone‑marrow derived dendritic cells (BMDCs) when challenged with C. albicans could provide a reason for the resistance behavior. The absence of galectin‑3 leads to enhanced release of the cytokines IL-6, which promote the differentiation of Th17 response in mice,Citation87 and IL-23, required for Th17 cell expansion and effector functions.Citation88 The Th17 adaptive immunity appears to be primarily responsible for protection against fungal infections at the mucosa.Citation89 In fact, it has been recently reported that adoptive transfer of C. albicans galectin-3−/− dendritic cells induce higher Th17 responses and promote greater fungal clearance than galectin-3+/+ dendritic cells.Citation64 This has also been observed in the infection caused by Histoplasma capsulatum where galectin‑3 negatively regulates the ability of the host to clear this dimorphic fungus from target organs during the early phase of infection. This is potentially explained by the enhanced production of IL‑23 cytokines observed in infected galectin-3−/− dendritic cells.Citation90 A C. albicans strain defective in the β‑1,2‑mannosyltransferase Bmt1 and a strain with no β-M (bmt1 bmt2 bmt5 mutant) exhibit a similar or reduced virulence (respectively) in galectin-3−/− mice compared to its wild type counterpart.Citation82 We demonstrate here that the avirulent phenotype of cek1 mutantsCitation36 is not caused by the absence of galectin‑3, despite the significant reorganization of the cell wall which is reflected in the epitopes that are recognized by this receptor in the surface of the cell. This result, while apparently contradictory, emphasizes the multifactorial nature of the recognition of fungi by mammalian cells,Citation65 where different receptors (PRRs) are involved and their corresponding PAMPs are probably also influenced by the SVG pathway.
In conclusion, we show in this work that C. albicans cell wall remodeling is controlled by the SVG signaling pathway which regulates the exposure of essential epitopes involved in host recognition. These cell wall alterations are essential during host interaction and therefore, support the relevant role of this pathway as a target in antifungal research.
Materials and methods
Strains and growth conditions
The C. albicans wild type strain CAF2 Citation91 was used in this study, as well as mutant strains cek1,Citation36 mkc1,Citation26 hog1 Citation92 and hst7,Citation93 and CAF2 and cek1 strains constitutively expressing the RFP protein.Citation94 Cells were grown at 37ºC in YEPD medium (1% yeast extract, 2% peptone and 2% dextrose) unless otherwise stated. The generation times for the indicated strains growing at 37ºC in rich YEPD medium were the following: 78 min (CAF2), 82 min (cek1), 88 min (mkc1), 88 min (hog1) and 83 min (hst7).
Cell lines and culture conditions
The cell lines used in this work for interaction analysis with C. albicans were routinely incubated at 37ºC and 5% CO2 with controlled humidity. Peritoneal macrophages were obtained by stimulation with 1 mL thioglycollate. 4 days after injection in the peritoneal cavity, mice were sacrificed in CO2 atmosphere and peritoneal cells were harvested by lavage with cold PBS. The haematopoietic mother cells were extracted from the femurs and tibias of mice sacrificed in CO2 atmosphere. A syringe with a 25G 7/8 needle was used to flush out the bone marrow into a tube and clumps were broken with a pipette. Red blood cells (RBCs) were lysed by ACK lysis buffer for 1 min. and the reaction was stopped with R10 medium. Cells were then passed through a cell strainer (70 μm) to eliminate debris, centrifuged (1200 rpm/5 min.), and re-suspended in BMDC (for bone‑marrow derived dendritic cells differentiation) or BMM (for bone‑marrow derived macrophages differentiation) medium. The cells´ suspension was transferred to tissue culture dishes and incubated at 37ºC and 5% CO2. Differentiated cells were used between day 7 and 14 for interaction assays.
Reagents and antibodies
All reagents were obtained from Sigma (Sigma Aldrich Chimie, Saint Quentin Fallavier, France). The anti‑β‑1,2‑mannoside monoclonal antibody (5B2), a rat IgM, was developed in Dr. Poulain's laboratory.Citation95 The mouse monoclonal anti‑β‑1,3‑glucan IgG was provided by Biosupplies (Australia Pty) while 2G8 mAb was a gift from Dr. Cassone to Dr. Poulain. The anti‑α‑1,2‑mannoside monoclonal antibody EB‑CA1, a rat IgM, was supplied by Bio-Rad SA (Marnes la Coquette, France). mAb 16B1, a mouse IgG, described as specific to C. albicans hyphal forms.Citation58,96 Fluorescein isothiocyanate (FITC) - and phycoerythrin (PE)-conjugated anti-rat IgM, anti-mouse IgG or anti-rabbit IgG were obtained from Southern Biotechnology Laboratories (Birmingham, AL, USA).
RNA isolation, cDNA synthesis, and chip hybridization
Total RNA was isolated from exponentially growing cells by the "mechanical disruption protocol" using the RNeasy MIDI kit (Qiagen, Hilden, Germany) following the instructions of the manufacturer. RNA purity and integrity was assessed using RNA Nano Labchips in an Agilent 2100B Bioanalyzer (Agilent Technologies, Palo Alto, CA) following the manufacturer's instructions. cDNA synthesis, chip hybridization and Microarray image analysis were performed as previously reported.Citation97 The printed microarrays used in this study were provided by Operon, the Candida albicans AROS V1.2 is the combination of 2 oligo sets: (a) Candida albicans AROS V1.1 and (b) Candida albicans AROS V1.1 upgrade set (www.operon.com). For transcription under basal growth conditions and tunicamycin treatment (0.5 μg/mL, 2h), the total RNA from three different cultures was analyzed; additionally, for each sample, two different hybridizations were performed, including fluorochrome swapping in order to minimize transcriptional changes due to technical variability. Six DNA microarrays were therefore analyzed for each condition. The data were processed and treated using the statistical methods previously reported.Citation97 Significance analysis of the results was conducted using a Student's t test (GeneSight). Genes with p values of less than 0.05 were considered to be significantly differentially expressed. Gene ontology analysis was performed using the web tool Genecodis (genecodis.dacya.ucm.es/). Heat map was obtained using the MeV 4.6 software and shows gene expression ratios comparing the transcriptional response of cek1 vs CAF2 strain under non stress conditions. The degree of color saturation represents the log2 value of gene expression ratio, as indicated by the scale bar.
Transmission electron microscopy (TEM)
Stationary phase yeast cells grown in YEPD were diluted in pre‑warmed fresh media and incubated for 2 hours at 37ºC, in the presence or absence of tunicamycin (TUN) at 5 μg/mL. Cells were harvested (3000 rpm for 10 min at 4ºC) and immediately fixed in 2.5% glutaraldehyde (Serva), 0.1M cacodylate buffer, pH 7.2 – 7.4 for 1h at 4ºC. Cells were then post fixed in 1% OsO4 and embedded in Epon prior sectioning on a Reichert Jung ultra-microtome. Ultrathin sections were stained with uranyl acetate and lead citrate, and examined with a Zeiss EM10C electron microscope.
Analysis of exposure of cell wall epitopes by flow cytometry
The levels of α‑1,2‑mannosides, β‑1,2‑mannosides, β‑1,3‑glucans and chitin detected by specific antibodies on the yeast cell wall was analyzed by flow cytometry. For staining, 106 yeast cells were washed with PBS containing 2% FCS and incubated for 15 min. with 1/200 dilution of the mAb 5B2 (for β‑1,2‑mannosides), 1/50 dilution of the mAb anti‑β‑1,3‑glucan, EB‑CA1 (for α‑1,2‑mannosides) or FITC-WGA for chitin detection. For antibody detection, cells were incubated for 15 min with 1/100 dilution of FITC‑labeled anti‑rat IgM or PE‑labeled anti‑mouse IgG secondary Ab. A negative control was performed by adding to the samples solely the labeled secondary Ab at the same concentration. All processes were conducted at 4°C. After washing, cells were fixed with 0.4% paraformaldehyde and analyzed by FACS. Flow cytometry was performed by an EPICS XLMCL4 (Beckman Coulter) equipped with an argon ion laser, excitation power of 15 mW at 488 nm or by the Guava EasyCyte cytometer. Forward and side scatter were analyzed on linear scales, while green and red fluorescence intensity was determined on logarithmic scales. Gates were set around debris and intact cells were set on a forward and side scatter dot plot. The fluorescence histograms of 5000 cells were generated using gated data. Data acquisition and analysis were performed using WINMDI software (available from http://facs. scripps.edu) or InCyte software by Millipore. Mean fluorescence intensities were obtained by subtracting values of negative controls from the values given by each epitope.
Cell wall preparation, proteins solubilization and protein gel blotting
Pre‑cultured cells were inoculated into YEPD broth at a density of 106 cells/mL and grown at 37°C with continual shaking at 150 rpm until the culture reached an OD600 of 10 (for stationary growing cells) or 1 (for exponentially growing cells). Cells were harvested and washed twice with 50 mM Tris buffer, pH 8.0. Cells were suspended in lysis buffer (50 mM Tris, pH 8.0, 10 mM EDTA and protease inhibitors; Protease Inhibitor cocktail Set IV, Calbiochem), lysed and homogenized by vortex with acid-washed glass beads (0.5 mm, Sigma) for 30 min at 4°C. Cell walls were recovered by centrifugation at 3000 rpm at 4°C and washed 5 times with 1M NaCl to remove bound cytoplasmic proteins. After two final washes in 10 mM Tris pH 8.0, cell walls were suspended in 10 mM Tris, pH 8.0, and mannoproteins were solubilized sequentially. The procedure was derived from those described previously.Citation58 Briefly, cell walls were heat‑treated twice for 15 min at 100°C. The pellet was then extracted with 15 mM dithiothreitol (DTT) in 10 mM Tris, pH 8.0, for 1h at 56°C. The remaining pellet was divided into 2 fractions, one hydrolysed with 15 mM NaOH for 16 h at 4°C and neutralized with acetic acid, and the second digested with 5 mg/mL zymolyase 20T (MP Biomedicals) in 50 mM Tris pH 7.5, containing 10 mM 2‑mercaptoethanol and protease inhibitors for 1h at 37°C. These solubilized CWMPs corresponded, respectively, to non‑covalently bound proteins, proteins bound via disulphide bonds, PIR proteins or proteins directly linked to β-1,3 glucans, and GPI anchor-derived proteins linked to β-1,6 glucans. Cell wall protein content was estimated by the BCA (bicinchoninic acid) Protein Assay (Pierce). 5 μg of protein was loaded in yeast samples while 1 μg protein was used for hyphal samples.
Protein extraction from Candida-macrophage interaction was performed as previously described.Citation98 Anti‑phospho‑p44/42 (Thr202/Tyr204) antibody (New England Biolabs) was used to detect dually phosphorylated C. albicans MAPKs Mkc1 and Cek1 and Erk1/Erk2 from macrophages, anti‑galectin‑3 rat mAb antibody (supplied by Santa Cruz Biotechnology) was used to detect galectin‑3, and anti‑actin (mAB ICN) and anti‑tubulin (AbD serotec) were used as loading controls. Western blots were developed according to the manufacturer's instructions using the Hybond ECL kit (AmershamPharmacia Biotech).
C. albicans binding assays
For analysis of binding of fungal cells to macrophages and inhibition by lactose, yeast cells from overnight growing cultures were washed twice with PBS and re‑suspended in PBS containing 0.1 mg/mL FITC (Sigma-Aldrich; 1h, RT). After extensive washing, FITC-labeled yeasts were re‑suspended in PBS and counted. FITC‑labeled C. albicans yeast cells were added immediately to a confluent monolayer of macrophages from cell line Raw 264.7 at MOI 10:1 (yeast:macrophage). Interaction was allowed for 30 min at 4ºC. After extensive washing with PBS to remove unbound yeasts, cells were fixed with 4% paraformaldehyde (1h, RT), washed, and assessment of immune yeast bound to macrophages was performed by flow cytometry with a FACS guava easyCyte (Millipore). When indicated, macrophage cells were pre‑treated with 100 mM lactose for 30 min at 37ºC / 5% CO2.
To analyze the phagocytosis of C. albicans to macrophages, a differential staining protocol was used to quantify the phagocytic process.Citation99 C. albicans yeasts were pre‑labeled with Oregon Green 488 (1 M) (MolecularProbes). Labeling was performed in the dark with gentle shaking (30°C) for 1 h. Cells were washed twice with PBS, 100 mM glycine and re‑suspended in PBS at the desired density. Raw 264.7 macrophages (5 × 105 cells/well) were allowed to ingest Oregon Green-labeled yeasts on sterile glass coverslips for 3 h at 37ºC in 5% CO2 atmosphere using a fungus-macrophage ratio of 1:1. Cells were then washed with ice-cold PBS and fixed in 4% paraformaldehyde for 30 min. To distinguish between internalized and attached/non-ingested yeast, C. albicans cells were counter stained with calcofluor white M2R (Sigma) (2.5 μM) for 15 min in the dark. After several washes, coverslips were mounted with specific mounting medium (DakoCytomation Denmark A/S). The number of ingested cells (containing green fluorescence) and/or adhered/non-ingested (calcofluor white blue fluorescence) were quantified by phase-contrast and fluorescence microscopy using a Carl Zeiss Axioplan-2 microscope (Carl Zeiss AG) with a mercury HBO/100-watt lamp fitted with FITC (excitation/emission BP 480/30 and BP 535/40, respectively) and UV filters (4_,6-diamidino-2-phenylindole, dihydrochloride excitation/emission BP 365/12 and long pass 397, respectively) and equipped with a digital camera, Spot-2 (Diagnostic Instruments). Metamorph5.0 (Universal Imaging Corp.) and ImageJ version 1.35h (available at rsb.info.nih.gov/ij/index.html) software were used to analyze images. Data are expressed as the percentage of cells internalized by macrophages of at least three different experiments.
Experimental animal models and virulence assay
Galectin‑3 knockout mice were developed as previously describedCitation100 and were kindly provided by Dr. Fu‑Tong Liu. galectin-3+/+ and galectin-3−/− littermates obtained from heterozygous breeders were used to obtain peritoneal macrophages, to prepare BMDCs and BMMs and for the virulence assay. The mice used in the virulence assay were maintained in the animal facility from UC Davis Medical School, California, USA. All the experiments were approved by the Institutional Animal Care and Use Committee of the University of California, Davis (Sacramento, CA).
For the virulence assay 11 male and 7 female C57BL/6J mice (10 galectin-3+/+ and 10 galectin-3−/−), with age between 5.3 and 7.7 weeks and weight between 17 and 26 grams were used. C. albicans strains were grown overnight in YEPD at 37ºC, washed 2 times with sterile PBS and counted in the Neubauer chamber. 0.66 × 106 CAF2 and 0.59 × 106 cek1 cells were used to infect galectin-3+/+ and galectin-3−/− mice via the tail vein. Mice weight loss was measured every day and after death, organs were weighted, homogenized via the WhirlBag method (Whirl Pak Bags® from Fisher Scientific), diluted in PBS and cultured in YEPD plates for 24h for CFU assessment.
Immunofluorescence galectin assay
The interaction between C. albicans and murine macrophages to reveal galectin expression, was visualized through indirect immunofluorescence. Raw 264.7 were grown on glass coverslips in 24‑well plates until confluent monolayers were formed. The immune cells were then infected with yeast cells constitutively expressing a red fluorescent protein (RFP) at a MOI 1:1 (yeast:MΦ). Upon 45 min. and 3h interaction at 37ºC and 5% CO2, the samples were washed with PBS, fixed with paraformaldehyde 4% for 30 min. at 4ºC, and washed again. Cell membranes were permeabilized for 15 min with PBS containing 0.2% Tween-20 at RT. After two washes with PBS, the macrophage Fc receptors were blocked with IgG from mouse serum (Sigma-Aldrich) overnight (1:500) in RPMI containing 10% BSA and 0.2% Saponin. After this, the coverslips were washed twice for 10 min. in gentle shacking and overlaid with Rat anti‑galectin‑3 antibody, diluted 1:1000, for 1h. The samples were washed with PBS, overlaid with anti-Rat FITC antibody (Sigma-Aldrich, diluted 1:1000), incubated for 1h and finally washed with PBS in the dark and mounted with anti-fading solution for fluorescence microscopy.
Fluorescence microscopy images were obtained from a Nikon Eclipse TE2000-U coupled with a Hamamatsu ORCA-ER CCD camera. The filter used was Nikon B-2E/C for green (Alexa488) and Nikon G-2A for red (RFP). All pictures from each experiment were taken (gain and exposure) and processed (brightness and contrast) equally, using the Aquacosmos 1.3 software and Adobe Photoshop CS5.
Statistical analysis
One‑way ANOVA followed by Dunnett's correction for multiple comparisons was applied to evaluate differences among mutants as indicated in the figures. Data are expressed as the mean of three experiments ± SD, unless otherwise stated. Statistics were performed using the GraphPad software. In all cases * indicates p ≤ 0.05, ** p ≤ 0.01, and *** p < 0.001. Survival data was analyzed by Kaplan-Meier log rank (Mantel-Cox) test statistics.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KVIR_S_1163458.zip
Download Zip (538.5 KB)Acknowledgments
We thank M. Whiteway for sharing strains. We are grateful to L. Yu and M. Alvarez for technical assistance.
Funding
Work in our laboratory is supported by Grants PCIN-2014-052 (INFECT-ERANET), BIO2015-64777-P from Ministerio de Economía y Competitividad and Grant S2010-BMD2414 from Comunidad Autónoma de Madrid.
References
- Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence 2013; 4:119-28; PMID:23302789; http://dx.doi.org/10.4161/viru.22913
- Gow NA, Hube B. Importance of the Candida albicans cell wall during commensalism and infection. Curr Opin Microbiol 2012; 15:406-12; PMID:22609181; http://dx.doi.org/10.1016/j.mib.2012.04.005
- Poulain D, Jouault T. Candida albicans cell wall glycans, host receptors and responses: elements for a decisive crosstalk. Curr Opin Microbiol 2004; 7:342-9
- Chaffin WL. Candida albicans cell wall proteins. Microbiol Mol Biol Rev 2008; 72:495-544; http://dx.doi.org/10.1128/MMBR.00032-07
- Lenardon MD, Munro CA, Gow NA. Chitin synthesis and fungal pathogenesis. Curr Opin Microbiol 2010; 13:416-23; PMID:20561815; http://dx.doi.org/10.1016/j.mib.2010.05.002
- Fradin C, Poulain D, Jouault T. ß-1,2-linked oligomannosides from Candida albicans bind to a 32-kilodalton macrophage membrane protein homologous to the mammalian lectin galectin-3. Infect Immun 2000; 68:4391-8; PMID:10899835; http://dx.doi.org/10.1128/IAI.68.8.4391-4398.2000
- Jouault T, El Abed-El BM, Martinez-Esparza M, Breuilh L, Trinel PA, Chamaillard M, Trottein F, Poulain D. Specific recognition of Candida albicans by macrophages requires galectin-3 to discriminate Saccharomyces cerevisiae and needs association with TLR2 for signaling. J Immunol 2006; 177:4679-87; PMID:16982907; http://dx.doi.org/10.4049/jimmunol.177.7.4679
- Hall RA, Gow NA. Mannosylation in Candida albicans: role in cell wall function and immune recognition. Mol Microbiol 2013; 90:1147-61; PMID:24125554; http://dx.doi.org/10.1111/mmi.12426
- Fradin C, Bernardes ES, Jouault T. Candida albicans phospholipomannan: a sweet spot for controlling host response/inflammation. Semin Immunopathol 2015; 37:123-30; PMID:25394861; http://dx.doi.org/10.1007/s00281-014-0461-5
- Romani L. Immunity to fungal infections. Nat Rev Immunol 2011; 11:275-88; PMID:21394104; http://dx.doi.org/10.1038/nri2939
- Netea MG, Ferwerda G, Van der Graaf CA, Van der Meer JW, Kullberg BJ. Recognition of fungal pathogens by toll-like receptors. CurrPharmDes 2006; 12:4195-201
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 2010; 11:373-84; PMID:20404851; http://dx.doi.org/10.1038/ni.1863
- Sato K, Yang XL, Yudate T, Chung JS, Wu J, Luby-Phelps K, Kimberly RP, Underhill D, Cruz PD Jr, Ariizumi K. Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor gamma chain to induce innate immune responses. J Biol Chem 2006; 281:38854-66; PMID:17050534; http://dx.doi.org/10.1074/jbc.M606542200
- Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, Fujikado N, Kusaka T, Kubo S, Chung SH, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity 2010; 32:681-91; PMID:20493731; http://dx.doi.org/10.1016/j.immuni.2010.05.001
- Netea MG, Gow NA, Munro CA, Bates S, Collins C, Ferwerda G, Hobson RP, Bertram G, Hughes HB, Jansen T, et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest 2006; 116:1642-50
- Cambi A, Netea MG, Mora-Montes HM, Gow NA, Hato SV, Lowman DW, Kullberg BJ, Torensma R, Williams DL, Figdor CG. Dendritic cell interaction with Candida albicans critically depends on N-linked mannan. J Biol Chem 2008; 283:20590-9; PMID:18482990; http://dx.doi.org/10.1074/jbc.M709334200
- Brouwer N, Dolman KM, van Houdt M, Sta M, Roos D, Kuijpers TW. Mannose-binding lectin (MBL) facilitates opsonophagocytosis of yeasts but not of bacteria despite MBL binding. J Immunol 2008; 180:4124-32; PMID:18322223; http://dx.doi.org/10.4049/jimmunol.180.6.4124
- Esteban A, Popp MW, Vyas VK, Strijbis K, Ploegh HL, Fink GR. Fungal recognition is mediated by the association of dectin-1 and galectin-3 in macrophages. PNAS USA 2011; 108:14270-5; PMID:21825168; http://dx.doi.org/10.1073/pnas.1111415108
- Brown GD, Gordon S. Immune recognition. A new receptor for beta-glucans. Nature 2001; 413:36-7; PMID:11544516; http://dx.doi.org/10.1038/35092620
- Sundblad V, Croci DO, Rabinovich GA. Regulated expression of galectin-3, a multifunctional glycan-binding protein, in haematopoietic and non-haematopoietic tissues. Histol Histopathol 2011; 26:247-65; PMID:21154238
- Rabinovich GA, Toscano MA. Turning 'sweet' on immunity: galectin-glycan interactions in immune tolerance and inflammation. Nat Rev Immunol 2009; 9:338-52; PMID:19365409; http://dx.doi.org/10.1038/nri2536
- Liu FT, Patterson RJ, Wang JL. Intracellular functions of galectins. Biochim Biophys Acta 2002; 1572:263-73; PMID:12223274; http://dx.doi.org/10.1016/S0304-4165(02)00313-6
- Ochieng J, Furtak V, Lukyanov P. Extracellular functions of galectin-3. Glycoconj J 2004; 19:527-35; PMID:14758076; http://dx.doi.org/10.1023/B:GLYC.0000014082.99675.2f
- Kohatsu L, Hsu DK, Jegalian AG, Liu FT, Baum LG. Galectin-3 induces death of Candida species expressing specific beta-1,2-linked mannans. J Immunol 2006; 177:4718-26; PMID:16982911; http://dx.doi.org/10.4049/jimmunol.177.7.4718
- Román E, Arana DM, Nombela C, Alonso-Monge R, Pla J. MAP kinase pathways as regulators of fungal virulence. Trends Microbiol 2007; 15:181-90; http://dx.doi.org/10.1016/j.tim.2007.02.001
- Navarro-García F, Sanchez M, Pla J, Nombela C. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol Cell Biol 1995; 15:2197-206; http://dx.doi.org/10.1128/MCB.15.4.2197
- Navarro-García F, Alonso-Monge R, Rico H, Pla J, Sentandreu R, Nombela C. A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans. Microbiology 1998; 144:411-24; http://dx.doi.org/10.1099/00221287-144-2-411
- Navarro-García F, Eisman B, Fiuza SM, Nombela C, Pla J. The MAP kinase Mkc1p is activated under different stress conditions in Candida albicans. Microbiology 2005; 151:2737-49; http://dx.doi.org/10.1099/mic.0.28038-0
- Díez-Orejas R, Molero G, Navarro-García F, Pla J, Nombela C, Sánchez-Pérez M. Reduced virulence of Candida albicans MKC1 mutants: a role for a mitogen-activated protein kinase in pathogenesis. Infect Immun 1997; 65:833-7
- Kumamoto CA. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. PNAS USA 2005; 102:5576-81; PMID:15800048; http://dx.doi.org/10.1073/pnas.0407097102
- Alonso-Monge R, Navarro-García F, Molero G, Díez-Orejas R, Gustin M, Pla J, Sánchez M, Nombela C. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J Bacteriol 1999; 181:3058-68; PMID:10322006
- Alonso-Monge R, Navarro-García F, Román E, Negredo AI, Eisman B, Nombela C, Pla J. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryotic Cell 2003; 2:351-61; PMID:12684384; http://dx.doi.org/10.1128/EC.2.2.351-361.2003
- Smith DA, Nicholls S, Morgan BA, Brown AJ, Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell 2004; 15:4179-90; PMID:15229284; http://dx.doi.org/10.1091/mbc.E04-03-0181
- Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, Quinn J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell 2006; 17:1018-32; PMID:16339080; http://dx.doi.org/10.1091/mbc.E05-06-0501
- Eisman B, Alonso-Monge R, Román E, Arana DM, Nombela C, Pla J. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryotic Cell 2006; 5:347-58; PMID:16467475; http://dx.doi.org/10.1128/EC.5.2.347-358.2006
- Csank C, Schröppel K, Leberer E, Harcus D, Mohamed O, Meloche S, Thomas DY, Whiteway M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun 1998; 66:2713-21; PMID:9596738
- Lee BN, Elion EA. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. PNAS USA 1999; 96:12679-84; PMID:10535982; http://dx.doi.org/10.1073/pnas.96.22.12679
- Román E, Nombela C, Pla J. The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol Cell Biol 2005; 25:10611-27; http://dx.doi.org/10.1128/MCB.25.23.10611-10627.2005
- Román E, Cottier F, Ernst JF, Pla J. Msb2 signaling mucin controls activation of Cek1 mitogen-activated protein kinase in Candida albicans. Eukaryotic Cell 2009; 8:1235-49; http://dx.doi.org/10.1128/EC.00081-09
- Herrero de Dios C, Román E, Diez C, Alonso-Monge R, Pla J. The transmembrane protein Opy2 mediates activation of the Cek1 MAP kinase in Candida albicans. Fungal Genet Biol 2013; 50:21-32; PMID:23149115; http://dx.doi.org/10.1016/j.fgb.2012.11.001
- Ernst JF, Pla J. Signaling the glycoshield: maintenance of the Candida albicans cell wall. Int J Med Microbiol 2011; 301:378-83; PMID:21555242; http://dx.doi.org/10.1016/j.ijmm.2011.04.003
- Román E, Alonso-Monge R, Gong Q, Li D, Calderone R, Pla J. The Cek1 MAPK is a short-lived protein regulated by quorum sensing in the fungal pathogen Candida albicans. FEMS Yeast Research 2009; 9:942-55; http://dx.doi.org/10.1111/j.1567-1364.2009.00545.x
- Galan-Diez M, Arana DM, Serrano-Gomez D, Kremer L, Casasnovas JM, Ortega M, Cuesta-Domínguez A, Corbí AL, Pla J, Fernández-Ruiz E. Candida albicans beta-glucan exposure is controlled by the fungal CEK1-mediated mitogen-activated protein kinase pathway that modulates immune responses triggered through dectin-1. Infect Immun 2010; 78:1426-36; PMID:20100861; http://dx.doi.org/10.1128/IAI.00989-09
- Guhad FA, Jensen HE, Aalbaek B, Csank C, Mohamed O, Harcus D, Thomas DY, Whiteway M, Hau J. Mitogen-activated protein kinase-defective Candida albicans is avirulent in a novel model of localized murine candidiasis. FEMS Microbiol Lett 1998; 166:135-9; http://dx.doi.org/10.1111/j.1574-6968.1998.tb13194.x
- Arana DM, Alonso-Monge R, Du C, Calderone R, Pla J. Differential susceptibility of mitogen-activated protein kinase pathway mutants to oxidative-mediated killing by phagocytes in the fungal pathogen Candida albicans. Cell Microbiol 2007; 9:1647-59; PMID:17346314; http://dx.doi.org/10.1111/j.1462-5822.2007.00898.x
- Román E, Alonso-Monge R, Miranda A, Pla J. The Mkk2 MAPKK regulates cell wall biogenesis in cooperation with the Cek1-pathway in Candida albicans. PLoS One 2015; 10:e0133476; http://dx.doi.org/10.1371/journal.pone.0133476
- Chapman R, Sidrauski C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu Rev Cell Dev Biol 1998; 14:459-85; PMID:9891790; http://dx.doi.org/10.1146/annurev.cellbio.14.1.459
- Cantero PD, Ernst JF. Damage to the glycoshield activates PMT-directed O-mannosylation via the Msb2-Cek1 pathway in Candida albicans. Mol Microbiol 2011; 80:715-25; PMID:21375589; http://dx.doi.org/10.1111/j.1365-2958.2011.07604.x
- Cantero P, Lengsfeld C, Subanovic M, Román E, Pla J, Ernst J. Transcriptional and physiological adaptation to defective protein-O-mannosylation in Candida albicans. Mol Microbiol 2007; 64:1115-28; PMID:17501932; http://dx.doi.org/; http://dx.doi.org/10.1111/j.1365-2958.2007.05723.x
- Wheeler RT, Fink GR. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog 2006; 2:e35; PMID:16652171; http://dx.doi.org/10.1371/journal.ppat.0020035
- Torosantucci A, Bromuro C, Chiani P, De BF, Berti F, Galli C, Norelli F, Bellucci C, Polonelli L, Costantino P, et al. A novel glyco-conjugate vaccine against fungal pathogens. JExpMed 2005; 202:597-606; http://dx.doi.org/10.1084/jem.20050749
- Lavigne LM, Albina JE, Reichner JS. Beta-glucan is a fungal determinant for adhesion-dependent human neutrophil functions. J Immunol 2006; 177:8667-75; PMID:17142767; http://dx.doi.org/10.4049/jimmunol.177.12.8667
- Li RK, Cutler JE. Chemical definition of an epitope/adhesin molecule on Candida albicans. J Biol Chem 1993; 268:18293-9; PMID:7688744
- Fradin C, Jouault T, Mallet A, Mallet JM, Camus D, Sinay P, Poulain D. ß-1,2-linked oligomannosides inhibit Candida albicans binding to murine macrophage. JLeukocBiol 1996; 60:81-7
- Jacquinot PM, Plancke Y, Sendid B, Strecker G, Poulain D. Nature of Candida albicans-derived carbohydrate antigen recognized by a monoclonal antibody in patient sera and distribution over Candida species. FEMS Microbiol Lett 1998; 169:131-8; http://dx.doi.org/10.1111/j.1574-6968.1998.tb13309.x
- Trinel PA, Faille C, Jacquinot PM, Cailliez JC, Poulain D. Mapping of Candida albicans oligomannosidic epitopes by using monoclonal antibodies. Infect Immun 1992; 60:3845-51; PMID:1379989
- Chauhan N, Inglis D, Román E, Pla J, Li D, Calera JA, Calderone R. Candida albicans response regulator gene SSK1 regulates a subset of genes whose functions are associated with cell wall biosynthesis and adaptation to oxidative stress. Eukaryotic Cell 2003; 2:1018-24; PMID:14555484; http://dx.doi.org/10.1128/EC.2.5.1018-1024.2003
- Fradin C, Slomianny MC, Mille C, Masset A, Robert R, Sendid B, Ernst JF, Michalski JC, Poulain D. ß-1,2 oligomannose adhesin epitopes are widely distributed over the different families of Candida albicans cell wall mannoproteins and are associated through both N- and O-glycosylation processes. Infect Immun 2008; PMID:18644880
- Martinez-Esparza M, Sarazin A, Jouy N, Poulain D, Jouault T. Comparative analysis of cell wall surface glycan expression in Candida albicans and Saccharomyces cerevisiae yeasts by flow cytometry. J Immunol Methods 2006; 314:90-102
- Shibata N, Ichikawa T, Tojo M, Takahashi M, Ito N, Okubo Y, Suzuki S. Immunochemical study on the mannans of Candida albicans NIH A-207, NIH B-792, and J-1012 strains prepared by fractional precipitation with cetyltrimethylammonium bromide. Arch Biochem Biophys 1985; 243:338-48; PMID:3002275; http://dx.doi.org/10.1016/0003-9861(85)90511-9
- Trinel PA, Jouault T, Cutler JE, Poulain D. Beta-1,2-mannosylation of Candida albicans mannoproteins and glycolipids differs with growth temperature and serotype. Infect Immun 2002; 70:5274-8; PMID:12183581; http://dx.doi.org/10.1128/IAI.70.9.5274-5278.2002
- Sharkey LL, McNemar MD, Saporito-Irwin SM, Sypherd PS, Fonzi WA. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J Bacteriol 1999; 181:5273-9; PMID:10464197
- Kasai K, Hirabayashi J. Galectins: a family of animal lectins that decipher glycocodes. J Biochem 1996; 119:1-8; PMID:8907168; http://dx.doi.org/10.1093/oxfordjournals.jbchem.a021192
- Fermin Lee A, Chen HY, Wan L, Wu SY, Yu JS, Huang AC, Miaw SC, Hsu DK, Wu-Hsieh BA, Liu FT. Galectin-3 modulates Th17 responses by regulating dendritic cell cytokines. Am J Pathol 2013; 183:1209-22; PMID:23916470; http://dx.doi.org/10.1016/j.ajpath.2013.06.017
- Netea MG, Brown GD, Kullberg BJ, Gow NA. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol 2008; 6:67-78; PMID:18079743; http://dx.doi.org/10.1038/nrmicro1815
- Conti HR, Gaffen SL. Host responses to Candida albicans: Th17 cells and mucosal candidiasis. Microbes and Infection 2010; 12:518-27; PMID:20381638; http://dx.doi.org/10.1016/j.micinf.2010.03.013
- Kozel TR, MacGill RS, Percival A, Zhou Q. Biological activities of naturally occurring antibodies reactive with Candida albicans mannan. Infect Immun 2004; 72:209-18; PMID:14688098; http://dx.doi.org/10.1128/IAI.72.1.209-218.2004
- Li D, Williams D, Lowman D, Monteiro MA, Tan X, Kruppa M, Fonzi W, Roman E, Pla J, Calderone R. The Candida albicans histidine kinase Chk1p: signaling and cell wall mannan. Fungal Genet Biol 2009; 46:731-41; PMID:19563901; http://dx.doi.org/10.1016/j.fgb.2009.06.008
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 2000; 101:249-58; PMID:10847680; http://dx.doi.org/10.1016/S0092-8674(00)80835-1
- Wimalasena TT, Enjalbert B, Guillemette T, Plumridge A, Budge S, Yin Z, Brown AJ, Archer DB. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet Biol 2008; 45:1235-47; PMID:18602013; http://dx.doi.org/10.1016/j.fgb.2008.06.001
- Back SH, Schroder M, Lee K, Zhang K, Kaufman RJ. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods 2005; 35:395-416; PMID:15804613; http://dx.doi.org/10.1016/j.ymeth.2005.03.001
- Kimata Y, Kimata YI, Shimizu Y, Abe H, Farcasanu IC, Takeuchi M, Rose MD, Kohno K. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol Biol Cell 2003; 14:2559-69; PMID:12808051; http://dx.doi.org/10.1091/mbc.E02-11-0708
- Hsu CL, Prasad R, Blackman C, Ng DT. Endoplasmic reticulum stress regulation of the Kar2p/BiP chaperone alleviates proteotoxicity via dual degradation pathways. Mol Biol Cell 2012; 23:630-41; PMID:22190740; http://dx.doi.org/10.1091/mbc.E11-04-0297
- Saraswat D, Kumar R, Pande T, Edgerton M, Cullen PJ. Signaling Mucin Msb2 Regulates Adaptation to Thermal Stress in Candida albicans. Mol Microbiol 2016; PMID:26749104
- Staab JF, Bahn YS, Tai CH, Cook PF, Sundstrom P. Expression of transglutaminase substrate activity on Candida albicans germ tubes through a coiled, disulfide-bonded N-terminal domain of Hwp1 requires C-terminal glycosylphosphatidylinositol modification. J Biol Chem 2004; 279:40737-47; PMID:15262971; http://dx.doi.org/10.1074/jbc.M406005200
- Prill SK, Klinkert B, Timpel C, Gale CA, Schroppel K, Ernst JF. PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol Microbiol 2005; 55:546-60; PMID:15659169; http://dx.doi.org/10.1111/j.1365-2958.2004.04401.x
- Bates S, MacCallum DM, Bertram G, Munro CA, Hughes HB, Buurman ET, Brown AJ, Odds FC, Gow NA. Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J Biol Chem 2005; 280:23408-15; PMID:15843378; http://dx.doi.org/10.1074/jbc.M502162200
- Tamai R, Kiyoura Y. Candida albicans and Candida parapsilosis rapidly up-regulate galectin-3 secretion by human gingival epithelial cells. Mycopathologia 2014; 177:75-9; PMID:24436012; http://dx.doi.org/10.1007/s11046-013-9725-1
- Linden JR, Kunkel D, Laforce-Nesbitt SS, Bliss JM. The role of galectin-3 in phagocytosis of Candida albicans and Candida parapsilosis by human neutrophils. Cell Microbiol 2013; 15:1127-42; PMID:23279221; http://dx.doi.org/10.1111/cmi.12103
- Baum LG, Garner OB, Schaefer K, Lee B. Microbe-Host Interactions are Positively and Negatively Regulated by Galectin-Glycan Interactions. Front Immunol 2014; 5:284; PMID:24995007; http://dx.doi.org/10.3389/fimmu.2014.00284
- Linden JR, De Paepe ME, Laforce-Nesbitt SS, Bliss JM. Galectin-3 plays an important role in protection against disseminated candidiasis. Med Mycol 2013; 51:641-51; PMID:23488971; http://dx.doi.org/10.3109/13693786.2013.770607
- Courjol F, Jouault T, Mille C, Hall R, Maes E, Sendid B, Mallet JM, Guerardel Y, Gow NA, Poulain D, et al. beta-1,2-mannosyltransferases 1 and 3 participate in yeast and hyphae O- and N-linked mannosylation and alter Candida albicans fitness during infection. Open Forum Infect Dis 2015; 2:ofv116; PMID:26389126
- Vankrunkelsven A, De Ceulaer K, Hsu D, Liu FT, De Baetselier P, Stijlemans B. Lack of galectin-3 alleviates trypanosomiasis-associated anemia of inflammation. Immunobiology 2010; 215:833-41; PMID:20605052; http://dx.doi.org/10.1016/j.imbio.2010.05.028
- Li Y, Komai-Koma M, Gilchrist DS, Hsu DK, Liu FT, Springall T, Xu D. Galectin-3 is a negative regulator of lipopolysaccharide-mediated inflammation. J Immunol 2008; 181:2781-9; PMID:18684969; http://dx.doi.org/10.4049/jimmunol.181.4.2781
- Ferraz LC, Bernardes ES, Oliveira AF, Ruas LP, Fermino ML, Soares SG, Loyola AM, Oliver C, Jamur MC, Hsu DK, et al. Lack of galectin-3 alters the balance of innate immune cytokines and confers resistance to Rhodococcus equi infection. Eur J Immunol 2008; 38:2762-75; PMID:18825751; http://dx.doi.org/10.1002/eji.200737986
- Quattroni P, Li Y, Lucchesi D, Lucas S, Hood DW, Herrmann M, Gabius HJ, Tang CM, Exley RM. Galectin-3 binds Neisseria meningitidis and increases interaction with phagocytic cells. Cell Microbiol 2012; 14:1657-75; PMID:22827322; http://dx.doi.org/10.1111/j.1462-5822.2012.01838.x
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 2006; 24:179-89; PMID:16473830; http://dx.doi.org/10.1016/j.immuni.2006.01.001
- Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med 2005; 201:233-40; PMID:15657292; http://dx.doi.org/10.1084/jem.20041257
- Hernandez-Santos N, Gaffen SL. Th17 cells in immunity to Candida albicans. Cell Host Microbe 2012; 11:425-35
- Wu SY, Yu JS, Liu FT, Miaw SC, Wu-Hsieh BA. Galectin-3 negatively regulates dendritic cell production of IL-23/IL-17-axis cytokines in infection by Histoplasma capsulatum. J Immunol 2013; 190:3427-37; PMID:23455499; http://dx.doi.org/10.4049/jimmunol.1202122
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics 1993; 134:717-28; PMID:8349105
- San José C, Alonso-Monge R, Pérez-Díaz RM, Pla J, Nombela C. The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J Bacteriol 1996; 178:5850-2
- Leberer E, Harcus D, Broadbent ID, Clark KL, Dignard D, Ziegelbauer K, Schmidt A, Gow NA, Brown AJ, Thomas DY. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. PNAS USA 1996; 93:13217-22; PMID:8917571; http://dx.doi.org/10.1073/pnas.93.23.13217
- Prieto AD, Román E, Correia I, Pla J. The HOG pathway is critical for the colonization of the mouse gastrointestinal tract by Candida albicans. PLoS One 2014; 9:e87128; PMID:24475243; http://dx.doi.org/10.1371/journal.pone.0087128
- Poulain D, Deveaux M, Cailliez JC, Hossein-Foucher C, Dutoit E, Camus D, Van Cutsem J, Marchandise X. Imaging of systemic Candida albicans infections with a radioiodinated monoclonal antibody: experimental study in the guinea pig. Int J Rad Appl Instrum B 1991; 18:677-86
- Marot-Leblond A, Grimaud L, David S, Sullivan DJ, Coleman DC, Ponton J, Robert R. Evaluation of a rapid immunochromatographic assay for identification of Candida albicans and Candida dubliniensis J Clin Microbiol 2004; 42:4956-60; PMID:15528679; http://dx.doi.org/10.1128/JCM.42.11.4956-4960.2004
- Garcia R, Bermejo C, Grau C, Perez R, Rodriguez-Pena JM, Francois J, Nombela C, Arroyo J. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J Biol Chem 2004; 279:15183-95; PMID:14739279; http://dx.doi.org/10.1074/jbc.M312954200
- Martin H, Rodriguez-Pachon JM, Ruiz C, Nombela C, Molina M. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae J Biol Chem 2000; 275:1511-9; PMID:10625705; http://dx.doi.org/10.1074/jbc.275.2.1511
- Fernandez-Arenas E, Cabezon V, Bermejo C, Arroyo J, Nombela C, Díez-Orejas R, Gil C. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol Cell Proteomics 2007; 6:460-78; http://dx.doi.org/10.1074/mcp.M600210-MCP200
- Hsu DK, Yang RY, Pan Z, Yu L, Salomon DR, Fung-Leung WP, Liu FT. Targeted disruption of the galectin-3 gene results in attenuated peritoneal inflammatory responses. Am J Pathol 2000; 156:1073-83; PMID:10702423; http://dx.doi.org/10.1016/S0002-9440(10)64975-9