ABSTRACT
FipB, an essential virulence factor in the highly virulent Schu S4 strain of F. tularensis subsp. tularensis, shares sequence similarity with Disulfide Bond formation (Dsb) proteins, which can have oxidoreductase, isomerase, or chaperone activity. To further explore FipB's role in virulence potential substrates were identified by co-purification and 2D gel electrophoresis, followed by protein sequencing using mass spectrometry. A total of 119 potential substrates were identified. Proteins with predicted enzymatic activity were prevalent, and there were 19 proteins that had been previously identified as impacting virulence. Among the potential substrates were IglC, IglB, and PdpB, three components of the Francisella Type Six Secretion System (T6SS), which is also essential for virulence. T6SS are widespread in Gram-negative pathogens, but have not been reported to be dependent on Dsb-like proteins for assembly or function. The presented results suggest that FipB affects IglB and IglC substrates differently. In a fipB mutant there were differences in free sulfhydryl accessibility of IglC, but not IglB, when compared to wild-type bacteria. However, for both proteins FipB appears to act as a chaperone that facilitates proper folding and conformation. Understanding the role FipB plays the assembly and structure in this T6SS may reveal critical aspects of assembly that are common and novel among this widely distributed class of secretion systems.
KEYWORDS:
Introduction
Francisella tularensis subsp. tularensis is a tier 1 select agent, which is a biological agent determined by the US. Dept. of Health & Human Services to be a potentially severe threat to human or animal health. Concerns over documented and potential use of F. tularensis as a biological weapon, and reports of the development of weaponized strains that are resistant to antibiotics and vaccines have led to increased interest in defining mechanisms of virulence as a means of identifying new targets for therapy and immune protection.Citation1,2
There are several subspecies of F. tularensis that vary in virulence. Francisella tularensis subsp. tularensis causes the most severe disease, while F. tularensis subsp holarctica strains cause a milder disease. Commonly used lower virulence model strains include the Live Vaccine Strain (LVS), an attenuated F. holarctica strain, and Francisella novicida, a closely related species that has low virulence in humans, but is virulent in mice. Although they vary significantly in virulence, all of the subspecies are quite similar at the genomic level (>98 %).Citation3 There are few recognizable virulence factors encoded in the Francisella genome, but several have been identified that are required for virulence and phagosomal escape.Citation4-8 Included in this group of virulence factors are FipB, as well as components of the Francisella Type Six Secretion System (T6SS).
FipB is an essential virulence factor that is required for phagosomal escape, and intracellular survival.Citation9 FipB mutants are also completely avirulent in mice.Citation9 FipB protein has similarity to DsbA proteins, which are periplasmic oxidoreductases that catalyze disulfide bond formation in nascent proteins.Citation10,11 E. coli, as well as many other Gram-negative bacteria, also contain a separate but related protein DsbC, which is an isomerase that refolds incorrectly folded proteins.Citation12 However, a separate DsbC protein has not been identified in Francisella strains. We, and others have found that FipB has both oxidoreductase and isomerase activities.Citation13,14 Recombinant FipB has also been demonstrated to have chaperone activity using an in vitro assay.Citation15
T6SSs are quite widespread among Gram-negative bacteria.Citation16 Some are key for host-pathogen interactions, but others function more in bacteria-bacteria encounters, and may be important for colonization of an environmental niche. The Francisella T6SS is clearly an outlier T6SS sharing only five out of 13 core conserved T6SS proteins leading to some speculation as to whether the Francisella T6SS functions as a secretion system.Citation17 However, demonstration of a contractile sheath by cryo-electron microscopy Citation18 and secretion of effector proteins into the host cytoplasm have largely confirmed that the genomes of Francisella species encode a functional T6SS.Citation19-21 A model of the Francisella T6SS structure, which resembles other T6SSs and is supported by the atomic structure,Citation18 proposes that two proteins, IglA and IglB, form heterodimers and polymerize to assemble the contractile sheath. A third protein, IglC, forms hexamers and assembles as a nanotube inside the IglA/IglB sheath.Citation22 This model for IglC is based on the structure of Hemolysin coregulated protein, (Hcp), a conserved component of T6SSs.Citation22 It has been suggested that IglC serves as the functional equivalent of Hcp, though they share little sequence similarity. IglC, like Hcp, is also secreted.Citation19,20 Mutations in iglA, iglB and iglC are severely attenuating, and mutant bacteria fail to escape into the cytoplasm.Citation18,22,23
DsbA activity is important for the function of many virulence factors in other bacteria including the assembly and function of Type Three Secretion Systems.Citation24-26 We therefore reasoned that the avirulence of a ΔfipB mutant was due to improper folding of key Francisella virulence factors. To better understand the function and role of FipB in virulence we sought to identify its substrates. Among the identified substrates were three components of the T6SS, IglB, IglC, and PdpB. Although this makes sense phenotypically, because both ΔfipB and mutants in the T6SS are highly attenuated and fail to escape the phagosome, in other organisms T6SS have not been reported to require DsbA for function, and the crystalized structure of IglC does not suggest intramolecular disulfide bonds.Citation27 Our data suggest that FipB is required for proper protein folding of the T6SS components. The dependence of the T6SS on FipB may be an adaptation to the loss of some of the conserved elements found in other T6SSs.
Results
Identification of FipB substrates
Functional and mutational analyses of the FipB protein and gene support a role for the FipB protein in disulfide bond formation, including an essential dependence on the two cysteines found in the conserved active site of DsbA proteins, CXXC (Cysteine- any amino acid- any amino acid-Cysteine).Citation14,28 We hypothesize that the avirulence of the ΔfipB mutant is due to the inability to produce functional critical virulence factors. To identify factors that were dependent on FipB, we sought to co-purify FipB with its substrates. In E. coli it was reported to be difficult to co-purify DsbA with its substrates. Kadokura et al. speculated that the interaction of E. coli DsbA with its substrates was rapid and transient, making co-purification difficult.Citation29 They performed a mutational analysis to identify mutations that would slow down the oxidation reaction or trap substrates. Mutation of the conserved cis-proline residue P151, which is located in the substrate-binding pocket of E. coli DsbA,Citation30 enhanced the interaction of E. coli DsbA with its substrates and facilitated their identification.Citation29 Mutation of the second cysteine in conserved active site of DsbA proteins, CXXC to alanine Citation29 also resulted in substrate trapping.Citation31
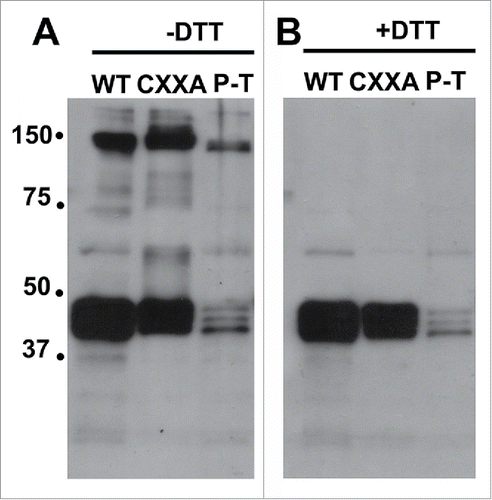
FipB has a cis-proline residue located nearly the same distance (just one 1 amino acid farther) from the conserved enzymatic CXXC site as in E. coli DsbA. The structure of FipB has not been solved, but we reasoned that mutations in these corresponding residues, C167A and P286T would similarly trap FipB substrates. His-tagged versions of wild-type, C167A (CXXA), and P286T FipB proteins were introduced in trans via a plasmid encoding these proteins into a ΔfipB mutant of Schu S4, a virulent F. tularensis subsp. tularensis strain. His-tagged FipB was purified using a metal binding column, and then the purified material was separated on SDS-PAGE using non-denaturing or denaturing sample buffer, transferred to a nylon membrane, and incubated with anti-FipB antibody (). High molecular sized complexes containing FipB were observed under non-denaturing conditions for wild-type, CXXA and P286T versions of FipB, although there were fewer complexes detected with FipB-P286T. Note that FipB alone migrated as a triplet between the 37 and 50 kDa markers. This is a typical pattern of FipB migration on Western blots; these isoforms are likely due to differences in glycosylation.Citation32
Figure 1. FipB migrated as high molecular weight complexes that were sensitive to reduction. FipB protein was purified from strains expressing His-tagged wild-type FipB, FipB-CXXA, or FipB P286T, using a metal affinity column. FipB was eluted from the column and run on separate SDS-PAGEs using sample buffer without (−DTT, Panel A) or with DTT (+ DTT, Panel B), and then transferred to a nylon membrane for Western blotting. FipB complexed with co-purified substrates were visualized with anti-FipB antibody.
Two methods were used to identify the proteins that co-purified with FipB, 2D gel electrophoresis followed by mass spectrometry sequencing of individual spots (Fig. S1), and mass spectrometry sequencing of purified His-tagged FipB complexes without further separation. The sequencing of seven individual spots from the 2D gel identified 52 proteins (Table S1). Samples for direct sequencing of co-purified material included His-tagged FipB-P286T, and FipB-CXXA as well as a His-tagged FipB-AXXA as a negative control. Overall 107 proteins were identified by co-purification with FipB-CXXA or FipB-P286T in this analysis. Only 29 proteins were identified from the FipB-P286T sample. Three proteins were unique to FipB-P286T, but these proteins were only identified by one or two peptides. We failed to detect any protein that co-purified with the His-tagged FipB-AXXA mutant, supporting a requirement for a disulfide bond interaction between FipB and its substrates. In total 119 proteins were identified by total sequencing and 2D gel analyses (Table S1). Forty out of the 52 proteins (77%) detected by 2D gel separation were also identified by sequencing the total co-purified material.
A summary of predicted subcellular location and Clusters of Orthologous Genes (COG) categories classification of all 119 proteins detected by 2D gel or total sequencing of His-tagged purified FipB-CXXA and FipB-P286T is shown in . The subcellular location of many of the proteins (45%, 53/119) could not be predicted. Twenty-nine percent (35/119) were predicted to be cytoplasmic, and 26% (31/119) in the outer membrane, secreted, or found in multiple locations. A diverse set of 20 COG categories was represented. The top six categories, ranging from 16 to 9%, were: i) Unknown, ii) Energy production and conversion, iii) Translation, ribosome structure, and biogenesis, iv) Cell envelope biogenesis and outer membrane, v) Amino acid transport and metabolism, and vi) Posttranslational modification, protein turnover, chaperones. Eighty-two percent of the proteins had two or more cysteines. Eleven proteins had one cysteine and 11 had no cysteines, which was unexpected. Since we failed to detect any co-purifying proteins with the AXXA mutant of FipB, we anticipated that there would have been at least one cysteine in the protein (see Table S1). It is possible that these proteins interact with FipB indirectly though interaction with a substrate or through a non-specific interaction with a substrate. Five out of the 22 had a predicted cytoplasmic location, and another three had significant similarity to translation machinery, supporting the later possibility. Ten were identified by only one or two peptides; however, others were identified by greater than 15 peptides.
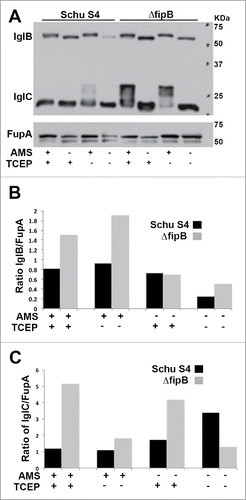
Figure 2. Accessibility of free sulfhydryls in IglB and IglC in wild-type and ΔfipB mutant bacteria. Panel A) Total bacterial lysates were labeled with AMS, a reagent that reacts with free sulfhydryls and adds 500 Da. Some samples were first treated with TCEP, to reduce existing disulfide bonds. Samples were separated on 4–15% SDS gel before transfer to PVDF membranes for immunoblots. Proteins were visualized with anti-IglC and IglB monoclonal antibodies. The same blot was stripped and then rehybridized with anti-FupA antibody. Blots are representative of at least three blots. Panels B& C) Blots were scanned by densitometry, and the amount of IglB (Panel B) or IglC (Panel C) was compared to the loading control FupA.
To narrow the list of putative substrates to those with greater confidence we applied a more stringent filter. Proteins that were identified in at least two out of the three samples, CXXA, P286T samples, or 2-D gel, or by four or more peptides in at least one sample are listed in . We also eliminated any gene that was likely essential. Essentiality was judged based on the failure to isolate a transposon mutant in that gene in a saturated comprehensive library of F. novicida (Francisella.org) Citation33 and bioinformatics. Thirteen proteins that fell into this category were annotated to have roles in such functions as in translation, transcription and various cytosolic metabolic functions. These proteins are highlighted in Table S1. While this filter is not foolproof, and may eliminate valid substrates or retain some nonphysiological interactions, it allowed for selection of higher confidence FipB-substrate interactions. Using these criteria, 37 putative substrates were identified. With one exception all had two or more cysteines, and only two were predicted to have a cytoplasmic location. Proteins with predicted enzymatic activity were prevalent (14/37). Of particular note were 19 putative FipB substrates that have been previously identified as impacting virulence in various screens for attenuated mutants in mice, or for defects in intracellular growth in various cells types. Among the known virulence factors identified were DipA, FopA, KatG, EmrA1, and three components of the T6SS.
Table 1. List of putative FipB substrates
FipB is required for the proper folding of T6SS components
Intramolecular disulfide bond interactions among T6SS components have not been explored, though the solved crystal structure of IglC does not suggest any intramolecular disulfide bonds.Citation27 IglB, which has six cysteines, interacts with IglA to form the contractile sheath through the interdigitation of strands from both proteins.Citation18 PdpB, which has five cysteines, is proposed to be an inner membrane component.Citation34 IglC has four cysteines, and is predicted to comprise the core of the sheath and is also secreted into the host cytoplasm.Citation19,20,34
If these T6SS components were FipB substrates, then one would expect that the oxidation state of their cysteines would be altered in a FipB mutant. To examine the oxidation state of the cysteines of IglB and IglC, TCA-precipitated cell lysates were labeled with the sulfhydryl-reactive reagent AMS. AMS irreversibly reacts with free sulfhydryls, and adds 500 Da to the molecular weight of the protein. PdpB was not analyzed by this method because even if all 5 cysteines were labeled, this would only add 2.5 kDa to a 127 kDa protein, which would be difficult to detect with this gel system.
IglB was labeled with AMS, with or without treatment with the reducing agent TCEP, as evidenced by the slight decrease in band migration in AMS treated samples in both in wild-type and ΔfipB strains, indicating that IglB has free sulfhydryl groups that are not influenced by FipB (). However, in samples that were not treated with AMS and TCEP there was less IglB detected in both strains (). IglB polymerizes with IglA to form the T6SS sheath, so one might expect that as part of the polymerized sheath IglB would not be able to enter the gel.Citation18 Since the amount of IglB was less under nonreducing conditions this could suggest that the formation of the sheath is sensitive to reducing agents. Another related possibility is that epitope recognized by the IglB-specific monoclonal antibody is more accessible when the protein is labeled with AMS or reduced. We were unable to detect IglB with this IglB monoclonal in any wild-type lysate on SDS-PAGE when nonreducing loading buffer was used, but we could detect some IglB in ΔfipB lysates (data not shown). Of note was that more IglB was detected in all ΔfipB lysates compared to wild-type, lending support to the idea that FipB influences the conformation of IglB ().
Since IglC has free cysteines then one might expect to see up to four additional bands corresponding to AMS-labeled IglC. There was an increase in the amount of IglC with 3 or 4 AMS- labeled cysteines in the ΔfipB mutant compared to wild-type bacteria (), indicating that there were more free accessible cysteines in IglC in the ΔfipB mutant. However, we also observed an unexpected result; when lysates from wild-type bacteria were first reduced with TCEP before labeling with AMS, a treatment that should result in the labeling of all cysteine residues, there was no AMS labeling (). This result was also seen when DTT was used as the reducing agent (data not shown). This result suggests that IglC forms a tightly folded, reduction-resistant protein that is dependent on FipB, and that reduction with TCEP also helps to protect free cysteines from labeling. We also observed that, similar to IglB, there was overall more IglC protein detected in most of ΔfipB mutant lysates, (). The exception was the lysate that had not been treated with AMS or TCEP. As mentioned above a possible explanation for this result is that the epitope recognized by the anti-IglC monoclonal antibody is more exposed in the ΔfipB mutant. Take all together these data support the conclusion that in the absence of FipB IglB and IglC do not assume their native conformations.
To further explore the effect of FipB on IglC His-tagged recombinant IglC and FipB were purified and used in an in vitro assay to examine the ability of FipB to affect the oxidation or reduction of IglC's cysteines (). Recombinant His-IglC alone formed multiple higher molecular weight forms that were not influenced by the presence of up to 50 μM DTT. However, when FipB was added the number of higher molecular weight bands decreased and there was increased AMS labeling. Although this in an in vitro assay, it demonstrated the ability of FipB to directly reduce IglC intermolecular disulfide bonds.
Figure 3. FipB resolved higher MW complexes of IglC and prevented higher MW complexes of FopA. Panel A) His-IglC was incubated with AMS and increasing concentrations of DTT in the presence or absence of His-FipB. Panel B) Western blot of total cell lysates of wild-type, ΔfopA, and ΔfipB strains. The same blot was incubated with anti-FopA antibody, then stripped and incubated with anti-FipB, and then stripped again and incubated with anti-FupA antibody.
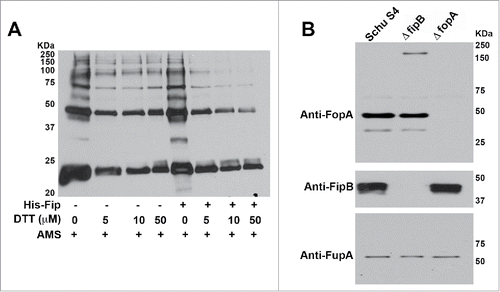
We looked at another FipB substrate, FopA, and detected a higher molecular weight band corresponding to FopA only in ΔfipB mutant lysates (), suggesting that prevention of incorrect disulfide bonds can occur in vivo.
Growth in KCl is deleterious to Schu S4 strains
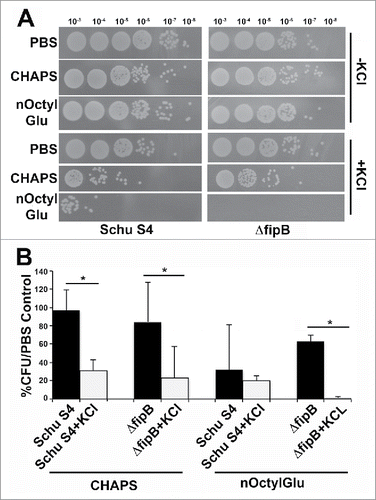
If IglC, IglB or PdpB were FipB substrates, then one would predict that the T6SS would not be functional in a ΔfipB mutant. Testing whether the T6SS is functional in a ΔfipB mutant is challenging because in Francisella the secretion of effectors had only been detected in the cytoplasm of host cells.Citation19-21 Since the ΔfipB mutant does not grow intracellularly this cannot be directly assessed. However, Clemens et al. found that secretion of T6SS effectors could be induced in F. novicida by adding 5% KCl to the growth media.Citation18 When we tried this protocol with Schu S4 we found that the addition of 5% KCl to the growth media drastically inhibited growth. Schu S4 and the ΔfipB mutant did grow when 2.5% KCl was to the added to the growth media, though cultures failed to reach a similar OD595 in stationary phase as bacteria grown without added KCl (Fig. S3). Attempts were made to determine if T6SS effectors were secreted under this condition. Culture supernatants were examined for presence of secreted effector IglC in the presence of added KCl (). Non-secreted T6SS component IglB, FopA, an outer membrane protein, and LPS were used as controls for cell lysis or decreased membrane integrity. IglC was detected in KCl-treated culture supernatants from both wild-type and ΔfipB strains. However, we also detected IglB, and FopA in the same supernatants, indicating that the addition of KCl increased cell lysis or affected membrane integrity. This situation was even more acute for the ΔfipB mutant because we could detect LPS in the supernatant. There was also indication of loss of membrane integrity without added KCl in the ΔfipB mutant. To test this further bacteria were grown overnight in TSB/c ± KCl, incubated with different concentrations of detergents CHAPS or nOctylGlu, diluted, and then spotted on MHA/c plates. The results for the 0.25% detergent condition are shown in . Both wild-type and ΔfipB strains were more sensitive to killing by detergents when grown with KCl. Schu S4 exhibited a decrease in CFU even in the absence of detergent, suggesting that KCl alone has a toxic effect on Schu S4. There was also a decrease in CFUs in the KCl-grown ΔfipB mutant, though not to the same extent. This may be because the difference in the final OD595 in stationary phase, ± KCl was less for the ΔfipB mutant (Fig. S3). In sum, these results indicate that in Schu S4 strain background growth in KCl is not a viable means of assessing T6SS-mediated secretion and therefore we were unable to directly show that T6SS-mediated secretion was disrupted in a ΔfipB mutant.
Figure 4. Detection of nonsecreted T6SS component and outer membrane constituents in the supernatants of KCl-grown bacteria. IglB, a nonsecreted T6SS component, outer membrane protein FopA, and LPS were detected in culture supernatants of KCl-grown bacteria. Bacterial cultures were grown overnight with or without 2.5 % KCl, and adjusted to the same OD595. Whole cell lysates and culture supernatants were prepared as described in material and methods. The Western blot was incubated with anti-IglB and IglC antibodies, then stripped and incubated with an anti-FopA antibody. The same blot was stripped again and incubated with an anti-LPS antibody. Statistical significance was measured using an ANOVA and Dunn's multiple comparison tests (* p value <0 .05).
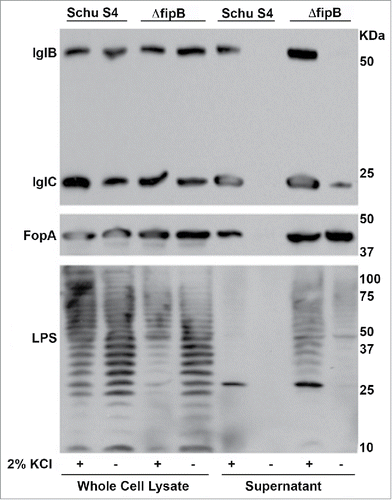
Figure 5. Growth in 2.5% KCl increases sensitivity to detergents. Schu S4 or ΔfipB strains were grown overnight in TSB/c with or without 2.5% of KCl. Cultures were adjusted to an OD595 of one, then incubated with 0.25% CHAPS or n-Octyl glucoside (nOctylGlu) for 90 min at RT. Serial dilutions, (by a factor of ten), were spotted on MHA/c plates and incubated at 37 ˚C for two d (Panel A). Quantitation of the detergent sensitivity was determined by comparing the number of recovered CFUs compared to PBS-treated controls from at least three independent experiments (Panel B). Statistical significance was measured using an ANOVA and Dunn's multiple comparison tests (* p value <0 .05).
Discussion
Among the potential FipB substrates identified in this paper were three components of the Francisella T6SS IglB, IglC, and PdpB. The data presented in this paper provide evidence that the T6SS of Francisella requires the FipB protein for proper conformation of IglB and IglC, and likely impacts the assembly of the T6SS apparatus. Both fipB and iglC mutants are highly attenuated and fail to escape the phagosome, so the avirulence of fipB mutants may largely be attributed to the loss of a functional T6SS.
The FipB protein has been shown to have both oxidoreductase, and isomerase activities, and is presumed to catalyze disulfide bonds in substrate proteins and also rearrange incorrectly formed disulfide bonds.Citation13,15,28 The FipB ortholog in the LVS strain of F. tularensis holarctica has been shown to have chaperone activity in vitro using the citrate synthase assay.Citation15 The results from the experiments presented in this paper support an isomerase or chaperone role for FipB that may or may not involve a disulfide bond in the final conformation of the protein.
Our data support a role for FipB in the formation of the T6SS structure. We observed an apparent increase in the amount of IglB and IglC protein in the ΔfipB mutant. The most likely explanation for this is that the epitopes recognized by the monoclonal antibodies used to detect these proteins are more exposed in the ΔfipB mutant, which is an indication that these proteins are not in the correct conformation. While there was a difference in AMS labeling of IglC in ΔfipB mutant, for IglB, there was no detectable difference in free sulfhydryls between the wild-type and the ΔfipB mutant, suggesting that for some substrates FipB may reduce incorrect disulfide bonds or prevent them from forming. The ClpV ATPase, a conserved component found in other T6SSs, but apparently absent in Francisella T6SS, has been shown to have a chaperone function that is required for the assembly of the IglA/IglB-equivalent protein sheath; Citation35 FipB could be supplying this aspect of the ClpV ATPase.
It is possible that the changes in IglB and IglC conformations are indirect, and the result the effect of FipB on another substrate, which may also have chaperone activity. However, there were no likely candidates identified by our analysis. We were able to show in an in vitro assay that recombinant FipB was able to reduce multimers of IglC to monomer form. While this supports the model, it should be noted that these assays have low substrate specificity.Citation36
The specific mechanism of interaction between FipB and IglB and IglC needs to be further explored, but it must rely on the CXXC enzymatic active site of FipB; no substrates were purified with an AXXA mutant of FipB, and previous work has shown that the enzymatic active site of FipB is essential for its role in virulence.Citation28 The T6SS of Francisella is atypical, lacking many of the conserved proteins characteristic of T6SSs.Citation17 The requirement of FipB may be an adaptation to the loss of some of these conserved components and has assumed the role of chaperone. Further studies of the interaction of FipB and the T6SS components will help to define its structure and assembly, which can then be contrasted and compared to other T6SSs.
We attempted to determine if T6SS was operating in the ΔfipB mutant by inducing secretion using growth in 5% KCl, a method that was used by Clemens et al. to induce T6SS effectors in F. novicida. However, we found that Schu S4 did not grow in 5% KCl. The bacteria tolerated growth in 2.5% KCl, but when we looked for secretion of effectors in the supernatant, we detected IglC, which was expected, but also found structural component IglB, outer membrane protein FopA, and LPS, which should not have been present (). Bacteria grown in the presence of 2.5% KCl were also more sensitive to detergents (). These results indicated that in Schu S4, and likely other F. tularensis subsp tularensis strains, growth in KCl compromises the membrane integrity and unlike F. novicida, this method cannot be used to assess T6SS in Schu S4. These results also suggest that regulation or control of the secretion of T6SS effectors differs between F. novicida and the more virulent F. tularensis subsp. tularensis strain Schu S4. In nature F. tularensis subsp. tularensis and F. novicida reside in different environmental niches.Citation37 F. tularensis subsp. tularensis is a vector borne, zoonotic bacteria, while F. novicida has never been isolated in wild animals or arthropods, but associated with brackish or salt water, and soil, so different environmental inducers might be expected.
We identified 119 potential FipB substrates in the Schu S4 strain using a combination of 2D gel electrophoresis and tandem mass spectrometry. These substrates need to be validated through other methods, in part because proteins that are likely found only in the cytoplasm were identified, as well as several proteins that. are predicted to be essential. The ΔfipB mutant grows in culture, so substrates that are essential proteins would not be expected. Many of these essential proteins are also cytoplasmic, but it is possible that FipB may have some noncritical chaperone or disulfide bond formation role for extracytoplasmic essential proteins.
FipB substrates have also been identified in the LVS strain of F. tularensis holarctica,Citation14,38 (see Table S1). Straskova et al. used 2D gels and the sensitive protein labeling technique, ITRAQ®, to compare protein abundance in wild-type LVS and an LVS ΔfipB (dsbA) strain.Citation14 This was based on the assumption that if FipB were necessary for protein folding or disulfide bond formation, then its substrates would be unstable and degraded. Hiniker and Bardwell used this assumption to identify a number of E. coli DsbA substrates by comparing the abundance of specific proteins in wild-type and dsbA strains using 2D gels.Citation39 Straskova et al. were able to identify 11 proteins with altered abundance in a dsbA (fipB) LVS mutant using ITRAQ®, however, nine had increased levels, and only two were decreased. The two proteins that had decreased abundance corresponded to FipB, and FipA. FipA is a small polypeptide encoded just upstream of FipB, in the same transcriptional unit.Citation28 We identified four out nine of these proteins (Table S1). Ren et al. used a method similar to ours, co-purifying proteins with a CXXA mutant bait.Citation38 We identified 25 out of the 52 substrates identified by Ren et al. Of note, Ren et al. failed to identify IglC, or IglB. We used the virulent Schu S4 strain, while both other investigators used LVS, but experimental conditions may also account for the differences in the identified putative substrates.
FipB is a novel member of the DsbA family of proteins that can participate in multiple roles in disulfide bond formation and protein folding. The ability of one protein to perform these multiple functions may be an adaptation to its reduced genome. FipB's essential requirement for virulence and its ability to affect the functions of multiple proteins, including components of the T6SS, make it an attractive target for antimicrobial therapy.
Materials and methods
Bacterial strains, media and reagents
Francisella strains were grown on cysteine-supplemented Mueller-Hinton agar (MHA/c) plates, or in Trypticase Soy broth (TSB/c) and with kanamycin (Kan) (15 μg/ml) when required. Studies involving Schu S4 and derivatives of this strain were carried out in an approved Biosafety Level 3 laboratory. Escherichia coli strains were grown in Luria-Bertani (LB) broth, or on LB agar plates with kanamycin (50 μg/ml) as required. Monoclonal antibodies specific for IglB, IglC, and PdpB were obtained from BEI Resources. Antibodies to FipB, FopA, and LPS were generated in house. Anti-FupA was gift from Girija Ramakrishnan.Citation40
Construction of mutant strains
Mutation and plasmid constructions were performed as previously described.Citation9 DNA was prepared and purified using a commercial kit (Qiagen). Oligonucleotides were synthesized by Integrated DNA Technologies Inc. Restriction endonucleases and ligase were purchased from New England Biolabs. HotStart® Taq (Qiagen) was used for routine PCR. FastStartH High fidelity PCR system (Roche) was used for construction of plasmids. All cloning products were verified by DNA sequencing. Site direct mutagenesis was accomplished with a site-directed mutagenesis kit (QuikChange®). Expression of fipB and mutant genes was verified by Western blot with rabbit anti-FipB antibody.Citation9 DNA transformation was performed as previously described.Citation4
Purification of his-tag FipB with substrates
ΔfipB complemented in trans with genes expressing variant His-tagged FipB (CXXA, AXXA or P286T), were grown in TSB/c with 15 μg/ml Kan for overnight at 37 ˚C, 200 rpm shaking. Bacteria were pelleted by centrifugation at 3,000 rpm for 30 min and then resuspended in 10 mM N-Ethylmaleimide (NEM) [pH 6], and incubated at room temperature (RT) for 5 min. One hundred % TCA was added to a 5% final concentration to precipitate proteins at 4 ˚C overnight. Precipitated proteins were harvested by centrifugation at 8,000 rpm for 20 min at 4 ˚C. Protein pellets were washed twice with cold acetone. Air dried pellets were alkylated by 10 mM NEM, 100 mM Tris-HCl [pH 6.0], containing 1% SDS, and protease inhibitor cocktail (Sigma) at RT for 5 min. The alkylated lysate was diluted four times with 50 mM Tris-HCl, [pH 8.0], containing 300 mM NaCl and centrifuged at 10,000 rpm for 20 min at 4 ˚C. Supernatants were transferred to tube containing TALON® resin pre-equilibrated with buffer A (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 0.1% of SDS) and incubated at 4 ˚C overnight with rocking prior to loading on a Poly-Prep Chromatography column (Bio-Rad). Columns were washed with 20X column volume of Buffer A and 2 ml Buffer A containing 10 mM imidazole. Co-purified proteins were eluted with Buffer A containing 300 mM imidazole. Elutions were precipitated with TCA (10% final), then acetone washed and air-dried as described above. For samples where FipB and FipB complexes were visualized by Western Blots pellets were resuspended in 150 μl of 1X SDS-PAGE loading buffer (50 mM Tris-HCl (pH 6.8), 2% (w/v) SDS (sodium dodecyl sulfate; electrophoresis grade), 0.1% (w/v) bromophenol blue and 10% (v/v) glycerol) and with or without or 100 mM dithiothreitol (DTT), separated on a 10% SDS-PAGE and then transferred to PVDF membranes for Western immunoblotting with anti-FipB antibody followed by corresponding HRP-conjugated secondary antibodies. Blots were developed using Pierce ECL Western blotting substrate and exposed to X-ray film.
Two-dimensional (2D) gel electrophoresis
2D gel electrophoresis was performed as described by Jameson-Lee et al.Citation41 Briefly, eluted samples were boiled with SDS-PAGE loading buffer and resolved on Bio-Rad Criterion Precast gel (12 well, 4–12% acrylamide) under nonreducing conditions. The lanes were excised and treated in warm 100 mM DTT for 15 min. Free sulfhydryls were alkylated by 100 mM iodoacetamide in Laemmli loading buffer without bromophenol blue for 5 min. The gel strip was then placed in a 1-well Criterion Precast gel (6–16% acrylamide). The strip was locked in place by addition of an agarose overlay (2-D starter Kit, Bio-Rad) before gel electrophoresis. Proteins were visualized with silver stain.
Mass spectrometry sequencing
Sequencing was performed at the University of Virginia Biomolecular Research Facility. Samples were analyzed by LC-MS consisting of a Thermo Electron Obritrap Velos ETD mass spectrometer system with a Prottan nanospray ion source interfaced to a self-packed 8 cm × 75 μm id Phenomenex Jupiter 10 μm C18 reversed-phase capillary column. The data were analyzed by database searching using the Sequest search algorithm against F. tularensis.
4-acetoamido-4′-maleimidylstilbene 2,2′-disulfonate (AMS) labeling
The in vivo redox statuses of IglC and IglB were analyzed by alkylation of free thiol groups by AMS (Molecular Probes). An overnight bacterial culture grown in Chamberlain's defined media (CDM) was precipitated with 10% final concentration of TCA on ice for 1 hr. CDM was used in these experiments because this was the media used by Ren et al. to label FipB (DsbA) substrates in LVS.Citation38 However, we also performed the same experiments with bacteria grown in TSB/c and did not observe any discernable difference. Total protein was pelleted by centrifugation at 14,000 rpm for 15 min at 4 ˚C. The pellet was washed twice with cold acetone then resuspended in 200 μl of 100 mM Tris-HCl [pH8.0], 1% SDS, 1 mM EDTA with or without 5 mM Tris (2-carboxyethyl) phosphine (TCEP) and incubated at 37 ˚C for 1 hr. AMS was added to a final concentration of 10 mM and then incubated at RT for 2 hrs. Extra AMS was removed by TCA precipitation as described above. The pellet was resuspended in 50 μl 1% SDS, 100 mM Tris-HCl [pH8.0]. An equal volume of 2X SDS-PAGE loading buffer (100 mM Tris-HCl (pH 6.8), 4% (w/v) SDS (sodium dodecyl sulfate; electrophoresis grade), 0.2% (w/v) bromophenol blue, 20% (v/v) glycerol) was added to each sample, which was then boiled, separated on a Criterion™ Precast Gel (4–15% Tris-HCl, 1.0 mm Bio-RAD), and then transferred to a PVDF membrane for Western blot analysis with specific antibodies followed by corresponding HRP-conjugated secondary antibodies. The blot was developed using Pierce ECL Western blotting substrate and exposed to X-ray film.
Oxidioreduction assay
Recombinant FipB was purified as previously described.Citation13 Recombinant IglC was purified from E. coli BL21 containing pQE60-iglC-his (gift from Tom Kawula) grown in Luria broth with 100 μg/ml ampicillin to 0.5 OD600, then induced by 0.1 mM IPTG for additional 4 hrs. Bacteria were lysed by the addition of lysis buffer (6M urea, 0.5 M NaCl, 50 mM NaH2PO4, 10 U DNase and EDTA-free protease inhibitor cocktail, pH 8.0) and incubated with Talon resin (Clontech) at 4°C overnight. The resin was washed with wash buffer (6M urea, 0.5 M NaCl, 50 mM NaH2PO4 and protease inhibitor cocktail pH 8.0). His-tagged IglC was eluted with 300 mM imidazole, and dialyzed against 35 mM HEPES, pH 7.3. Purity was analyzed by SDS-PAGE, followed by GelCode Blue Stain (ThermoFisher). Concentration was assessed using the BCA protein kit (Pierce).
His-tagged IglC (4 μM in 50mM HEPES-NaOH, pH7.3) was incubated for 1 hr at 25°C with various concentrations of DTT (0 to 500 μM), and in the presence or absence of 4 μM His-tagged FipB. The reaction was stopped by TCA precipitation. The pellet was washed 3 times with cold acetone, air-dried, dissolved in a freshly prepared solution containing 1% SDS, 100 mM Tris-HCl, pH = 7.5, and 5 mM AMS, then incubated in the dark at 37°C for 1 hr. Samples were mixed with nonreducing 5X SDS-PAGE loading buffer and boiled for 5 min. Proteins were separated on a 12% SDS/PAGE, transferred to a nylon membrane and incubated with anti-IglC antibody.
Preparation of KCl treated culture supernatants
Ten ml bacterial cultures were grown in TSB/c with or without 2.5 % KCl supplement for 24 hrs and then each strain was adjusted to the same OD. Fifty μl of the culture was removed and an equal volume of 2X SDS loading buffer was added, and then boiled to create the whole lysates. Bacteria were pelleted by centrifugation, and the supernatant was filtered by 0.22 μm filter, precipitated with TCA, and resuspended in 100 μl of 100 mM Tris-HCl, pH 7.5, 1% SDS. An equal volume of 2X sample buffer containing 2% 2-Mercaptoethanol was added to the tube, and then boiled. Fifty μl was applied to a 10% SDS-PAGE for protein separation, and transfer to immunoblots as described above.
Measurement of detergent sensitivity
Schu S4 or ΔfipB strains were grown overnight in TSB/c with or without 2.5% of KCl. Cultures were adjusted to an OD595 of one, then incubated with 0.25% of 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate (CHAPS) or n-Octyl glucoside (nOctylGlu) for 30 and 90 min at RT. Factor of ten serial dilutions were spotted on MHA/c plates and incubated at 37 ˚C for two days. Statistical significance was measured using an ANOVA and Dunn's multiple comparison tests.
Abbreviations
| AMS | = | 4-acetoamido-4′-maleimidylstilbene 2,2′-disulfonate |
| CDM | = | Chamberlain's defined media |
| COG | = | Clusters of Orthologous Genes |
| CXXC | = | Cysteine- any amino acid- any amino acid-Cysteine |
| Dsb | = | disulfide bond |
| DTT | = | 1,4-Dithiothreitol |
| Hcp | = | Hemolysin coregulated protein |
| IPTG | = | Isopropyl β-D-1-thiogalactopyranoside |
| LVS | = | Live Vaccine Strain |
| MAL-PEG | = | Methoxypolyethylene glycol maleimide |
| MW | = | molecular weight |
| nOctylGlu | = | n-Octyl glucoside |
| T6SS | = | Type six secretion system |
| TCEP | = | Tris (2-carboxyethyl) phosphine |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KVIR_S_1168550.zip
Download Zip (1.4 MB)Acknowledgments
We thank the W.M. Keck Biomedical Mass Spectrometry Laboratory and Nick Sherman for protein sequencing support. We also thank Thomas Kawula for the plasmid encoding His-tagged IglC, Carol Gilchrist and Girija Ramakrishnan for critical reading of the manuscript.
Funding
The Keck lab is funded by a grant from the University of Virginia's School of Medicine. This work was supported by R56 AI091746 to BJM, and T32 AI 055432 to MEC and GBM.
References
- Alibek K. Biohazard. Random House: New York, 1999.
- Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, et al. Tularemia as a biological weapon: medical and public health management. Jama 2001; 285:2763-73; PMID:11386933; http://dx.doi.org/10.1001/jama.285.21.2763
- Rohmer L, Fong C, Abmayr S, Wasnick M, Larson Freeman TJ, Radey M, Guina T, Svensson K, Hayden HS, Jacobs M, et al. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol 2007; 8:R102; PMID:17550600; http://dx.doi.org/10.1186/gb-2007-8-6-r102
- Qin A, Mann BJ. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol 2006; 6:69; PMID:16879747; http://dx.doi.org/10.1186/1471-2180-6-69
- Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KK, Roberts MJ, Ludu JS, Letendre GW, Meierovics AI, Stephens G, et al. A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol 2004; 186:6430-6; PMID:15375123; http://dx.doi.org/10.1128/JB.186.19.6430-6436.2004
- Lauriano CM, Barker JR, Yoon SS, Nano FE, Arulanandam BP, Hassett DJ, Klose KE. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc Natl Acad Sci U S A 2004; 101:4246-9; PMID:15010524; http://dx.doi.org/10.1073/pnas.030769-0101
- Gray CG, Cowley SC, Cheung KK, Nano FE. The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol Lett 2002; 215:53-6; PMID:12393200; http://dx.doi.org/10.1111/j.1574-6968.2002.tb11369.x
- Sjostedt A. Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes Infect 2006; 8:561-7; PMID:16239121; http://dx.doi.org/10.1016/j.micinf.2005.08.001
- Qin A, Scott DW, Thompson JA, Mann BJ. Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect Immun 2009; 77:152-61; PMID:18981253; http://dx.doi.org/10.1128/IAI.01113-08
- Bessette PH, Qiu J, Bardwell JC, Swartz JR, Georgiou G. Effect of sequences of the active-site dipeptides of DsbA and DsbC on in vivo folding of multidisulfide proteins in Escherichia coli. J Bacteriol 2001; 183:980-8; PMID:112-08797; http://dx.doi.org/10.1128/JB.183.3.980-988.2001
- Bardwell JC, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell 1991; 67:581-9; PMID:1934062; http://dx.doi.org/10.1016/0092-8674(91)90532-4
- Collet JF, Bardwell JC. Oxidative protein folding in bacteria. Mol Microbiol 2002; 44:1-8; PMID:11967064; http://dx.doi.org/10.1046/j.1365-2958.2002.02851.x
- Qin A, Zhang Y, Clark ME, Rabideau MM, Millan Barea LR, Mann BJ. FipB, an essential virulence factor of Francisella tularensis subspecies tularensis, has dual roles in disulfide bond formation. J Bacteriol 2014; 196(20):3571-81.
- Straskova A, Pavkova I, Link M, Forslund AL, Kuoppa K, Noppa L, Kroca M, Fucikova A, Klimentova J, Krocova Z, et al. Proteome analysis of an attenuated Francisella tularensis dsbA mutant: identification of potential DsbA substrate proteins. J Proteome Res 2009; 8:5336-46; PMID:19799467; http://dx.doi.org/10.1021/pr900570b
- Schmidt M, Klimentova J, Rehulka P, Straskova A, Spidlova P, Szotakova B, Stulik J, Pavkova I. Francisella tularensis subsp. holarctica DsbA homologue: a thioredoxin-like protein with chaperone function. Microbiology 2013; 159:2364-74; PMID:24014665; http://dx.doi.org/10.1099/mic.0.070516-0
- Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and regulation of the type VI secretion system. Annu Rev Microbiol 2012; 66:453-72; PMID:22746332; http://dx.doi.org/10.1146/annurev-micro-121809-151619
- Broms JE, Sjostedt A, Lavander M. The role of the francisella tularensis pathogenicity island in Type VI secretion, intracellular survival, and modulation of host cell signaling. Front Microbiol 2010; 1:136; PMID:21687753; http://dx.doi.org/10.3389/fmicb.2010.00136
- Clemens DL, Ge P, Lee BY, Horwitz MA, Zhou ZH. Atomic Structure of T6SS reveals interlaced array essential to function. Cell 2015; 160:940-51; PMID:25723168; http://dx.doi.org/10.1016/j.cell.2015.02.005
- Broms JE, Meyer L, Sun K, Lavander M, Sjostedt A. Unique substrates secreted by the type VI secretion system of Francisella tularensis during intramacrophage infection. PLoS One 2012; 7:e50473; PMID:23185631; http://dx.doi.org/10.1371/journal.pone.0050473
- Hare RF, Hueffer K. Francisella novicida pathogenicity island encoded proteins were secreted during infection of macrophage-like cells. PLoS One 2014; 9:e105773; PMID:25158041; http://dx.doi.org/10.1371/journal.pone.0105773
- Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, Liu J, Celli J, Arulanandam BP, Klose KE. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol 2009; 74:1459-70; PMID:20054881; http://dx.doi.org/10.1111/j.1365-2958.2009.06947.x
- de Bruin OM, Ludu JS, Nano FE. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol 2007; 7:1; PMID:17233889; http://dx.doi.org/10.1186/1471-2180-7-1
- Lindgren H, Golovliov I, Baranov V, Ernst RK, Telepnev M, Sjostedt A. Factors affecting the escape of Francisella tularensis from the phagolysosome. J Med Microbiol 2004; 53:953-8; PMID:15358816; http://dx.doi.org/10.1099/jmm.0.45685-0
- Miki T, Okada N, Danbara H. Two periplasmic disulfide oxidoreductases, DsbA and SrgA, target outer membrane protein SpiA, a component of the Salmonella pathogenicity island 2 type III secretion system. J Biol Chem 2004; 279:34631-42; PMID:15169785; http://dx.doi.org/10.1074/jbc.M402760200
- Ellermeier CD, Slauch JM. RtsA coordinately regulates DsbA and the Salmonella pathogenicity island 1 type III secretion system. J Bacteriol 2004; 186:68-79; PMID:14679226; http://dx.doi.org/10.1128/JB.186.1.68-79.2004
- Yu J, Kroll JS. DsbA: a protein-folding catalyst contributing to bacterial virulence. Microbes Infect 1999; 1:1221-8; PMID:10580278; http://dx.doi.org/10.1016/S1286-4579(99)00239-7
- Sun P, Austin BP, Schubot FD, Waugh DS. New protein fold revealed by a 1.65 A resolution crystal structure of Francisella tularensis pathogenicity island protein IglC. Protein Sci 2007; 16:2560-3; PMID:17905833; http://dx.doi.org/10.1110/ps.073177307
- Qin A, Scott DW, Rabideau MM, Moore EA, Mann BJ. Requirement of the CXXC motif of novel Francisella infectivity potentiator protein B FipB, and FipA in virulence of F tularensis subsp. tularensis. PLoS One 2011; 6:e24611; PMID:21931773; http://dx.doi.org/10.1371/jour-nal.pone.0024611
- Kadokura H, Tian H, Zander T, Bardwell JC, Beckwith J. Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science 2004; 303:534-7; PMID:14739460; http://dx.doi.org/10.1126/science.109-1724
- Paxman JJ, Borg NA, Horne J, Thompson PE, Chin Y, Sharma P, Simpson JS, Wielens J, Piek S, Kahler CM, et al. The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes. J Biol Chem 2009; 284:17835-45; PMID:19389711; http://dx.doi.org/10.1074/jbc.M109.01-1502
- Depuydt M, Leonard SE, Vertommen D, Denoncin K, Morsomme P, Wahni K, Messens J, Carroll KS, Collet JF. A periplasmic reducing system protects single cysteine residues from oxidation. Science 2009; 326:1109-11; PMID:19965429; http://dx.doi.org/10.1126/science.117-9557
- Thomas RM, Twine SM, Fulton KM, Tessier L, Kilmury SL, Ding W, Harmer N, Michell SL, Oyston PC, Titball RW, et al. Glycosylation of DsbA in Francisella tularensis subspecies tularensis. J Bacteriol 2011; 5498-509; PMID:21803994; http://dx.doi.org/10.1128/JB.00438-11
- Gallagher LA, Ramage E, Jacobs MA, Kaul R, Brittnacher M, Manoil C. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci U S A 2007; 104:1009-14; PMID:17215359; http://dx.doi.org/10.1073/pnas.0606713104
- de Bruin OM, Duplantis BN, Ludu JS, Hare RF, Nix EB, Schmerk CL, Robb CS, Boraston AB, Hueffer K, Nano FE. The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiol 2011; 157:3483-91; http://dx.doi.org/10.1099/mic.0.052308-0
- Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J 2009; 28:315-25; PMID:19131969; http://dx.doi.org/10.1038/emboj.2008.269
- Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem 1979; 254:9627-32; PMID:385588
- Kingry LC, Petersen JM. Comparative review of Francisella tularensis and Francisella novicida. Front Cell Infect Microbiol 2014; 4:35; PMID:24660164; http://dx.doi.org/10.3389/fcimb.2014.00035
- Ren G, Champion MM, Huntley JF. Identification of disulfide bond isomerase substrates reveals bacterial virulence factors. Mol Microbiol 2014; 94(4):926-44.
- Hiniker A, Bardwell JC. In vivo substrate specificity of periplasmic disulfide oxidoreductases. J Biol Chem 2004; 279:12967-73; PMID:14726535; http://dx.doi.org/10.1074/jbc.M311391200
- Ramakrishnan G, Sen B, Johnson R. Paralogous outer membrane proteins mediate uptake of different forms of iron and synergistically govern virulence in Francisella tularensis tularensis. J Biol Chem 2012; 287:25191-202; PMID:22661710; http://dx.doi.org/10.1074/jbc.M112.37-1856
- Jameson-Lee M, Garduno RA, Hoffman PS. DsbA2 (27 kDa Com1-like protein) of Legionella pneumophila catalyses extracytoplasmic disulphide-bond formation in proteins including the Dot/Icm type IV secretion system. Mol Microbiol 2011; 80:835-52; PMID:21375592; http://dx.doi.org/10.1111/j.1365-2958.2011.07615.x
- Su J, Yang J, Zhao D, Kawula TH, Banas JA, Zhang JR. Genome-wide identification of Francisella tularensis virulence determinants. Infect Immun 2007; 75:3089-101; PMID:17420240; http://dx.doi.org/10.1128/IAI.01865-06
- Mohapatra NP, Balagopal A, Soni S, Schlesinger LS, Gunn JS. AcpA is a Francisella acid phosphatase that affects intramacrophage survival and virulence. Infect Immun 2007; 75:390-6; PMID:17060465; http://dx.doi.org/10.1128/IAI.01226-06
- Mohapatra NP, Soni S, Rajaram MV, Strandberg KL, Gunn JS. Type A Francisella tularensis acid phosphatases contribute to pathogenesis. PLoS One 2013; 8:e56834; PMID:23457625; http://dx.doi.org/10.1371/journal.pone.0056834
- Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A 2007; 104:6037-42; PMID:17389372; http://dx.doi.org/10.1073/pnas.0609675104
- Wehrly TD, Chong A, Virtaneva K, Sturdevant DE, Child R, Edwards JA, Brouwer D, Nair V, Fischer ER, Wicke L, et al. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol 2009; 11:1128-50; PMID:19388904; http://dx.doi.org/10.1111/j.1462-5822.2009.01316.x
- Chong A, Wehrly TD, Child R, Hansen B, Hwang S, Virgin HW, Celli J. Cytosolic clearance of replication-deficient mutants reveals Francisella tularensis interactions with the autophagic pathway. Autophagy 2012; 8:1342-56; PMID:22863802; http://dx.doi.org/10.4161/auto.20808
- Lindgren H, Shen H, Zingmark C, Golovliov I, Conlan W, Sjostedt A. Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect Immun 2007; 75:1303-9; PMID:17210667; http://dx.doi.org/10.1128/IAI.01717-06
- Mahawar M, Atianand MK, Dotson RJ, Mora V, Rabadi SM, Metzger DW, Huntley JF, Harton JA, Malik M, Bakshi CS. Identification of a novel Francisella tularensis factor required for intramacrophage survival and subversion of innate immune response. J Biol Chem 2012; 287:25216-29; PMID:22654100; http://dx.doi.org/10.1074/jbc.M112.367672
- Asare R, Akimana C, Jones S, Abu Kwaik Y. Molecular bases of proliferation of Francisella tularensis in arthropod vectors. Environ Microbiol 2010; 12:2587-612; PMID:20482589; http://dx.doi.org/10.1111/j.1462-2920.2010.02230.x
- Brotcke A, Weiss DS, Kim CC, Chain P, Malfatti S, Garcia E, Monack DM. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect Immun 2006; 74:6642-55; PMID:17000729; http://dx.doi.org/10.1128/IAI.01250-06
- Enstrom M, Held K, Ramage B, Brittnacher M, Gallagher L, Manoil C. Genotype-phenotype associations in a nonmodel prokaryote. MBio 2012; 3:e00001-12.
- Mohapatra NP, Soni S, Bell BL, Warren R, Ernst RK, Muszynski A, Carlson RW, Gunn JS. Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect Immun 2007; 75:3305-14; PMID:17452468; http://dx.doi.org/10.1128/IAI.00351-07
- Ma Z, Banik S, Rane H, Mora VT, Rabadi SM, Doyle CR, Thanassi DG, Bakshi CS, Malik M. EmrA1 membrane fusion protein of Francisella tularensis LVS is required for resistance to oxidative stress, intramacrophage survival and virulence in mice. Mol Microbiol 2014; 91:976-95; PMID:24397487; http://dx.doi.org/10.1111/mmi.12509
- Tempel R, Lai XH, Crosa L, Kozlowicz B, Heffron F. Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect Immun 2006; 74:5095-105; PMID:16926401; http://dx.doi.org/10.1128/IAI.00598-06
- Twine S, Bystrom M, Chen W, Forsman M, Golovliov I, Johansson A, Kelly J, Lindgren H, Svensson K, Zingmark C, et al. A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58-kgdalton protein is attenuated for virulence and is an effective live vaccine. Infect Immun 2005; 73:8345-52; PMID:16299332; http://dx.doi.org/10.1128/IAI.73.12.8345-8352.2005
- Kadzhaev K, Zingmark C, Golovliov I, Bolanowski M, Shen H, Conlan W, Sjostedt A. Identification of genes contributing to the virulence of Francisella tularensis SCHU S4 in a mouse intradermal infection model. PLoS One 2009; 4:e5463; PMID:19424499; http://dx.doi.org/10.1371/journal.pone.0005463
- Meibom KL, Dubail I, Dupuis M, Barel M, Lenco J, Stulik J, Golovliov I, Sjostedt A, Charbit A. The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol Microbiol 2008; 67:1384-401; PMID:18284578; http://dx.doi.org/10.1111/j.1365-2958.2008.06139.x
