ABSTRACT
Salmonella-infected cells are characterized by the presence of intra-cellular membranous tubules that emerge from bacterial vacuoles and extend along microtubules. The formation of Salmonella-induced tubules depends on the Salmonella pathogenicity island 2-encoded type III secretion system (T3SS-2) that translocates bacterial effector proteins inside host cells. Effector proteins have enzymatic activities or allow for hijacking of cellular functions. The role of Salmonella-induced tubules in virulence remains unclear but their absence is correlated with virulence defects. This study describes the presence of inter-cellular tubules that arise between daughter cells during cytokinesis. Inter-cellular tubules connect bacterial vacuoles originally present in the parent cell and that have been apportioned between daughters. Their formation requires a functional T3SS-2 and effector proteins. Our data establish a correlation between the formation of inter-cellular tubules and the asymmetric distribution of bacterial vacuoles in daughters. Thus, by manipulating the distribution of bacteria in cytokinetic cells, Salmonella T3SS-2 effector proteins may increase bacterial spreading and the systemic character of the infection.
Introduction
Salmonella Typhimurium (Salmonella enterica subsp. enterica serovar Typhimurium) is a Gram-negative bacterial pathogen and a prominent cause of human gastroenteritis and systemic infection in immunocompromised individuals. This is an intracellular bacterium that survives in a variety of tissues and cells.Citation1 The capacity to replicate intracellularly is essential for Salmonella virulence. Intracellular Salmonella resides in a membrane bound compartment named the Salmonella-containing vacuole (SCV). It also replicates in the cytosol of epithelial cellsCitation2 but the physiological relevance of the presence of fast replicating cytosolic Salmonella in epithelia is not understood.
Growing within a SCV presents important challenges that are supported by Salmonella type 3 secretion systems (T3SS). T3SSs direct the secretion and translocation of effector proteins (effectors) from the bacterial to the eukaryotic cytosol. Effectors are capable of influencing host functions. T3SS-1 effectors manipulate the cortical actin cytoskeleton, trigger the formation of membrane ruffles and facilitate the uptake of bacteria by non-phagocytic cells. T3SS-2 translocates more than 20 effectors that are collectively required for intracellular replication and virulence in mice (for review see ref.Citation3).
Salmonella vacuoles have several unique morphological characteristics. A SCV contains a single bacterium and divides when the bacterium undergoes cell division.Citation4 This trait requires the function of host proteins. Abnormal vacuoles enclosing several bacteria have been observed upon inhibition of dynein functionCitation4 or in the absence Plekhm1, a host protein that interacts with the T3SS-2 effector SifA.Citation5 The SCVs are connected to each other by membrane tubules that elongate with the mechanical support of the microtubule cytoskeleton and motors. Tubules were originally described in infected epithelial cellsCitation6 but also form in macrophagesCitation7-9 though they are more difficult to observe. Different kinds of tubules have been describedCitation10 and are together referred to as Salmonella-induced tubules (SIT). At least 6 T3SS-2 effectors are involved in the formation (SifA, SopD2, SteA),Citation11-13 elongation (PipB2)Citation14 or structure (SseF, SseG)Citation15 of SITs. The precise functions of SITs remain elusive but their absence is associated with a virulence defect.
We observed that Salmonella-infected cells, like non-infected cells, enter mitosis and apportion bacteria between daughter cells. During the process of cytokinesis the clusters of bacterial vacuoles destined to one or the other daughter cell remain connected by a membrane tubule that passes through the midbody. The present study investigates the mechanism of formation and the function of these tubules in relation with the host cell division and bacterial spreading.
Results
Salmonella-infected cells are occasionally connected by inter-cellular membrane tubules
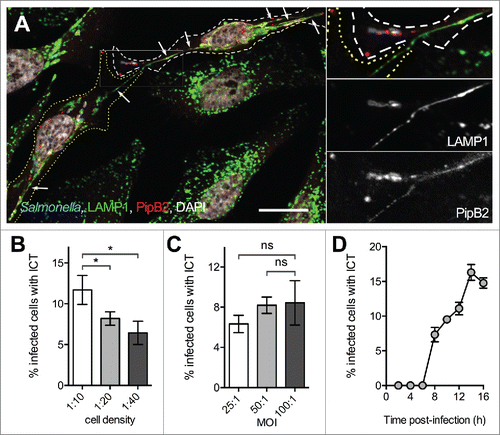
SITs are intra-cellular tubules frequently seen in Salmonella-infected epithelial cells.Citation6 We observed in HeLa cells the presence of membrane tubules emerging from one infected cell and appearing in continuity with similar tubules present in the adjacent infected cells (). These tubules were comparable to SITs as they were 1) not observed in non-infected cells nor in cells infected with a T3SS-2 deficient strain (ΔssaV); 2) connected to SCVs; 3) enriched in lysosomal glycoproteins such as LAMP1 (), in various T3SS-2 effectors (Fig. S1) and in SCAMP3 (Fig. S2). We examined the cell culture and infection conditions favoring the formation of these inter-cellular tubules, which we will refer to as ICTs, and analyzed the kinetics of their formation (). ICTs were first observed at 8 h post-infection (p.i.) and in a maximum of 15-20 % infected HeLa cells at 14 h p.i..
Figure 1. Observation of inter-cellular tubules (ICT). (A) HeLa cells were infected with wild-type Salmonella expressing CFP and PipB2-2HA. Cells were fixed at 16 h p.i., immunostained for LAMP-1 and HA, and imaged by confocal microscopy for CFP (blue), LAMP1 (green), HA (red) and nuclei (white). White and yellow dotted lines delineate 2 neighboring Salmonella-infected cells. Magnified insets in 4 colors or showing grayscale images for LAMP1 and PipB2 are presented. A LAMP1- and PipB2-positive Salmonella-induced tubule emerges from one cell and is in continuity with a similar tubule present in the adjacent cell. Arrows point Salmonella-induced tubules. Bar, 20 µm or 10 µm for the magnified insets. (B) Influence of the cell density on the formation of ICTs. HeLa cells were seeded at various surface ratio and infected 24 h later with GFP-expressing wild-type Salmonella at a MOI of 100:1. Cells were fixed at 16 h p.i. (C) Influence of the multiplicity of infection (MOI) on the formation of ICTs. HeLa cells were seeded at a surface ratio of 1:10 and infected 24 h later with various MOI of GFP-expressing wild-type Salmonella. Cells were fixed at 16 h p.i. (D) Kinetics of formation of ICTs. HeLa cells were seeded at a surface ratio of 1:10 and infected 24 h later with GFP-expressing wild-type Salmonella at a MOI of 100:1. Cells were fixed at different times p.i. (B - D) Infected cells presenting ICTs were enumerated by fluorescence microscopy. Data are the mean ± SD or representative (D) of 3 independent experiments. (B & C) Multiple t-tests were used to compare the mean values.
ICTs do not form between independently infected HeLa cells
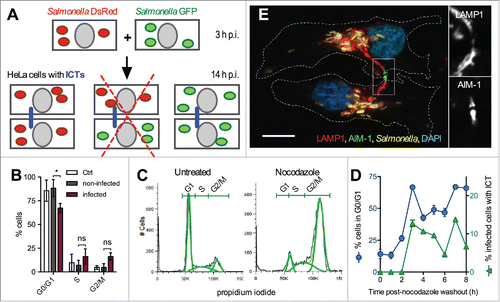
To define the conditions that lead to the formation of ICTs, we examined if these tubules could arise upon encounter of 2 infected cells. For this, we trypsinized and then co-cultured 2 batches of HeLa cells previously infected for 3 h with Salmonella strains expressing either GFP or DsRed. ICTs were almost exclusively observed between cells enclosing Salmonella expressing the same color protein (). ICTs connecting cells enclosing GFP and DsRed bacteria were very rare (less than 1% of infected cells presenting ICTs) and, in most cases, both cells contained the 2 types of bacteria, strongly suggesting a secondary infection. We conclude that ICTs do not form upon encounter of 2 infected cells.
Figure 2. ICTs form between cytokinetic cells. (A) ICTs do not exist between cells that have been infected independently. Schematic representation of the experiment. HeLa cells were infected with GFP- or DsRed-expressing Salmonella. Cells of the 2 independent groups were trypsinized at 3 h p.i., mixed and co-cultured. Cells were fixed at 14 h p.i. and immunostained for LAMP-1. ICTs (blue line) were observed by fluorescence microscopy. ICTs were not observed between cells containing Salmonella not expressing the same color protein (redcross) (B) Salmonella-infected cells continue their progression through the cell cycle. HeLa cells were mock-infected (Ctrl) or infected with Salmonella for 14 h, DNA stained with propidium iodide and analyzed by flow cytometry. Mock-infected cells and the 2 populations of infected (GFP positive) and non-infected (GFP negative) cells were analyzed for their DNA content. Results are the means ± SD of 3 independent experiments. Multiple t-tests were used to compare the mean values. (C) Nocodazole treatment arrests cells in G2/M phase. HeLa cells were treated with nocodazole (0.4 µg / ml for 16 h) or left untreated, DNA stained with propidium iodide and analyzed by flow cytometry. FlowJo 8.3 was used to delineate population (green curves) on histogram plots and to quantify the percentage of cells with 2N (G1), 2 to 4N (S), and 4N (G2/M) DNA content. (D) Formation of ICTs and entry in G1 phase are concomitant. HeLa cells were infected with wild-type Salmonella expressing GFP for 6.5 h and further treated with nocodazole (0.4 µg / ml) for 12h. Cells were fixed at different times post nocodazole washout and stained with propidium iodide for flow cytometry analysis or immunostained for LAMP1. The percentages of infected cells in G1 phase (blue line) or with ICT (green line) are plotted function of time post-washout. Results presented in (C, D) are representative of 3 independent experiments. (E) ICTs pass through the midbody of cytokinetic Salmonella-infected cells. HeLa cells were infected with GFP-expressing Salmonella for 14 h, fixed, immunostained for AIM-1 and LAMP1. Cells were imaged for GFP (yellow), LAMP1 (red), AIM-1 (green) and DAPI (blue) using confocal microscopy. Magnified insets showing single labeling for LAMP1 and AIM-1 are presented on the right. White dotted lines delineate 2 Salmonella-infected daughter cells at the end of the cytokinesis process. Bar, 20 µm or 10 µm for the magnified insets.
ICTs form during mitosis of an infected cell
Another possibility was that ICTs form during the cytokinesis of an infected cell. As Salmonella has been reported to perturb the cell cycle,Citation16 we first evaluated the consequences of an infection on the capacity of HeLa cell to divide. We infected cells with GFP-expressing bacteria for 14 h and analyzed by flow cytometry the DNA content of the Salmonella-infected (GFP positive) and non-infected (GFP negative) cell populations (). As compared to mock-infected and to non-infected cells we found a lower percentage of infected cells in the G0/G1 phase but small and non-significant increases for the S and G2/M populations (). Thus, within our experimental time frame, Salmonella does not substantially affect the capacity of HeLa cells to go through their regular cell cycle. Next, we examined whether a correlation exists between the cell cycle and the formation of ICTs. Cytokinesis is the last step of the M phase and this process immediately precedes the G1 phase. Therefore, we checked whether entry in G1 phase and the formation of ICTs were concomitant. We treated Salmonella-infected cells with the microtubule destabilizing agent nocodazole that arrests the cell cycle prior mitosis. After 16 h treatment, more than 70 % of HeLa cells had a G2/M-amount of DNA while the number of cells in the G0/G1 fraction was reduced to 14% (). Following nocodazole washout, cells resumed their progression through the cell cycle. The number of cells in G0/G1 phase started to increase 2 h post-washout and peaked at 67 % after 3 and 7 h (). A parallel microscopic analysis of immunostained cells showed that the fraction of infected cells with ICTs followed a very similar profile (). These results associate the cytokinesis that is observed at the transition between M and G0/G1 phases and the presence of ICTs, suggesting these tubules could form during cytokinesis.
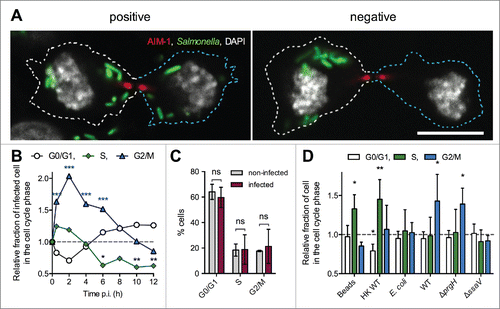
The protein AIM-1 (Aurora and Ipl1-like midbody-associated protein) is concentrated in the midbody, which is the central region of the thin inter-cellular cytoplasmic bridge formed between daughter cells at the end of the cytokinesis.Citation17 AIM-1 immunostaining of infected cells showed that ICTs are passing through the midbody ( and S1). We found 67.4 ± 2.8 % of infected and cytokinetic cells with ICTs. These results strongly suggest that ICTs form during the cytokinesis of Salmonella-infected cells. We propose that these tubules are SITs still connecting the 2 groups of bacterial vacuoles upon their apportioning in daughter cells.
Salmonella does not block cell cycle progression of macrophages
Since macrophages are key players in salmonellosis,Citation1,18 we investigated the cell cycle and the presence of ICTs in mouse macrophages. RAW 264.7 cells were infected with Salmonella for 14 h. AIM-1 immunostaining was used to identify dividing cells (). We found similar percentages of mitotic cells in the infected and non-infected populations of macrophages (5.8 ± 0.7 % and 5.2 ± 0.4 %, respectively). It suggests that Salmonella do not alter the capacity of RAW 264.7 macrophages to go through mitosis. We also analyzed infected macrophages for the presence of tubules after immunostaining for LAMP-1 or PipB2-2HA, but we could not detect the presence of ICTs in these cells.
Figure 3. Cell cycle analysis of Salmonella-infected RAW 264.7 macrophages. (A) Salmonella-infected RAW 264.7 macrophages undergo mitosis. RAW 264.7 macrophages were infected with GFP-expressing wild-type Salmonella for 14 h. Fixed cells were immunostained for AIM-1 and imaged for GFP (green), AIM-1 (red), and DAPI (white) by confocal microscopy. Infected macrophages divide and bacteria are distributed in both daughters (left image) or remain all in one of the daughters (right image). Bar, 10 µm. (B) Kinetic analysis of infected vs. non-infected RAW 264.7 cell cycle. RAW 264.7 cells were incubated with GFP-expressing wild-type Salmonella, fixed at different time points, DNA stained with propidium iodide and analyzed by flow cytometry (t = 0 corresponds to non-infected cells, t = 30 min corresponds to the end of the incubation period with bacteria). GPF fluorescence was used to define the gates corresponding to infected and non-infected cell populations. For each time point, the ratio between the percentages of Salmonella-infected versus non-infected cells in each phase of cell cycle was determined. Data (mean) are from 3 independent experiments. A two-way ANOVA test was used to determine whether a mean value was significantly different from one. Significant P values are indicated: *, P<0.05; **, P<0.01; ***, P<0.001. (C) Percentages of infected and non-infected RAW 264.7 cell in G0/G1, S and G2/M phases 14 h p.i. Data (mean ± SD) are from 3 independent experiments. Multiple t-tests were used to compare the mean values. P values: ns, not significant. (D) Salmonella are preferentially phagocytosed by pre-mitotic or mitotic RAW 264.7 cells. RAW 264.7 cells were incubated in the presence of fluorescent beads (Fluoresbrite® YG), heat-killed Salmonella (HK WT) or GFP-expressing bacteria (E. coli, or wild-type (WT), ΔprgH or ΔssaV Salmonella strains) for 30 minutes to allow phagocytosis. Cells were fixed and DNA stained with propidium iodide. For heat killed Salmonella, cells were immunostained for LPS. Normalized data were calculated by the ratio between the percentages of particle-containing vs. control cells in each phase of cell cycle. Data (mean ± SD) are from 5 to 9 independent experiments. A one-sample t-test was used to determine whether a mean value was significantly different from one. Significant P values are indicated: *, P<0.05; **, P<0.01.
Next we followed the cell cycle of infected versus non-infected mouse macrophages cells. As compared to non-infected cells, we found 1.63 ± 0.15 and 2.04 ± 0.26 times more infected cells in the G2/M phase 30 min and 2 h p.i., respectively (), indicating that Salmonella are preferentially taken-up by pre-mitotic and/or mitotic macrophages. Between 8 and 10 h p.i the fraction of infected G2/M cells returned to a normal level whereas cells in S phase were slightly under-represented. At 14 h p.i., the infected and non-infected populations were not statistically different ().
T3SS-2-expressing bacteria are preferentially taken up by mitotic macrophages
A T3SS-1-dependent preferential invasion of G2/M HeLa cells has been previously reported.Citation19 In this study, we used Salmonella grown in minimal medium that favors the expression of T3SS-2 rather than of T3SS-1Citation20 in order to decrease the T3SS-1-mediated killing of macrophages.Citation21 Thus, T3SS-1 is probably not involved in the preferential uptake of Salmonella by G2/M RAW 264.7 cells. To verify this point, we incubated macrophages for 30 min with different types of live or inert particles, analyzed their DNA content by flow cytometry and determined the fractions of cells having taken particles in various phases of the cell cycle (). Inert particles (latex beads and heat killed Salmonella) were preferentially taken-up by cells in S phase. E. coli and a ΔssaV mutant that lacks a functional T3SS-2 were equally taken-up by cells in either phase, while bacteria that do not express T3SS-1 (ΔprgH) showed a profile similar to that of wild-type Salmonella. All together these data indicate that Salmonella expressing T3SS-2 are preferentially phagocytosed by G2/M macrophages.
T3SS-2 effectors support the formation of ICTs and the apportioning of SCVs between daughter cells
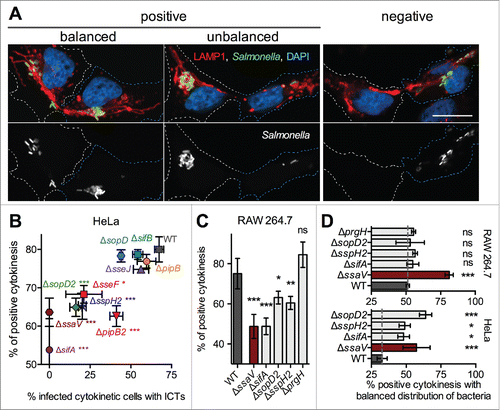
During cytokinesis the content of the parent cell is divided in 2 parts. Daughter cells receive equal amounts of genetic material and a part of the cytoplasmic content. We observed variable situations regarding the distribution of bacteria between daughters. It was observed that one daughter did not contain bacteria (right panels in ). The existence of daughters with a complete absence of Salmonella was confirmed by the lack of signal after immunostaining with an anti lipopolysaccharide antibody (Fig. S3). This kind of cytokinesis was referred to as "negative." Among the "positive" cytokinesis (bacteria are present in both daughters, left panels in ), we sometimes observed that one daughter contained far more bacteria than the other. The cytokinesis was termed as "unbalanced" if one daughter contained more than 2/3 of total bacteria. Other positive cytokinetic situations were referred to as "balanced" (). Having defined these observations, we asked whether ICTs could play a role in the apportioning of bacteria between daughter cells. As the formation of ICTs requires T3SS-2 function we infected HeLa cells with wild-type Salmonella or a ΔssaV strain. For each strain, 100 to 200 infected and mitotic cells were analyzed by microscopy for the presence of ICTs and the distribution of bacteria in daughter cells. ICTs were seen in 67.4 ± 2.8 % cytokinetic cells infected by the wild-type strain and absent from ΔssaV-infected cells. We scored 77 ± 0.7 % and 63.3 ± 1.4 % positive cytokinesis for wild-type and ΔssaV infected cells, respectively. However, we observed, both in wild-type and ΔssaV infected cells a correlation between the number of bacteria and the percentage of positive cytokinesis (Figure S4A), indicating that the lower percentage of positive cytokinesis for the ΔssaV mutant may be directly linked to its replication defect (83% of ΔssaV infected cells contained less than 10 bacteria as compared to 13.8% for wild-type infected HeLa cells, Figure S4A). To get more insight, we analyzed a selection of strains deleted of genes encoding for a T3SS-2 effector. In we plotted positive cytokinesis as a function of cytokinetic cells with ICTs. This representation revealed the presence of 2 groups of bacteria. Mutant strains belonging to the first group (ΔsopD, ΔsifB, ΔsseJ, ΔpipB) were similar to wild-type Salmonella. Mutants of the second group (ΔsifA, ΔsopD2, ΔsseF, ΔsspH2, ΔpipB2) were, like the ΔssaV mutant, characterized by the presence of few or the absence of ICTs and a low number of positive cytokinesis. While the latter defect might also be attributed to an intracellular replication defect for ΔsseFCitation22 and ΔsifACitation23 mutants, the deletions of pipB2,Citation7 sspH2Citation24 or sopD2Citation12 do not impact the intracellular replication of Salmonella in HeLa cells.
Figure 4. Distribution of Salmonella in cytokinetic cells. (A) Salmonella are diversely distributed in daughter cells during cytokinesis. HeLa cells were infected with CFP-expressing Salmonella for 14h, fixed, immunostained for LAMP1 and imaged for CFP (green), LAMP1 (red), and DAPI (blue) using confocal microscopy. Grayscale images showing single labeling for bacteria (CFP) are shown. Cytokinetic cells with different distribution of bacteria in daughters are presented. In the right images, bacteria are found in only one of the daughter cells. Cytokinesis was categorized as "negative." The left images present cytokinetic figures with bacteria in both daughters (categorized as "positive"). If one of the daughter cells contained up to 2/3 of total bacteria, the cytokinesis was categorized as "balanced." If one of the daughter cells contained more than 2/3 of total bacteria, the cytokinesis was categorized as "unbalanced." Bar, 20 µm (B - D) Influence of T3SS-2 effectors on the distribution of Salmonella in daughter cells. Cells were infected with GFP- and PipB2-2HA-expressing bacteria, fixed 14 h p.i. and stained for HA, AIM-1 and nuclei. (B) HeLa cells. For each strain, "positive" cytokinesis cells were plotted as a function of cells with ICTs. One-way ANOVA with Dunnett's post-test was used to compare the mean of positive cytokinesis of mutant strains with those of wild-type Salmonella. (C) RAW 264.7 cells. The fractions of "positive" cytokinesis in RAW 264.7 cells were determined for wild-type Salmonella and a selection of strains. (D) The fractions of cytokinetic HeLa and RAW 264.7 cells with a balanced distribution of bacteria were determined. (B - D) Data (mean ± SD) are from 3 to 6 independent experiments. (C - D) Unpaired t-tests were used to compare the mean of a strain with the mean of wild-type Salmonella or to compare the means of 2 strains. P values: ns, not significant; *, P<0.05; **, P<0.01; ***, P<0.001.
Strains of the second group are deleted of genes known to be involved in the formation of SITs except for sspH2. Yet, we found that a ΔsspH2 mutant is also defective for the formation of SITs in HeLa cells (Fig. S5). These results establish a correlation between the formation of ICTs and the apportioning of bacteria in both daughters.
Next, we tested if such a correlation exists in macrophages. SITs are barely observable in resting macrophages and we did not detect the formation of ICTs. Nevertheless, we analyzed the distribution of bacteria in daughters for a selection of T3SS-2 mutants and, as a control, a ΔprgH strain that lacks a functional T3SS-1. Like in HeLa cells, we observed less positive cytokinesis in macrophages infected with ΔssaV and ΔsifA mutants than in those infected with the wild-type strain (). Strains lacking SopD2 or SspH2 presented also a significantly lower number of positive cytokinesis (). A ΔprgH mutant was not different from the wild-type bacteria. Altogether, these results indicate that strains that do not form or produce altered SITs in HeLa cells are affected for the formation of ICTs and highlight an association between a reduced formation of ICTs in HeLa cells and a lower fraction of positive cytokinesis in HeLa and macrophages.
Lastly, we analyzed the influence of T3SS-2 on the even distribution of bacteria between daughter cells. As compared to a ΔssaV strain and to a selection of T3SS-2 effector mutants (ΔsifA, ΔsopD2, ΔsspH2), we found less mitotic HeLa cells with a balanced number of wild-type bacteria. This phenotype was independent of the number of wild-type or ΔssaV intracellular bacteria (Fig S4B). We found in mitotic mouse macrophages a similar difference between wild-type and a ΔssaV mutant. However, the selected mutants (ΔsifA, ΔsopD2, ΔsspH2 and ΔprgH) were not different from the wild-type strain. Together these data show that T3SS-2 sustains an asymmetric distribution of bacteria during cytokinesis.
Discussion
Bacterial effectors have a range of activities inside host cells that support pathogenesis.Citation25 This study adds new functions to the actions covered by the T3SS-2: the uptake of Salmonella by pre-mitotic and/or mitotic macrophages and the asymmetric apportioning of bacteria during mitosis.
T3SS-1 is required to invade non-phagocytic cells.Citation26 Invasion involves the translocon protein SipB that binds cholesterol.Citation27 As cell surface cholesterol is more abundant during mitosis, Salmonella invade preferentially mitotic epithelial cells.Citation19 The present work emphasizes a similar role for T3SS-2 in the uptake of Salmonella by G2/M macrophages. SseB, SseC and SseD are secreted by T3SS-2 and form a translocon.Citation28 Whether these proteins bind cholesterol is unknown.
Within the time frame of our experiments Salmonella does not block the cell cycle of infected cells. This is evidenced by the fact that: i) ICTs form between mitotic cells; ii) nocodazole treatment blocks both infected and non-infected cells in G2/M phase and cell cycle resumes upon nocodazole wash out; iii) infected and non-infected cells have similar cell cycle profiles. Yet, recently published data found a T3SS-2- and SpvB-dependent arrest of Salmonella-infected HeLa cells in G2/M phase.Citation16 SpvB possesses an ADP-ribosyl transferase activity. Its ectopic expression leads to a G1/S or G2/M arrest of the cell cycle of tumor cells.Citation29 The influence of T3SS-2 on the cell cycle was detected at very late time points (> 20 h p.i.) while spvB is induced rather early during infection.Citation30 Therefore, It is possible that SpvB is expressed but not immediately secreted or is not sufficient to induce a modification of the cell cycle. Our experiments were performed in a narrower time frame and this might explain why, in our experimental context, Salmonella did not modify the host cell cycle.
We initially observed the presence in infected epithelial cells of tubules seemingly passing from one cell to a neighbor cell. Our data suggest that these tubules are remnants of SITs that form during mitosis. As tubules connect SCVs, one may assume they still exist after SCVs were apportioned in daughter cells. These tubules pass trough the midbody and are probably closed at the time of abscission.
During cell division, the content of a mother cell is distributed between daughter cells. This mechanism has been extensively described for the genetic material. Organelles that are present in multiple dispersed copies will be randomly allocated more or less equally in each daughter cell. The Golgi apparatus, which is a single functional unit, fragments at the onset of mitosis thereby favoring its symmetrical random distribution in daughter cells. However the distribution is not necessarily symmetrical. For example mitochondria are asymmetrically distributed between daughters in human mammary stem-like cells. Old damaged mitochondria are segregated away from the daughter destined to become a new stem cell.Citation31 We found that T3SS-2 effectors influence the distribution of SCVs during mitosis by favoring the presence of bacteria in both daughter cells and their asymmetric apportioning. We detected a correlation between the number of intracellular bacteria and the presence of bacteria in both daughters, indicating that for some mutants this phenotype may only be a consequence of a replication defect. However, we also found less positive cytokinesis for mutants that, in HeLa cells, do not present any replication defect, and for which the effector may have a direct role in this observation (SopD2, PipB2). In contrast to the previous phenotype, the asymmetric distribution of bacteria in daughters is independent of the number of intracellular bacteria. It is also influenced by the T3SS-2 in epithelial cells and macrophages. Yet, the absence of a single effector (SifA, sopD2, SspH2) is sufficient to perturb the asymmetric distribution of Salmonella in HeLa cells but not in macrophages. Overall, these processes reduce the bacterial burden of one of the daughters and thereby contribute to its survival. In vivo, this asymmetric apportioning may promote the dissemination of the infection. Ultimately it will be important to investigate the consequence of an infection on the cell cycle during a systemic infection and to determine whether the apportioning of bacteria between daughter cells favor the bacterial spreading and the dissemination of the infection.
Materials and methods
Materials
HA-tagged proteins were detected using a mouse monoclonal anti HA (Covance, clone 16B12). The mouse anti human LAMP1 H4A3 antibody developed by J. T. August was obtained from Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development (NICHD) and maintained by the University of Iowa (Department of Biology). The mouse anti AIM-1 (#611082) was obtained from BD transduction Laboratories. The mouse anti lipopolysaccharide of Salmonella Typhimurium (clone 1E6) was from Meridian Life Science. The rabbit anti SCAMP3 antibody was generously provided by J. David Castle (University of Virginia, Charlottesville). Fluorescent Alexa secondary antibodies were obtained from Jackson ImmunoResearch. Fluoresbrite® YG Carboxylate Microspheres 1 µm were from PolySciences, Inc. (#15702-10). Propidium Iodide was from Sigma (P4170).
Statistical analyses
Statistical analyses were performed with Prism 6 software (GraphPad). with one-way ANOVA and Tukey post-test or 2-tailed unpaired Student's t test. P-values: ns, not significant; *, P<0.05; **, P<0.01; ***, P <0.0005.
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in . Strains were cultured in LB broth (Difco) or minimal medium (M9 pH 7.2, glycerol 0.2%, MgSO4 1 mM, CaCl2 200 mM, thiamine 1 mg / ml, casamino acids 1 mg / ml). Ampicillin (50 µg / ml), kanamycin (50 µg / ml), tetracycline (10 µg / ml) and chloramphenicol (50 µg / ml) were added when required.
Table 1. Salmonella strains.
Eukaryotic cells and culture conditions
RAW 264.7 mouse macrophages and human HeLa cells were grown in DMEM (GibcoBRL) supplemented with 10% foetal calf serum (FCS; GibcoBRL), 2 mM nonessential amino acids, and glutamine (GibcoBRL) at 37°C in 5% CO2. Nocodazole (1 mg / ml stock solution in DMSO) was added as indicated.
Bacterial infection
HeLa cells were seeded in 6-well plates with or without 12 mm diameter glass coverslips at a surface ratio of 1:10 unless otherwise indicated, 24 h before infection. Bacteria were grown in LB broth overnight at 37°C with shaking, diluted 1:33 in fresh LB broth, and incubated in the same conditions for 3.5 h. Bacteria were pelleted by centrifugation, resuspended in Earle's buffered salt solution (pH 7.4) and added to the cells at a multiplicity of infection of 100:1 unless otherwise indicated. The infection was allowed to proceed for 10 min at 37°C in 5% CO2. RAW 264.7 macrophages were routinely grown in Petri dishes normally used for microbiology. Twenty-four hours before infection, cells were resuspended, counted and seeded at a density of 105 cells per cm2 in 6-well tissue culture plates with glass coverslips. Bacteria were cultured overnight in minimal medium at 37°C with shaking, pelleted by centrifugation and opsonised in DMEM containing FCS and 10% normal mouse serum for 30 min on ice. Bacteria were diluted in cell growth medium and added to the cells at a multiplicity of infection of 100:1. Plates were centrifuged at 500 g for 5 min at 4°C and incubated for 30 min at 37°C in 5% CO2. Cells were washed 3 times with growth medium containing 100 µg / ml gentamicin and incubated in this medium for 1 h, after which the gentamicin concentration was decreased to 5 µg / ml for the remainder of the experiment.
Flow cytometry
For cell cycle analysis, cells were fixed with 3% paraformaldehyde and permeabilized in ice cold 70 % ethanol. DNA staining was performed for 30 min at 37 °C with 10 µg / ml propidium iodide and 0.2 mg ml / ml RNAse A (Sigma, R5503) in 20 mM HEPES, 160 mM NaCl, 1 mM EGTA. Cells were analyzed on a FACS LSR II UV apparatus (Becton Dickinson). Data analysis of the distribution of the different phases of cell cycle was performed using the FlowJo 8.3 software (Tree Star Inc.). Salmonella-infected GFP-positive cells and non-infetcted GFP-negative cells were gated based on the signal intensity in the FITC-A channel.
Immunofluorescence
Cells grown on coverslips were fixed with 3 % paraformaldehyde (pH 7.4) in PBS at room temperature for 10 min. Fixed cells were washed 3 times in PBS and permeabilized with 0.1 % saponin in PBS. Primary and secondary antibodies were diluted in PBS containing 0.1 % saponin and 5 % horse serum. Coverslips were incubated with primary antibodies for 60 min at room temperature, washed once in PBS containing 0.1 % saponin and then incubated with appropriate secondary antibodies. Coverslips were mounted onto glass slides using ProLong Gold with DAPI (Invitrogen). Cells were observed with an epifluorescence Axioplan2 microscope (Zeiss) or a LSM780 confocal laser scanning microscope (Zeiss).
Abbreviations
| ICT | = | inter-cellular tubules |
| p.i. | = | post-infection |
| SCV | = | Salmonella-containing vacuole |
| SIT | = | Salmonella-induced tubules |
| T3SS | = | Salmonella type 3 secretion system |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KVIR_S__1173298.docx
Download MS Word (8.7 MB)Acknowledgments
We thank Steve Garvis for the critical reading of the manuscript and David W. Holden for providing bacterial strains.
Funding
YZ was supported by the China Scholarship Council. We acknowledge financial supports from n° ANR-10-INBS-04-01 France Bio Imaging, LabEx INFORM, and from the Fondation pour la Recherche Médicale (FRM SC).
References
- Aussel L, Zhao W, Hébrard M, Guilhon A-A, Viala JPM, Henri S, Chasson L, Gorvel J-P, Barras F, Méresse S. Salmonella detoxifying enzymes are sufficient to cope with the host oxidative burst. Mol Microbiol 2011; 80:628-40; PMID:21362067; http://dx.doi.org/10.1111/j.1365-2958.2011.07611.x.
- Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci USA 2010; 107:17733-8; PMID:20876119; http://dx.doi.org/10.1073/pnas.1006098107.
- LaRock DL, Chaudhary A, Miller SI. Salmonellae interactions with host processes. Nat Rev Microbiol 2015; 13:191-205; PMID:25749450; http://dx.doi.org/10.1038/nrmicro3420.
- Eswarappa SM, Negi VD, Chakraborty S, Chandrasekhar Sagar BK, Chakravortty D. Division of the Salmonella-containing vacuole and depletion of acidic lysosomes in Salmonella-infected host cells are novel strategies of Salmonella enterica to avoid lysosomes. Infect Immun 2010; 78:68-79; PMID:19858305; http://dx.doi.org/10.1128/IAI.00668-09.
- McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, et al. PLEKHM1 Regulates Autophagosome-Lysosome Fusion through HOPS Complex and LC3/GABARAP Proteins. Mol Cell 2015; 57:39-54; PMID:25498145; http://dx.doi.org/10.1016/j.molcel.2014.11.006.
- Garcia-del Portillo F, Zwick MB, Leung KY, Finlay BB. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc Natl Acad Sci USA 1993; 90:10544-8; PMID:8248143; http://dx.doi.org/10.1073/pnas.90.22.10544.
- Knodler LA, Vallance BA, Hensel M, Jäckel D, Finlay BB, Steele-Mortimer O. Salmonella type III effectors PipB and PipB2 are targeted to detergent-resistant microdomains on internal host cell membranes. Mol Microbiol 2003; 49:685-704; PMID:12864852; http://dx.doi.org/10.1046/j.1365-2958.2003.03598.x.
- Rajashekar R, Liebl D, Seitz A, Hensel M. Dynamic remodeling of the endosomal system during formation of Salmonella-induced filaments by intracellular Salmonella enterica. Traffic 2008; 9:2100-16; PMID:18817527; http://dx.doi.org/10.1111/j.1600-0854.2008.00821.x.
- Krieger V, Liebl D, Zhang Y, Rajashekar R, Chlanda P, Giesker K, Chikkaballi D, Hensel M. Reorganization of the endosomal system in Salmonella-infected cells: the ultrastructure of Salmonella-induced tubular compartments. PLoS Pathog 2014; 10:e1004374; PMID:25254663; http://dx.doi.org/10.1371/journal.ppat.1004374.
- Schroeder N, Mota LJ, Méresse S. Salmonella-induced tubular networks. Trends Microbiol 2011; 19:268-77; PMID:21353564; http://dx.doi.org/10.1016/j.tim.2011.01.006.
- Stein MA, Leung KY, Zwick M, GarciadelPortillo F, Finlay BB. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol Microbiol 1996; 20:151-64; PMID:8861213; http://dx.doi.org/10.1111/j.1365-2958.1996.tb02497.x.
- Jiang X, Rossanese OW, Brown NF, Kujat-Choy S, Galán JE, Finlay BB, Brumell JH. The related effector proteins SopD and SopD2 from Salmonella enterica serovar Typhimurium contribute to virulence during systemic infection of mice. Mol Microbiol 2004; 54:1186-98; PMID:15554961; http://dx.doi.org/10.1111/j.1365-2958.2004.04344.x.
- Domingues L, Holden DW, Mota LJ. The Salmonella effector SteA contributes to the control of membrane dynamics of Salmonella-containing vacuoles. Infect Immun 2014; 82:2923-34; PMID:24778114; http://dx.doi.org/10.1128/IAI.01385-13.
- Knodler LA, Steele-Mortimer O. The Salmonella effector PipB2 affects late endosome/lysosome distribution to mediate Sif extension. Mol Biol Cell 2005; 16:4108-23; PMID:15987736; http://dx.doi.org/10.1091/mbc.E05-04-0367.
- Kuhle V, Hensel M. SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell Microbiol 2002; 4:813-24; PMID:12464012; http://dx.doi.org/10.1046/j.1462-5822.2002.00234.x.
- Maudet C, Mano M, Sunkavalli U, Sharan M, Giacca M, Förstner KU, Eulalio A. Functional high-throughput screening identifies the miR-15 microRNA family as cellular restriction factors for Salmonella infection. Nat Commun 2014; 5:4718; PMID:25146723; http://dx.doi.org/10.1038/ncomms5718.
- Terada Y, Tatsuka M, Suzuki F, Yasuda Y, Fujita S, Otsu M. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J 1998; 17:667-76; PMID:9450992; http://dx.doi.org/10.1093/emboj/17.3.667.
- Nix R, Altschuler S, Henson P, Detweiler C. Hemophagocytic Macrophages Harbor Salmonella enterica during Persistent Infection. PLoS Pathog 2007; 3:e193; PMID:18085823; http://dx.doi.org/10.1371/journal.ppat.0030193.
- Santos AJM, Meinecke M, Fessler MB, Holden DW, Boucrot E. Preferential invasion of mitotic cells by Salmonella reveals that cell surface cholesterol is maximal during metaphase. J Cell Sci 2013; 126:2990-6; PMID:23687374; http://dx.doi.org/10.1242/jcs.115253.
- Papezova K, Gregorova D, Jonuschies J, Rychlik I. Ordered expression of virulence genes in Salmonella enterica serovar typhimurium. Folia Microbiol (Praha) 2007; 52:107-14; PMID:17575908; http://dx.doi.org/10.1007/BF02932148.
- Brennan MA, Cookson BT. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol Microbiol 2000; 38:31-40; PMID:11029688; http://dx.doi.org/10.1046/j.1365-2958.2000.02103.x.
- Abrahams GL, Müller P, Hensel M. Functional dissection of SseF, a type III effector protein involved in positioning the salmonella-containing vacuole. Traffic 2006; 7:950-65; PMID:16800847; http://dx.doi.org/10.1111/j.1600-0854.2006.00454.x.
- Beuzón CR, Méresse S, Unsworth KE, Ruíz-Albert J, Garvis S, Waterman SR, Ryder TA, Boucrot E, Holden DW. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J 2000; 19:3235-49; http://dx.doi.org/10.1093/emboj/19.13.3235.
- Halici S, Zenk SF, Jantsch J, Hensel M. Functional analysis of the Salmonella pathogenicity island 2-mediated inhibition of antigen presentation in dendritic cells. Infect Immun 2008; 76:4924-33; PMID:18765734; http://dx.doi.org/10.1128/IAI.00531-08.
- Dean P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol Rev 2011; 35:1100-25; PMID:21517912; http://dx.doi.org/10.1111/j.1574-6976.2011.00271.x.
- Galán JE, Curtiss R. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA 1989; 86:6383-7; http://dx.doi.org/10.1073/pnas.86.16.6383.
- Hayward RD, Cain RJ, McGhie EJ, Phillips N, Garner MJ, Koronakis V. Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol Microbiol 2005; 56:590-603; PMID:15819617; http://dx.doi.org/10.1111/j.1365-2958.2005.04568.x.
- Nikolaus T, Deiwick J, Rappl C, Freeman JA, Schroder W, Miller SI, Hensel M. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J Bacteriol 2001; 183:6036-45; PMID:11567004; http://dx.doi.org/10.1128/JB.183.20.6036-6045.2001.
- Mesa-Pereira B, Medina C, Camacho EM, Flores A, Santero E. Novel tools to analyze the function of Salmonella effectors show that SvpB ectopic expression induces cell cycle arrest in tumor cells. PLoS ONE 2013; 8:e78458; PMID:24205236; http://dx.doi.org/10.1371/journal.pone.0078458.
- Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, Bongaerts RJM, Ahmad N, Rhen M, Hinton JCD. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol 2008; 10:958-84; PMID:18031307; http://dx.doi.org/10.1111/j.1462-5822.2007.01099.x.
- Katajisto P, Doehla J, Chaffer CL, Pentinmikko N, Marjanovic N, Iqbal S, Zoncu R, Chen W, Weinberg RA, Sabatini DM. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science 2015; 348:340-3; PMID:25837514; http://dx.doi.org/10.1126/science.1260384.
- Zhao W, Moest T, Zhao Y, Guilhon A-A, Buffat C, Gorvel J-P, Méresse S. The Salmonella effector protein SifA plays a dual role in virulence. Sci Rep 2015; 5:12979; PMID:26268777; http://dx.doi.org/10.1038/srep12979.
- Hensel, Shea JE, Waterman SR, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang FC, Holden DW. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol 1998; 30:163-74; PMID:9786193; http://dx.doi.org/10.1046/j.1365-2958.1998.01047.x.
- Henry T, Couillault C, Rockenfeller P, Boucrot E, Dumont A, Schroeder N, Hermant A, Knodler LA, Lecine P, Steele-Mortimer O, et al. The Salmonella effector protein PipB2 is a linker for kinesin-1. Proc Natl Acad Sci USA 2006; 103:13497-502; PMID:16938850; http://dx.doi.org/10.1073/pnas.0605443103.
