ABSTRACT
Dendritic cells (DCs) are critical for defense against a variety of pathogens and the formation of adaptive immune responses. The transcription factor Batf3 is critical for the development of CD103+CD11b− DCs, which promote IL-12–dependent protective immunity during viral and parasitic infections, dampen Th2 immunity during helminthic infection, and exert detrimental effects during bacterial infection. Whether CD103+ DCs modulate immunity during systemic or mucosal fungal disease remains unknown. Herein, we report that Batf3 is critical for accumulation of CD103+ DCs in the kidney and tongue at steady state, for their expansion during systemic and oropharyngeal candidiasis, and for tissue-specific production of IL-12 in kidney but not tongue during systemic and oropharyngeal candidiasis, respectively. Importantly, deficiency of CD103+ DCs does not impair survival or fungal clearance during systemic or oropharyngeal candidiasis, indicating that Batf3-dependent CD103+ DC accumulation mediates pathogen- and tissue-specific immune effects.
Introduction
Systemic candidiasis, caused by invasion of the commensal yeast Candida into the bloodstream, is a leading cause of nosocomial bloodstream infections in the United States.Citation1 Importantly, infected patients have a 30 – 40% mortality rate, despite antifungal therapy,Citation2 making systemic candidiasis an important unmet medical condition. Mucosal candidiasis, although not life-threatening, is more common than systemic candidiasis worldwide; oral thrush develops in the majority of AIDS patients,Citation3 and approximately 75% of all women will develop vaginal candidiasis at some point in their lifetime.Citation4 Furthermore, the emergence of azole- and echinocandin-resistant Candida strains makes treatment of these infections more difficult.Citation5 Therefore, it is important to understand cellular and molecular immunological factors that are necessary for protection against Candida infections in order to develop targeted immune-based therapies to improve patient outcomes.
Dendritic cells (DCs) are immune cells essential for the protection of the host against a wide variety of pathogens and for the generation of both innate and adaptive immune responses.Citation6 With regard to Candida, DCs are able to phagocytose and kill ingested Candida, produce cytokines in response to Candida stimulation, and present Candida-specific antigens in vitro.Citation7,8 However, the importance of DCs during C. albicans infection in vivo is not well-defined, as only recent studies have attempted to address their role during systemic and mucosal candidiasis.Citation9-11
There are several subsets of DCs, including classical, monocyte-derived, and plasmacytoid DCs. The classical DCs can be further subdivided, based on surface marker expression and/or function, with one of these subsets being CD103+ DCs, which correspond to CD8α+ DCs in lymphoid tissues.Citation6 CD103+ DCs are thought to be most important for cross-presentation of antigens and production of IL-12, and are dependent on several transcription factors, including Batf3, which is indispensable for their development.Citation6 CD103+ DC-dependent IL-12 production can be beneficial or detrimental during infection with the helminths Schistosoma mansoni or Heligmosomoides polygyrus, respectively.Citation12 Furthermore, CD103+ DCs are essential for protection against the parasitic protozoans Cryptosporidium parvum and Toxoplasma gondiiCitation13,14 and during West Nile and Sendai viral infections,Citation15,16 but detrimental during infection with the intracellular bacterium Listeria monocytogenes.Citation17 No studies to date have determined the in vivo role of CD103+ DCs on innate protection against fungal infections. Therefore, we sought to determine the impact of Batf3-deficiency on susceptibility to the commensal fungus, C. albicans, in both systemic and oral infection models in mice.
Results
We first investigated the role of CD103+ DCs in mediating control of systemic C. albicans infection by employing the well-established mouse model of systemic candidiasis in Batf3−/− mice. The kidney is the target organ in the model, which is thought to mimic skin-derived disease in humans.Citation2 Although CD103+ (or CD8α+) DCs are absent in several peripheral organs of Batf3−/− mice, including the lung, intestines, lymph nodes, dermis, spleen, thymus, and liver,Citation12,15,16 they have been shown to develop in the absence of Batf3 in the spleens and lungs during infection with intracellular pathogens, in the spleen upon IL-12 administration, or in the spleen and lymph nodes during short-term bone marrow reconstitution; Batf3-independent, Batf/Batf2-dependent mechanisms have been implicated in some of these settings.Citation18,19 Therefore, we first examined whether CD103+ DCs are present in the Batf3−/− kidney both prior to, and during C. albicans infection. We utilized flow cytometry and defined this population as CD45+MHCIIhighF4/80−CD11chighCD103+CD11b− cells (Figure S1A), as previously described.Citation20 CD103+ DCs were present in the Batf3+/+ kidney, and their number significantly increased during C. albicans infection (p < 0.001) (). In contrast, Batf3−/− kidneys were devoid of CD103+ DCs both under homeostatic conditions and during infection, indicating that CD103+ DC development is strictly Batf3-dependent in this organ in the model (); the paucity of CD103+ DCs was further highlighted by the decreased percentage of CD103+ DCs out of all DCs in uninfected and infected Batf3−/− kidneys (). In contrast, Batf3 deficiency did not significantly affect the accumulation of other myeloid cells (CD11b+CD103− DCs, neutrophils, macrophages, Ly6Chigh monocytes) in the infected kidney (Figure S2A). These data show that Batf3−/− kidneys lack CD103+ DCs both under homeostatic conditions and during systemic C. albicans infection.
Figure 1. The transcription factor Batf3 is critical for the accumulation of CD103+ DCs in the mouse kidney but dispensable for protection against systemic candidiasis. (A-D) Kidney leukocytes were isolated from Batf3+/+ and Batf3−/− mice left uninfected or infected intravenously with C. albicans 4 d prior to harvest, and analyzed using flow cytometry. (A-B) Representative zebra FACS plots from uninfected mice (A) or Candida-infected mice at day 4 p.i. (B) after gating on live CD45+MHCIIhighF4/80−CD11c+ DCs, and plotting based on CD103 and CD11b expression. (C) Total number of CD103+CD11b− DCs in the uninfected and Candida-infected kidney. (D) Percentage of CD103+CD11b− cells within all DCs in the uninfected and Candida-infected kidney. (E) Survival of Candida-infected Batf3+/+ and Batf3−/− mice. (F) Kidney fungal burden in Candida-infected Batf3+/+ and Batf3−/− mice determined at day 4 p.i. as the number of CFUs/gram of tissue. (G) Serum creatinine levels of Candida-infected Batf3+/+ and Batf3−/− mice at day 4 p.i. (H) Representative H&E stained kidney sections from Candida-infected Batf3+/+ and Batf3−/− mice at day 4 p.i. Data in (C, D, F, and G) were analyzed using unpaired t-tests or Mann-Whitney U tests, where appropriate. Data in (E) were analyzed using a log-rank (Mantel-Cox) test. Data in (A-D) are representative FACS plots or combined data from 2 independent experiments with a total n of 5–6 mice/group. Data in (E) are combined from 2 independent experiments with a total n of 19–21 mice/group. Data in (F-G) are combined from 2–3 independent experiments with a total n of 8–11 mice/group. Histology sections in (H) are representative images from 2 independent experiments with a total of 4–5 mice/group.
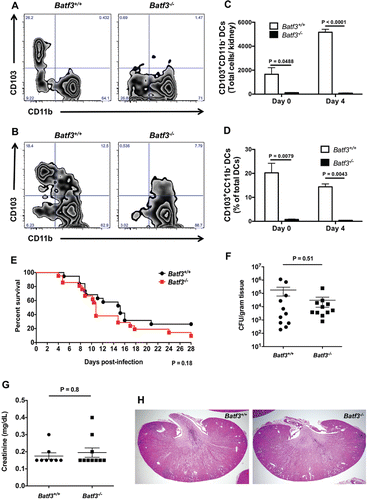
To determine whether lack of renal CD103+ DCs affects susceptibility to systemic candidiasis, mice were injected intravenously with 105 C. albicans and survival was monitored for 4 weeks. We found no difference between wild-type and Batf3−/− mice with regard to survival (), control of fungal proliferation in the kidney (), or serum creatinine levels (), which is an indicator of kidney damage. In addition, no differences were noted in the histological examination of C. albicans-infected Batf3+/+ and Batf3−/− kidneys (). Because Candida strain-specific differences in antifungal immune responses have been reported in vivo,Citation21 we infected Batf3+/+ and Batf3−/− with another strain isolated from a patient with systemic candidiasis (UC820),Citation20 and found no increased susceptibility of Batf3−/− mice, as shown by similar survival (Figure S3A) and CFUs at day 4 p.i (Figure S3B). Taken together, these data show that, although Batf3 is critical for renal accumulation of CD103+ DCs during systemic candidiasis, their deficiency does not impair the control of the infection in vivo.
We next sought to examine the in vivo role of Batf3-dependent CD103+ DCs during oropharyngeal candidiasis (OPC), as the immune factors that mediate protection during systemic versus mucosal Candida infections are typically non-overlapping.Citation22 As with the kidney, we used flow cytometry to first determine whether Batf3 is indispensable for accumulation of CD103+ DCs in the mouse tongue (see Figure S1B for gating strategy). Indeed, we found a small population of CD103+ DCs in the uninfected tongue of Batf3+/+ mice that was absent in Batf3−/− tongues (). We then used a previously reported sublingual C. albicans infection model,Citation23,24 and found a significant expansion in the number and percentage of CD103+ DCs out of all DCs in Batf3+/+ tongues (p < 0.01 and p < 0.0001, respectively), while CD103+ DCs remained absent in the tongue of Batf3−/− mice during fungal infection (). As with the kidney, Batf3 deficiency did not impair the accumulation of other myeloid cells (CD11b+CD103− DCs, neutrophils, macrophages, Ly6Chigh monocytes) in the infected tongue (Figure S2B). Collectively, these data show that, similar to the kidney, Batf3−/− tongues lack CD103+ DCs both under homeostatic conditions and during mucosal C. albicans infection.
Figure 2. Batf3 mediates CD103+ DC accumulation, but not protection from Candida infection, in the mouse oral mucosa. (A-D) Tongue leukocytes were harvested from Batf3+/+ and Batf3−/− mice left uninfected or sublingually infected with C. albicans 5 d prior to harvest, and analyzed using flow cytometry. (A-B) Representative zebra FACS plots from uninfected mice (A) or Candida-infected mice at day 5 p.i. (B) after gating on live CD45+MHCIIhighCD11chigh DCs, and plotting based on CD103 and CD11b expression. (C) Total number of CD103+CD11b− DCs in the uninfected and Candida-infected tongue. (D) Percentage of CD103+CD11b− cells within all DCs in the uninfected and Candida-infected tongue. (E) Fungal burden in Candida-infected Batf3+/+ and Batf3−/− tongue determined at days 1 and 5 p.i., expressed as the number of CFUs/gram of tissue. (F) Percentage of tongue epithelium with abscess formation at day 1 p.i. after oral Candida infection. (G-H) Representative images from H&E (G) and PAS (H) stained tongue sections are shown at days 1 (upper panels) and 5 (lower panels) p.i. Data in (C–F) were analyzed using unpaired t-tests or Mann-Whitney U tests, where appropriate. Data in (A-D) are representative FACS plots or combined data from 2 independent experiments with a total n of 4–6 mice/group. Data in (E) are combined from 2 independent experiments with a total n of 6–7 mice/group. Data in (F-H) are representative images or combined data from 2 independent experiments with a total n of 4 mice/group.
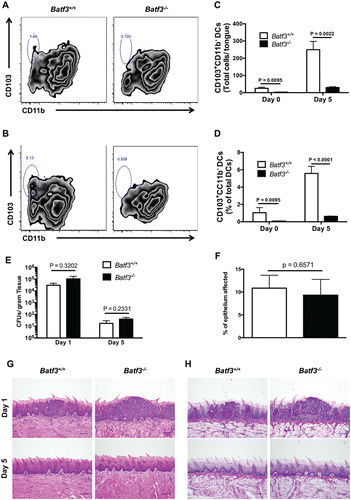
To determine whether lack of oral mucosal CD103+ DCs plays a role during OPC, Batf3+/+ and Batf3−/− mice were sublingually infected with C. albicans, and fungal load and histology were examined after infection. We found no difference in C. albicans growth at day 1 early post-infection (p.i.) between wild-type and Batf3−/− tongues. By day 5 p.i., the vast majority of C. albicans was cleared from both Batf3+/+ and Batf3−/− tongues (). In addition, the extent of epithelial involvement by abscess formation was similar between Batf3+/+ and Batf3−/− mice at day 1 p.i. (); abscesses were localized in the epithelial layer without penetration into deeper tissues and were cleared in both strains of mice by day 5 p.i. (). Furthermore, Batf3−/− mice did not exhibit increased fungal proliferation in the tongue at day 5 p.i., compared with Batf3+/+ mice, when we used the C. albicans strains 529L (Figure S4A) and Y72 (Figure S4B), which were obtained from patients with oral candidiasis.Citation23,25 Taken together, these data show that although Batf3 is critical for accumulation of CD103+ DCs in the oral mucosa, these cells are dispensable for protection against OPC in vivo.
As CD103+ DCs are known to produce a wide array of cytokines and chemokines upon recognition of microbial ligands, we measured levels of 13 cytokines and chemokines via Luminex array in kidney and tongue homogenates during systemic and mucosal infection with C. albicans, respectively. We found that CD103+ DCs are not major cellular sources for production of IL-1β, IL-4, IL-6, IL-17A, IL-22, CCL2, CCL3, CCL4, CCL5, CXCL1, or CXCL2 in the kidney and tongue during systemic and mucosal candidiasis, respectively (). Instead, in the tongue, the anti-inflammatory cytokine, IL-10, was ∼35% reduced in Batf3−/− mice after infection (). Importantly, production of both IL-17A and IL-22, which is essential for control of OPC,Citation26 was not impaired in Batf3-deficient tongues.
Table 1. CD103+ DCs are dispensable for the production of a variety of pro-inflammatory cytokines and chemokines in the kidney during systemic candidiasis.
Table 2. Induction of mucosal IL-17/IL-22 is Batf3-independent after oral C. albicans infection.
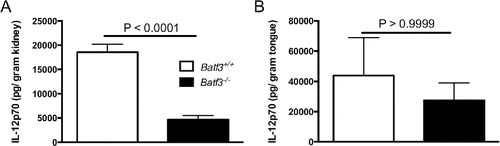
A cytokine that has been shown to be integrally dependent on the presence of CD103+ DCs is IL-12.Citation14,27 Interestingly, we found that IL-12p70 production was significantly reduced in the kidneys of systemically infected Batf3−/− mice, whereas IL-12p70 levels were similar in Batf3+/+ and Batf3−/− tongues during mucosal Candida infection (). Therefore, these data indicate that IL-12 production by CD103+ DCs, in the setting of Candida infection, is tissue-specific.
Figure 3. Batf3-dependent accumulation of CD103+ DCs mediates tissue-specific production of IL-12 during systemic, but not oropharyngeal candidiasis. IL-12 levels were measured in Batf3+/+ and Batf3−/− mice in the kidney at day 4 after intravenous C. albicans infection (A) or in the tongue at day 1 after sublingual C. albicans infection (B). Data were analyzed using unpaired t-test (A) or Mann-Whitney U test (B). Data are combined from 2 independent experiments with a total n of 6–8 mice/group.
Discussion
In the present study, we examined for the first time the in vivo role of Batf3-dependent CD103+ DCs in host defense during systemic and mucosal fungal infections in mice. During parasitic, viral and bacterial infection, CD103+ DCs have been reported to play crucial roles, whether protective or deleterious. Specifically, CD103+ DCs are essential for control of intestinal parasitic infections caused by Toxoplasma gondiiCitation14 and Cryptosporidium parvum,Citation13 for mediating protective immunity against viruses, and for control of syngeneic fibrosarcomas.Citation15,16 On the other hand, during infection with the intracellular bacterium Listeria monocytogenes, CD103+ DCs promote the development of disease by serving as an essential pathogen entry point.17 Interestingly, CD103+ DCs play differential roles during helminthic infections; they are protective during infection with Schistosoma mansoni, but deleterious during infection with Heligmosomoides polygyrus.Citation12 Our data demonstrate that CD103+ DCs are redundant for the control of mucosal and systemic infection with C. albicans, indicating that Batf3-dependent CD103+ DCs promote pathogen-specific immune responses. Future studies will be required to evaluate whether CD103+ DCs mediate antifungal host defense during infection caused by other fungi such as dimorphic fungi or ubiquitous inhaled molds.
It has been shown that protective IL-12 production is highly dependent on CD103+ DCs during infection with Toxoplasma gondiiCitation14 and Encephalitozoon cuniculi.Citation27 Furthermore, CD103+ DC-dependent production of IL-12 exacerbates chronic intestinal infection with Heligmosomoides polygyrus, and ameliorates IL-13-mediated liver fibrosis during infection with Schistosoma mansoni.Citation12 We show here that IL-12 production is dependent on CD103+ DCs during systemic, but not oral C. albicans infection. Decreased IL-12 production in the kidney during systemic candidiasis in Batf3−/− mice does not impair susceptibility to the infection, in agreement with the redundant role of IL-12 in the control of systemic candidiasis.Citation28 Instead, while IL-12 is known to mediate early control of OPC,Citation29 our data show that Batf3-dependent CD103+ DCs are dispensable for IL-12 production during OPC and infection control at the oral mucosa. These findings further highlight the dichotomy of immune factors that are necessary for control of mucosal vs. systemic candidiasis and underscore the organ-specific nature of the cellular immune response against Candida challenge. Future studies should aim to define the organ-specific microenvironment cues that dictate Batf3-dependent CD103+ DC-mediated cytokine production and determine the cellular source(s) of IL-12 in the Candida-infected oral mucosa.
It has been shown that CD103+ DCs can expand in a Batf3-independent manner in the spleens and lungs during intracellular bacterial infection, in the spleen upon IL-12 administration, or in the spleen and lymph nodes during short-term bone marrow reconstitution; Batf/Batf2 have been shown to promote Batf3-independent DC expansion in some of these conditions.Citation18,19 Our data show that CD103+ DC development is strictly Batf3-dependent in the kidney and tongue both at steady-state and during acute C. albicans infection. More work will be required to define whether generation of CD103+ DCs is Batf3-dependent in other Candida-infected tissues and during infection with other fungal pathogens, and to examine the tissue- and pathogen-specific cues that modulate Batf3-dependent and independent CD103+ DC development.
Besides their non-redundant roles in the innate control against infection, including systemic candidiasis,Citation9 DCs have well-characterized adaptive functions via presentation of antigens to T cells for the development of adaptive immune responses. Interestingly, on one hand, CD103+ DCs were not necessary for priming of Th17 cells during primary Candida infection.Citation10 Instead, a recent elegant study found that, during a secondary Candida infection, Batf3−/- mice had elevated Th17 responses and decreased CFUs in the skin upon epicutaneous priming and secondary epicutaneous challenge. This effect was not seen in the kidney upon epicutaneous priming and secondary systemic challenge,Citation11 underscoring the compartmentalized effects of CD103+ DCs during a secondary fungal infection. Therefore, future research should elucidate the role of CD103+ DCs in antifungal host defense during secondary and chronic Candida infection via oral, gastrointestinal, or systemic challenges.
In humans, autosomal recessive IRF8 deficiency predisposes to OPCCitation30 and is associated with complete lack of all monocyte and DC subsets, including CD141+ DCs, which correspond to mouse CD103+CD11b− DCs.Citation31 Since we show that Batf3−/− mice are not susceptible to OPC, our data suggest that monocytes and/or CD11b+ DC subsets, not CD103+CD11b− DCs, are likely important for control of OPC in humans. Mice lacking Irf8 have defective development of CD8α+ DCs, plasmacytoid DCs, and monocytes,Citation32 and studies should evaluate their susceptibility to OPC and define the mechanisms by which Irf8 promotes mucosal antifungal immunity.
In summary, we show that the transcription factor Batf3 is crucial for CD103+ DC accumulation in the oral mucosa and kidney at steady state and during fungal infection, and for promotion of tissue-specific IL-12 production during fungal challenge. Importantly, in contrast to bacterial, parasitic and viral infections, we demonstrate that Batf3-dependent tissue accumulation of CD103+ DCs is redundant for protective host defense during acute mucosal and systemic fungal disease. More work will be required to define the tissue- and pathogen-specific roles of Batf3-dependent CD103+ DCs in innate and adaptive immunity and to determine the differential roles of resident mononuclear phagocytes other than CD103+ DCs in mediating protection against mucosal and systemic fungal disease in mice and humans.Citation9,20
Materials and methods
Mice
Batf3−/− mice were a kind gift from Dr. Brian Kelsall (NIAID, NIH) and C57Bl/6 (Batf3+/+) mice were purchased from Jackson Laboratories. Batf3−/− mice were developed as previously described.Citation15 Age- and sex-matched mice were used and kept under specific pathogen-free conditions. All experiments were conducted in accordance with guidelines set forth by the Guide for the Care and Use of Laboratory Animals under a protocol approved by the Animal Care and Use Committee of the NIAID in an Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal facility.
Fungal strains and mouse models of C. albicans infection
C. albicans strain SC5314 was used in all experiments, unless stated otherwise. In some systemic infection experiments, the C. albicans strain UC820, which was isolated from a patient with systemic candidiasis, was used.Citation20 In some OPC infections, the C. albicans strains 529L or Y72, which were isolated from patients with oral candidiasis, were used.Citation23,25 C. albicans was grown in yeast extract (BD, Cat. #212750), peptone (BD, Cat. #211677), and dextrose (Avantor, Cat. #4912) media that contained penicillin and streptomycin (Corning, Cat. #30-002-Cl) in a shaking incubator at 30°C. Prior to infection, C. albicans was centrifuged, washed with PBS, and counted with a hemocytometer. For systemic C. albicans infections with SC5314, 105 C. albicans cells were injected into the lateral tail vein. For systemic C. albicans infections with UC820, 1.5 × 105 or 3 × 105 C. albicans cells were injected into the lateral tail vein for assessment of CFUs or survival, respectively. For OPC, sublingual infections were performed using cotton swabs saturated in media containing 107 C. albicans/ml, as previously described.Citation23,24
Determination of fungal colony forming units (CFUs)
To assess tissue fungal load after infection, the tongues or kidneys of mice were homogenized in PBS using an Omni tissue homogenizer. The homogenate was plated undiluted or after serial dilutions on yeast, peptone, and dextrose agar plates containing penicillin and streptomycin. The plates were incubated for 24–48 hours at 37°C, the colonies were counted, and the data were presented as the total number of CFUs per gram of tissue. To lower the limit of detection for fungal burden determination in the tongue, the entire homogenate was plated. If no colonies were counted, a value of 0 was assigned.
Single cell suspensions from the kidney and tongue
Single-cell suspensions were obtained from the kidney using a previously described technique.Citation33 Briefly, mice were perfused with PBS and kidneys were minced and incubated in a solution containing liberase TL (Roche Cat. #05989132001) and DNase I (Roche Cat. # 10104159001) for 20 min in a shaking water bath at 37°C. Digested tissue was passed through a 70 μm filter (Greiner Bio-One, Cat. # 542070) and red blood cells lysed with ACK lysis buffer (Quality Biological Cat. #118-156-721). Cells were then passed through a 40 μm filter (Greiner Bio-One, Cat. # 542040), resuspended in 40% Percoll (GE Healthcare, Cat. #17-0891-01), and overlaid onto 70% Percoll. The gradient was centrifuged at 872 g for 30 min at room temperature, and the cells at the interface between the 40% and 70% layers were collected and stained for flow cytometry. Single-cell suspensions were obtained from the tongue based on a previously described technique.Citation10 The tongue was finely minced and digested in a solution containing 385 U/ml collagenase type 4 (Worthington Cat. #LS004189), 2 U/ml dispase II (Gibco, Cat. # 17105-041), and 50 μg/ml DNase I in 10 ml RPMI for 45 min in a shaking water bath at 37°C. After 45 min, the solution was quenched with PBS + 5 mM EDTA (Quality Biological, Cat. # 351–027). The cells were serially filtered through a 70 μm filter and a 40 μm filter. The cells were washed, resuspended in 40% Percoll, and overlaid onto 70% Percoll. The gradient was centrifuged at 872 g for 30 min at room temperature, and the cells at the interface between the 40% and 70% layers were collected, washed with FACS buffer (PBS + 0.5% BSA + 0.01% NaN3), and resuspended in PBS for flow staining.
Flow cytometry
Single-cell suspensions from the kidney or tongue were stained with LIVE/DEAD fluorescent dye (Invitrogen, Cat. # L-23105) for 10 min in PBS at 4°C. Next, the cells were incubated with rat anti-mouse CD16/32 (eBioscience, Cat. #14–0161) for 10 min at 4°C to block Fc receptors, and stained for cell-surface markers for 30 min at 4°C. To identify CD103+ DCs and other myeloid populations, antibodies conjugated to eFluor 450, biotin/streptavidin BV570, PE, PerCP-Cy5.5, eFluor 605 NC, PE-Cy7, APC, AF700, or APC-Cy7 from eBioscience, Biolegend, or BD PharMingen were used. Antibodies were directed against MHC-II (Clone M5/114.15.2), CD103 (2E7), CD3 (17A2 or 145–2C11), B220 (RA3–6B2), CD19 (eBio1D3), NK1.1 (PK136), Ly6G (1A8), CD45 (30-F11), F4/80 (BM8), CD11c (N418), Ly6C (AL-21), and CD11b (M1/70). After washing with FACS buffer, cells were analyzed using a 5-laser LSR Fortessa (BD Biosciences, San Jose, CA), and data were analyzed using FlowJo software (FlowJo, Ashland, OR). Cell numbers were quantified using PE-conjugated counting beads (Spherotech, Cat. #ACFP-70-10), as previously described.Citation33
Determination of creatinine levels
Blood was collected from Batf3+/+ and Batf3−/− mice 4 d p.i. via cardiac puncture and placed in Serum Gel Z/1.1 tubes (Sarstedt, Cat. #41.1378.005), which were spun at ∼16,000 g for 10 minutes at 4°C. Serum was collected and stored at −80°C until analysis of creatinine levels by the NIH Clinical Chemistry Laboratory.
Histology
Kidneys were harvested at day 4 p.i., and tongues were harvested at days 1 and 5 p.i. following intravenous or sublingual C. albicans infection, respectively. The tissues were fixed in 10% formalin and embedded in paraffin. Longitudinal sections of the kidney or tongue were prepared for H&E or PAS staining (Histoserv, Inc., Germantown, MD). Sections were viewed under a microscope, and representative pictures were taken. For determination of the area affected by Candida infection, the entire tongue was photographed from the PAS-stained sections and the following formula was used: [(area of Candida abscesses/total area of the epithelium of the tongue) × 100%] using the Fiji image-processing program. This assessment was performed in a blinded manner with regard to the mouse genotype.
Determination of cytokine and chemokine protein concentrations
Batf3+/+ and Batf3−/− mice were infected with C. albicans intravenously or sublingually as described above, and kidneys or tongues were harvested at day 4 or day 1 p.i., respectively. Organs were homogenized with an Omni Tissue Homogenizer (Omni International) in PBS with 0.5% Tween-20 (Sigma-Aldrich Cat. #P7949) and a protease inhibitor cocktail (Roche Applied Science, Cat. #11836170001). The homogenate was then centrifuged at ∼16,000 g for 10 minutes at 4°C, supernatants were filtered through a 22μm filter (EMD Millipore, Cat. #SLGV033RS), and frozen at 80°C until use. To determine cytokine and chemokine protein concentrations, a multiplex bead array assay was utilized as previously described.Citation23 Antibodies and cytokine standards were purchased from R&D Systems or Peprotech as antibody pairs. Capture antibodies were coupled to individual Luminex bead sets for each cytokine or chemokine measured. Biotinylated antibodies were then added at twice the recommended concentration for ELISA, and all procedures were performed in PBS with 1% normal mouse serum (Gibco BRL), 1% normal goat serum (Gibco BRL), and 20 mM Tris-HCl (pH 7.4). The plates were read on a Luminex MAGPIX platform, and at least 50 beads were collected for each cytokine/chemokine per sample. The median fluorescence intensity for each bead was determined for analysis with the Milliplex software using a 5P regression algorithm.
Statistical analyses
For the survival studies, log-rank (Mantel-Cox) tests were used. For all other experiments, unpaired t-tests or Mann-Whitney U tests were performed, where appropriate. All statistical analyses were performed using Prism 6 software (GraphPad). Quantitative data are presented as mean ± standard errors of the mean (SEM), and a P value of <0.05 was considered significant.
Abbreviations
| CFUs | = | colony-forming units |
| DCs | = | Dendritic cells |
| OPC | = | oropharyngeal candidiasis |
| PAS | = | Periodic acid-Schiff |
| p.i. | = | post-infection |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KVIR_S_1186324.docx
Download MS Word (18.5 MB)Funding
This work was supported in part by the Intramural Research Program of the NIH, NIAID. JKL was supported by NIH grant 1R01AI108715.
References
- Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 2014; 370:1198-208; PMID:24670166; http://dx.doi.org/10.1056/NEJMoa1306801
- Lionakis MS. New insights into innate immune control of systemic candidiasis. Med Mycol 2014; 52:555-64; PMID:25023483; http://dx.doi.org/10.1093/mmy/myu029
- de Repentigny L, Lewandowski D, Jolicoeur P. Immunopathogenesis of oropharyngeal candidiasis in human immunodeficiency virus infection. Clin Microbiol Rev 2004; 17:729-59; PMID:15489345; http://dx.doi.org/10.1128/CMR.17.4.729-759.2004
- Sobel JD. Vaginitis. N Engl J Med 1997; 337:1896-903; PMID:9407158; http://dx.doi.org/10.1056/NEJM199712253372607
- Kanafani ZA, Perfect JR. Antimicrobial resistance: resistance to antifungal agents: mechanisms and clinical impact. Clin Infect Dis 2008; 46:120-8; PMID:18171227; http://dx.doi.org/10.1086/524071
- Mildner A, Jung S. Development and function of dendritic cell subsets. Immunity 2014; 40:642-56; PMID:24837101; http://dx.doi.org/10.1016/j.immuni.2014.04.016
- Newman SL, Holly A. Candida albicans is phagocytosed, killed, and processed for antigen presentation by human dendritic cells. Infect Immun 2001; 69:6813-22; PMID:11598054; http://dx.doi.org/10.1128/IAI.69.11.6813-6822.2001
- Romagnoli G, Nisini R, Chiani P, Mariotti S, Teloni R, Cassone A, Torosantucci A. The interaction of human dendritic cells with yeast and germ-tube forms of Candida albicans leads to efficient fungal processing, dendritic cell maturation, and acquisition of a Th1 response-promoting function. J Leukoc Biol 2004; 75:117-26; PMID:14525965; http://dx.doi.org/10.1189/jlb.0503226
- Whitney PG, Bar E, Osorio F, Rogers NC, Schraml BU, Deddouche S, LeibundGut-Landmann S, Reis e Sousa C. Syk signaling in dendritic cells orchestrates innate resistance to systemic fungal infection. PLoS Pathog 2014; 10:e1004276; PMID:25033445; http://dx.doi.org/10.1371/journal.ppat.1004276
- Trautwein-Weidner K, Gladiator A, Kirchner FR, Becattini S, Rulicke T, Sallusto F, LeibundGut-Landmann S. Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis. PLoS Pathog 2015; 11:e1005164; PMID:26431538; http://dx.doi.org/10.1371/journal.ppat.1005164
- Kashem SW, Igyarto BZ, Gerami-Nejad M, Kumamoto Y, Mohammed J, Jarrett E, Drummond RA, Zurawski SM, Zurawski G, Berman J, et al. Candida albicans morphology and dendritic cell subsets determine T helper cell differentiation. Immunity 2015; 42:356-66; PMID:25680275; http://dx.doi.org/10.1016/j.immuni.2015.01.008
- Everts B, Tussiwand R, Dreesen L, Fairfax KC, Huang SC, Smith AM, O'Neill CM, Lam WY, Edelson BT, Urban JF, Jr., et al. Migratory CD103+ dendritic cells suppress helminth-driven type 2 immunity through constitutive expression of IL-12. J Exp Med 2016; 213:35-51; PMID:26712805; http://dx.doi.org/10.1084/jem.20150235
- Lantier L, Lacroix-Lamande S, Potiron L, Metton C, Drouet F, Guesdon W, Gnahoui-David A, Le Vern Y, Deriaud E, Fenis A, et al. Intestinal CD103+ dendritic cells are key players in the innate immune control of Cryptosporidium parvum infection in neonatal mice. PLoS Pathog 2013; 9:e1003801; PMID:24367259; http://dx.doi.org/10.1371/journal.ppat.1003801
- Mashayekhi M, Sandau MM, Dunay IR, Frickel EM, Khan A, Goldszmid RS, Sher A, Ploegh HL, Murphy TL, Sibley LD, et al. CD8alpha(+) dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity 2011; 35:249-59; PMID:21867928; http://dx.doi.org/10.1016/j.immuni.2011.08.008
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 2008; 322:1097-100; PMID:19008445; http://dx.doi.org/10.1126/science.1164206
- Edelson BT, Kc W, Juang R, Kohyama M, Benoit LA, Klekotka PA, Moon C, Albring JC, Ise W, Michael DG, et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med 2010; 207:823-36; PMID:20351058; http://dx.doi.org/10.1084/jem.20091627
- Edelson BT, Bradstreet TR, Hildner K, Carrero JA, Frederick KE, Kc W, Belizaire R, Aoshi T, Schreiber RD, Miller MJ, et al. CD8alpha(+) dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes. Immunity 2011; 35:236-48; PMID:21867927; http://dx.doi.org/10.1016/j.immuni.2011.06.012
- Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Kc W, Albring JC, Satpathy AT, Rotondo JA, Edelson BT, Kretzer NM, et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 2012; 490:502-7; PMID:22992524; http://dx.doi.org/10.1038/nature11531
- Seillet C, Jackson JT, Markey KA, Brady HJ, Hill GR, Macdonald KP, Nutt SL, Belz GT. CD8alpha+ DCs can be induced in the absence of transcription factors Id2, Nfil3, and Batf3. Blood 2013; 121:1574-83; PMID:23297132; http://dx.doi.org/10.1182/blood-2012-07-445650
- Lionakis MS, Swamydas M, Fischer BG, Plantinga TS, Johnson MD, Jaeger M, Green NM, Masedunskas A, Weigert R, Mikelis C, et al. CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. J Clin Invest 2013; 123:5035-51; PMID:24177428; http://dx.doi.org/10.1172/JCI71307
- Marakalala MJ, Vautier S, Potrykus J, Walker LA, Shepardson KM, Hopke A, Mora-Montes HM, Kerrigan A, Netea MG, Murray GI, et al. Differential adaptation of Candida albicans in vivo modulates immune recognition by dectin-1. PLoS Pathog 2013; 9:e1003315; PMID:23637604; http://dx.doi.org/10.1371/journal.ppat.1003315
- Netea MG, Joosten LA, van der Meer JW, Kullberg BJ, van de Veerdonk FL. Immune defence against Candida fungal infections. Nat Rev Immunol 2015; 15:630-42; PMID:26388329; http://dx.doi.org/10.1038/nri3897
- Break TJ, Jaeger M, Solis NV, Filler SG, Rodriguez CA, Lim JK, Lee CC, Sobel JD, Netea MG, Lionakis MS. CX3CR1 is dispensable for control of mucosal Candida albicans infections in mice and humans. Infect Immun 2015; 83:958-65; PMID:25547797; http://dx.doi.org/10.1128/IAI.02604-14
- Solis NV, Filler SG. Mouse model of oropharyngeal candidiasis. Nat Protoc 2012; 7:637-42; PMID:22402633; http://dx.doi.org/10.1038/nprot.2012.011
- Rahman D, Mistry M, Thavaraj S, Challacombe SJ, Naglik JR. Murine model of concurrent oral and vaginal Candida albicans colonization to study epithelial host-pathogen interactions. Microbes Infect 2007; 9:615-22; PMID:17383212; http://dx.doi.org/10.1016/j.micinf.2007.01.012
- Huppler AR, Bishu S, Gaffen SL. Mucocutaneous candidiasis: the IL-17 pathway and implications for targeted immunotherapy. Arthritis Res Ther 2012; 14:217; PMID:22838497; http://dx.doi.org/10.1186/ar3893
- Moretto MM, Harrow DI, Hawley TS, Khan IA. Interleukin-12-Producing CD103+ CD11b- CD8+ Dendritic Cells Are Responsible for Eliciting Gut Intraepithelial Lymphocyte Response against Encephalitozoon cuniculi. Infect Immun 2015; 83:4719-30; PMID:26416905
- Netea MG, Vonk AG, van den Hoven M, Verschueren I, Joosten LA, van Krieken JH, van den Berg WB, Van der Meer JW, Kullberg BJ. Differential role of IL-18 and IL-12 in the host defense against disseminated Candida albicans infection. Eur J Immunol 2003; 33:3409-17; PMID:14635050; http://dx.doi.org/10.1002/eji.200323737
- Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med 2009; 206:299-311; PMID:19204111; http://dx.doi.org/10.1084/jem.20081463
- Hambleton S, Salem S, Bustamante J, Bigley V, Boisson-Dupuis S, Azevedo J, Fortin A, Haniffa M, Ceron-Gutierrez L, Bacon CM, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med 2011; 365:127-38; PMID:21524210; http://dx.doi.org/10.1056/NEJMoa1100066
- Watchmaker PB, Lahl K, Lee M, Baumjohann D, Morton J, Kim SJ, Zeng R, Dent A, Ansel KM, Diamond B, et al. Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat Immunol 2014; 15:98-108; PMID:24292363; http://dx.doi.org/10.1038/ni.2768
- Yanez A, Ng MY, Hassanzadeh-Kiabi N, Goodridge HS. IRF8 acts in lineage-committed rather than oligopotent progenitors to control neutrophil vs monocyte production. Blood 2015; 125:1452-9; PMID:25597637; http://dx.doi.org/10.1182/blood-2014-09-600833
- Lionakis MS, Lim JK, Lee CC, Murphy PM. Organ-specific innate immune responses in a mouse model of invasive candidiasis. J Innate Immun 2011; 3:180-99; PMID:21063074; http://dx.doi.org/10.1159/000321157
