 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Talaromyces (Penicillium) marneffei is an emerging opportunistic pathogen associated with HIV infection, particularly in Southeast Asia and southern China. The rapid uptake and killing of T. marneffei conidia by phagocytic cells along with the effective induction of an inflammatory response by the host is essential for disease control. T. marneffei produces a number of different laccases linked to fungal virulence. To understand the role of the various laccases in T. marneffei, laccase-encoding genes were investigated. Targeted single, double and triple gene deletions of laccases encoding lacA, lacB, and lacC showed no significant phenotypic effects suggesting redundancy of function. When a fourth laccase-encoding gene, pbrB, was deleted in the ΔlacA ΔlacB ΔlacC background, the quadruple mutant displayed delayed conidiation and the conidia were more sensitive to H2O2, sodium dodecyl sulfate (SDS), and antifungal agents than wild-type and other transformants. Conidia of the quadruple mutant showed marked differences in their interaction with the human monocyte cell line, THP-1 such that phagocytosis was significantly higher when compared with the wild-type at one and 2 hours of incubation while the phagocytic index was significantly different from 15 to 120 minutes. In addition, killing of the quadruple mutant by THP-1 cells was more efficient at 2 and 4 hours of incubation. The levels of the proinflammatory cytokines TNF-α, IL-1β and IL-6 from THP-1 cells infected with the quadruple mutant were also significantly increased in comparison with wild-type. The results demonstrate that production of laccases by T. marneffei actually promotes the pathogen's resistance to innate host defenses.
Introduction
Talaromyces marneffei, formerly known as Penicillium marneffei, is a thermally dimorphic fungus and the etiologic agent of the human systemic disease penicilliosis marneffei, which affects individuals in endemic areas throughout Southeast Asia and southern China.Citation1-3 T. marneffei has emerged as an opportunistic fungal pathogen associated with HIV infection in Northern Thailand, where penicilliosis is the third most common AIDS-associated disease, after tuberculosis and cryptococcosis.Citation4 Although the majority of T. marneffei infections in immunocompromised hosts have been found in HIV-infected patients, the incidence of penicilliosis marneffei has recently increased among non-HIV patients with other underlying diseases such as diabetes, cancer or systemic lupus erythematosus as putative susceptibility conditions.Citation5 T. marneffei is a member of an evolutionary diverse group of fungi that are thermally dimorphic.Citation6 In its environmental form, the fungus grows as saprophytic mycelium that produce the infective propagules (conidia). Conidia are inhaled into the host's lungs where they subsequently transform into yeast cells that replicate by binary fission.Citation7 The initial interactions of the T. marneffei conidia with the host phagocytic cells and the degree of activation of the host's innate immune responses to the fungus are critical parameters determining the host's ability to control disease.Citation8 Macrophages appear to be the initial immune cells that phagocytose conidia and secrete the pro-inflammatory cytokines responsible for inducing responses that subsequently arrest infection.Citation7,9 Phagocytosis of conidia and subsequent maturation of the phagosome exposes the fungal cells to an oxidative burst, acidic condition (pH 4–5),Citation10 enzymatic attack and nutrient-limitation, all of which are critical mechanisms for the macrophage to kill ingested microbes.Citation11 T. marneffei yeast cells are also susceptible to being killed by human neutrophils stimulated with granulocyte macrophage colony-stimulating factor (GM-CSF).Citation12 In penicilliosis marneffei patients, phagocytosed conidia can germinate into yeast cells that reside as intracellular parasites within macrophages.Citation13 Previous studies have demonstrated that some fungal laccases are upregulated during H2O2-mediated oxidative stress and nutrient starvation, especially glucose limitation, in order to support fungal growth and stress defense.Citation14-17
To understand the pathogenesis of T. marneffei, studies of the fungal determinants associated with virulence are needed. The putative virulence attributes of T. marneffei include adhesion,Citation18,19 dimorphism,Citation20-22 production of melanin or melanin-like substanceCitation23,24 and metabolic factors such as the glyoxylate cycle.Citation25 Laccases have been previously linked to fungal development, morphogenesis, detoxification processes, stress resistance, pigment formation, nutrient acquisition and pathogenicity.Citation26-28 Laccase is an important virulence factor in many pathogenic microorganisms including fungi and has been best characterized in Cryptococcus neoformans.Citation15,29 Expression of fungal laccase has been shown to be induced by environmental stress such as oxidative stress, acidic environments, and nutrient deprivation.Citation14,28 A laccase-like activity has been detected in protein extracts of dimorphic fungi including T. marneffei,Citation23 Paracoccidioides brasiliensis,Citation30 and Histoplasma capsulatum,Citation31 and laccase has been implicated in the enzymatic synthesis of melanin in these fungi.
In this study we sought to understand the role of the various laccases encoded by T. marneffei and determine how these laccases affect vegetative growth and H2O2-mediated oxidative stress. We have previously shown that the pbrB gene of T. marneffei encodes a laccase specifically required for pigment formation in conidia.Citation32 Three additional laccase-encoding genes of interest were investigated for their expression and roles in stress response and during growth in host cells. Gene deletion strains were generated that either lacked a single gene or combinations of genes. Notably, we only identified defects in the quadruple mutant that lacked all 4 laccase encoding genes (lacA, lacB, lacC and pbrB) suggesting significant redundancy, despite the fact that these genes show differential expression patterns. Conidiation in the ΔlacA ΔlacB ΔlacC ΔpbrB strain was delayed and the conidia were more susceptible to oxidative stress (H2O2), cell wall stress (SDS) and antifungal agents such as itraconazole, fluconazole and clotrimazole compared to wild-type and non-quadruple mutant conidia. Examination of the role of these laccases altered the fate of T. marneffei conidia in interactions with the THP-1 cells, a human monocytic cell line. The results demonstrate that laccase production significantly affects phagocytosis and intracellular survival of conidia in THP-1 cells as well as alters the release of cytokines by these host effector cells. Hence, we provide strong evidence for the role of laccases in the host immune response to T. marneffei.
Results
Talaromyces marneffei laccases are upregulated during oxidative stress at 37°C
T. marneffei produced and secreted laccase during vegetative growth at 28°C and 37°C (). We assessed whether oxidative stress and/or acidic condition could induce T. marneffei laccase expression by measuring enzymatic levels of cytoplasmic laccases. Cytoplasmic laccase activity was increased significantly (p < 0.01) after T. marneffei encountered with acidic (pH 5) or H2O2-mediated oxidative stress (1 mM H2O2 at pH 5; phagolysosome-like condition)Citation10 conditions (). However, the addition of glucose (5%) to the acidic and oxidative stress treatments reduced the level of T. marneffei laccase, particularly when coupled with oxidative stress conditions (p < 0.01). These data are consistent with the hypothesis that T. marneffei laccases are expressed in phagolysosomes of macrophages.
Figure 1. Expression of lac genes in response to oxidative stress in T. marneffei. A) Expression of lacA and lacC in 3-day-old yeast cells at 37°C before H2O2 treatment (0 min) and lacA and lacB during oxidative stress at 30 minute incubation (H2O2 30 min). The quantity of RNA sample used to amplified lac transcripts at 30 minutes was 2-fold lower than untreated sample and amplification of lac at 28°C. B) Transcripts of lacA and lacC presented in 3 day old hyphae at 28°C. C) Amplification of 18S RNA transcript as a control.
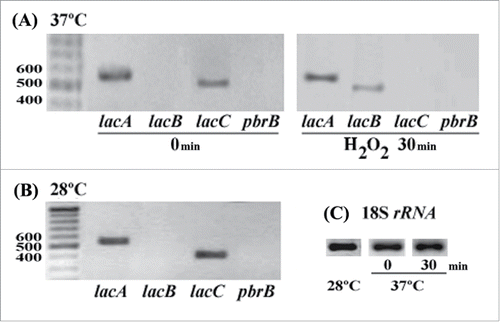
Table 1. Measurement of laccase activity in normal and stress conditions.
In this study, we focused on the expressions of 3 genes (lacA; PMAA072680, lacB; PMAA085520, and lacC; PMAA055370) because of their similarityCitation32 with Cryptococcus neoformans lac1; a defined virulence factor.Citation15 Amplification of lac transcripts in RNA extracts indicated different patterns of lac expression during growth under standard and oxidative stress conditions. During vegetative growth, lacA and lacC mRNA was readily detectable while lacB expression was not detected at either temperature. At 37°C lacA expression was about 2-fold higher than that of lacC but there was no difference at 28°C (, C). When T. marneffei was subjected to exogenous H2O2 for 30 minutes, lacB expression was induced. An induction of lacA and lacB due to exogenous H2O2 was verified by decreasing cDNA template (2-fold lower). LacA, but not lacC, transcripts were readily detectable. Also noteworthy was the expression of the lacB mRNA under these conditions. Upregulation of lacA and lacB was consistent with accumulation of LacA::GFP and LacB::GFP fusion proteins within fungal cells found during H2O2-mediated oxidative stress at 37°C (data not shown).
T.marneffei laccases are essential for cell integrity and stress resistance
To assess the role of these laccases, deletion strains for the lacA, lacB and lacC genes were generated. Levels of cytoplasmic laccase activity were significantly decreased when compared with wild-type (p < 0.01) () whereas secreted laccase activity was not detected in ΔlacA and ΔlacC mutants at 28°C (Fig. S1) and 37°C (data not shown). In vitro growth testing of these strains during hyphal growth at 28°C and yeast growth at 37°C under various conditions including H2O2 oxidative stress failed to detect any discernible differences from the wild-type control (data not shown). Since we found that lacA and lacB were expressed during H2O2-mediated oxidative stress at 37°C and lacA and lacC were expressed at both 28°C and 37°C under standard growth conditions, we generated ΔlacA ΔlacB double mutant and ΔlacA ΔlacB ΔlacC triple mutant strains to assess its sensitivity to stress. Growth testing of these strains at 28°C showed no discernible differences on standard medium (). Conidia from 2 independent colonies of each deletion strain were tested under H2O2-mediated oxidative stress at acidic condition, sodium dodecyl sulfate (SDS)-mediated cell wall stress and antifungal agent-mediated growth inhibition (see Materials and Methods). Each stressor was added to BHI medium where higher level of laccase activity was detected in the wild-type (data not shown). None of these deletion strains showed any detectable difference compared to the wild-type ().
Figure 2. Stress susceptibility tests showing stress sensitive phenotype of ΔlacA ΔlacB ΔlacC ΔpbrB transformant. Ten-fold serial dilutions of conidia were pipetted onto BHI agar containing 2.1 mM H2O2 (oxidative stress), 20 µg/ml SDS (cell wall stress), or antifungal agents (0.1 µg/ml clotrimazole, 0.04 µg/ml itraconazole, and 40 µg/ml fluconazole). Plates were placed in 37°C incubator for 1 week. Growth of the ΔlacA ΔlacB ΔlacC ΔpbrB strain was only evident in spots containing the higher concentrations of conidia showing that this strain is more sensitive to certain stressors compared with wild-type G681 and other mutants.
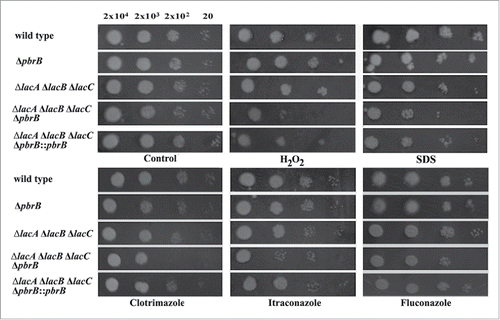
Table 2. Cytoplasmic laccase activities in the wild-type and mutants.
Table 3. Growth rate of transformants.
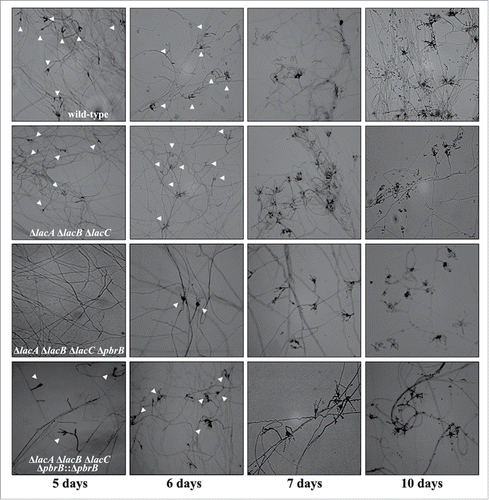
Conidia of triple deletion exhibited green color indicating the presence of DHN-melanin. We hypothesized that alteration of DHN-melanin synthesis in a ΔlacA ΔlacB ΔlacC background may increase the sensitivity of conidia to oxidative stress. To test this, we produced ΔlacA ΔlacB ΔlacC ΔpbrB quadruple deletion transformants. Macroscopic examination of ΔlacA ΔlacB ΔlacC ΔpbrB transformants resembled that of the other deletion mutant transformants and wild-type except for conidial coloration at 28°C (). In addition, it appeared that conidial production was reduced in the ΔlacA ΔlacB ΔlacC ΔpbrB strains (data not shown). To ensure these phenotypic observations were due to loss of pbrB in the triple lac deletion background, we generated a complemented strain in which the pbrB gene was reintroduced into the ΔlacA ΔlacB ΔlacC ΔpbrB transformant at the native locus. The resulting transformants (ΔlacA ΔlacB ΔlacC ΔpbrB::pbrB) produced green conidia similar to ΔlacA ΔlacB ΔlacC mutant and wild-type (). The conidial phenotype of the quadruple mutant could be due to a reduced growth rate, frequency of conidiophores or rate of conidial production on conidiophores. The radial growth rate data revealed that all of the strains had equivalent growth rates (). Colonies growing on the slides were observed microscopically for defects in conidiation and conidia production for 5, 6, 7, and 10 days at 28°C (). The conidiophores of all mutant strains were morphologically similar to those of the wild-type with respect to the various cell types and their abundance. In contrast, the density of conidiophores was greatly reduced in the ΔlacA ΔlacB ΔlacC ΔpbrB mutants, especially at the early time points. This suggested that conidiation of ΔlacA ΔlacB ΔlacC ΔpbrB transformant is delayed. Besides, we found that conidia of quadruple mutant (ΔlacA ΔlacB ΔlacC ΔpbrB) are more sensitive to various stressors. When compared with the other mutants, it was clear that the ΔlacA ΔlacB ΔlacC ΔpbrB mutant was more sensitive to H2O2, SDS and antifungal agents, including clotrimazole, itraconazole, and fluconazole (). Increasing of sensitivity to cell wall stressor (SDS) suggests that cell wall integrity may be affected. This is consistent with observations that show melanins are deposited at the cell wall and increase cell integrity, thus protecting fungal cells against various stressors.Citation37,38 Laccase activity in cytoplasmic extracts of mutants grown in BHI broth at 37°C for 3 days was determined and this showed a significant decrease in activity for the ΔlacA ΔlacB ΔlacC ΔpbrB strain compared to wild-type, single gene deletion and double gene deletion strains (p < 0.05) (). Statistic differences were shown significantly (p < 0.05) when compared between laccase activity of each mutant with wild-type or ΔpbrB mutant. Among triple mutant, quadruple mutant and pbrB-complemented quadruple mutant, their cytoplasmic laccase activities in 37°C cultures were not different (p > 0.05). ΔlacA ΔlacB ΔlacC ΔpbrB mutant could produce melanins detected by using anti-melanin monoclonal antibodyCitation23 developed for T. marneffei and confirmed by melanin extraction.Citation23
Figure 3. Phenotype of transformant colonies. Wild-type G681, ΔpbrB, ΔlacA ΔlacB ΔlacC ΔpbrB, ΔlacA ΔlacB ΔlacC, and ΔlacA ΔlacB ΔlacC ΔpbrB::pbrB cultured on ANM agar at 28°C for 2 weeks (A), and on BHI agar at 37°C for 5 days (B). Gel electrophoresis of lac amplification using genomic DNA of wild-type G681, ΔlacA ΔpbrB ΔlacC ΔpbrB, ΔlacA ΔpbrB ΔlacC ΔpbrB::pbrB strains as PCR templates.
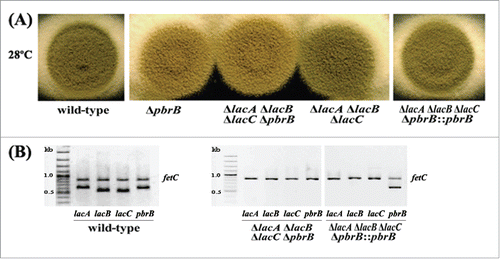
Figure 4. Representation of differences in density of conidiophores observed on slide cultures. Conidia of wild-type G681, ΔpbrB, ΔlacA ΔpbrB, ΔlacA ΔpbrB ΔlacC, ΔlacA ΔpbrB ΔlacC ΔpbrB, ΔlacA ΔpbrB ΔlacC ΔpbrB::pbrB were cultured on ANM agar and incubated in a moist chamber at 28°C for 5, 6, 7, and 10 days. Unlike other strains, ΔlacA ΔpbrB ΔlacC ΔpbrB produces very few conidiophores on day 5 to 6. Slide cultures were examined under the microscope for conidiophores. Images shown are at 100X magnification. White triangles point to conidiophores. The data represents the results of 2 independent experiments performed in duplicate.
Phagocytosis assay
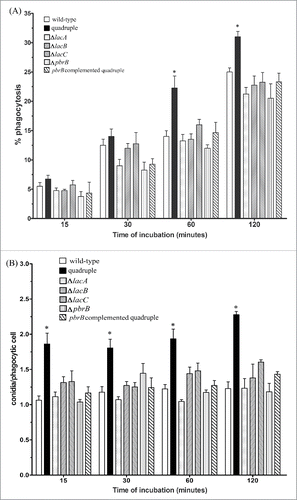
To assess whether any of these laccase-encoding genes play a role in the interaction of T. marneffei with host cells, the frequency with which T. marneffei wild-type, laccase mutant conidia and pbrB-complemented quadruple mutant are phagocytosed by THP-1 cells was measured by counting the number of conidia ingested over time. The percentages of conidia ingested increased over time for all strains examined (). For the early time points at 15 and 30 minutes post-inoculation, the percentages were similar (approximately 5–6 %) for all strains. However, at the later time points of 60 and 120 minutes post-inoculation there were significant differences between the quadruple lac gene deletion strain and the other strains. There were no significant differences in the wild-type conidia, the single lac gene mutants as well as the pbrB-complemented quadruple mutant at any of the intervals examined. Interestingly, the phagocytic index for each of the strains remained fairly constant over the different time intervals assessed (). The phagocytic index for the quadruple lac gene deletion (ΔlacA ΔlacB ΔlacC ΔpbrB) strain was significantly higher than the other strains at each time interval (p < 0.05). Phagocytosis rates of the pbrB-complemented quadruple mutant were similar to those of the wild-type and the single lac gene mutants at each time interval.
Figure 5. THP-1 macrophages more effectively ingested T. marneffei quadruple laccase mutant conidia. The percentages of phagocytosis (A) and phagocytic index (B) of THP-1 macrophages co-cultured with T. marneffei conidia from wild-type, quadruple lac gene deletion strain, the single gene disruptants, ΔlacA, ΔlacB, ΔlacC, ΔpbrB, and the pbrB complemented quadruple mutant at 15, 30, 60 and 120 min. Each bar represents the mean ± SEM of 3 sets of experiments, each performed in duplicate. The * denotes a p < 0.05 when comparing the wild-type and quadruple lac gene disruptant conidia.
Killing assay
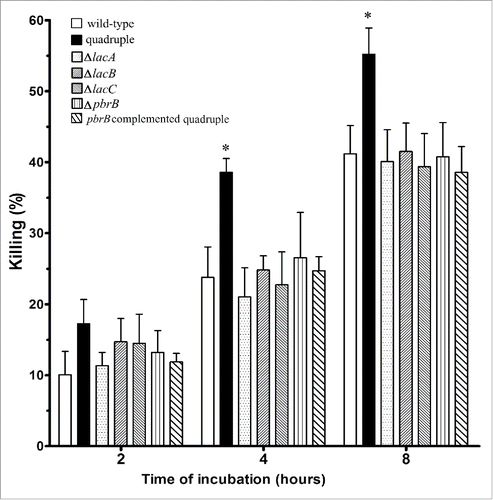
To compare the percentages of intracellular killing of T. marneffei wild-type and laccase mutant strains by THP-1 cells, CFUs were determined at different time intervals post-infection (). As early as 2 hours after inoculation, T. marneffei CFUs for the quadruple mutant dropped compared to conidia of the other strains, although the difference at this early time interval was not significant. This suggested that THP-1 cells killed quadruple lac gene mutant conidia more readily. At 4 and 8 hours, the THP-1 macrophages killed significantly more of the quadruple lac gene deletion conidia compared to any other strains. The percentages of single lac gene deletion strains and the reconstituted strain killed at each time interval were similar to the wild-type.
Figure 6. THP-1 macrophages kill T. marneffei quadruple laccase mutant conidia more effectively. The percentages of killing of conidia from T. marneffei wild-type, quadruple lac gene deletion strain, the single gene disruptants, ΔlacA, ΔlacB, ΔlacC, ΔpbrB, and the pbrB complemented quadruple mutant at 2, 4 and 8 hours. Each bar represents the mean ± SEM of 3 sets of experiments, each performed in duplicate. The * denotes a p < 0.05 when comparing the wild-type and quadruple lac gene disruptant conidia.
Cytokine response to wild-type and laccase deleted mutants of T. marneffei conidia
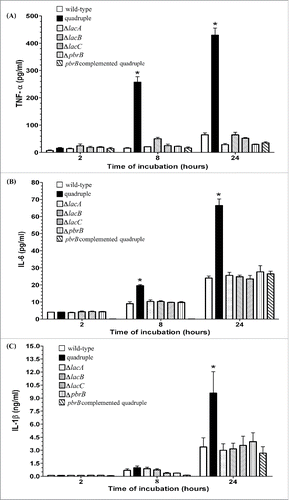
To determine the role of laccases on cytokine production, we examined the capacity of the different mutant conidia to induce TNF-α, IL-6 and IL-1β after incubation with THP-1 cells. The quadruple lac gene deletion strain elicited a significantly higher release of TNF-α as compared to the wild-type, any single lac gene deletion strains and the reconstituted strain after 8 and 24 hours of incubation (). Similarly, the quadruple lac gene deletion strain elicited a significantly stronger IL-6 and IL-1β response at 8 and 24 hours of incubation (, 7C). There were no significant differences in the production of these cytokines when comparing the wild-type to the single lac gene mutants as well as the pbrB-complemented quadruple mutant.
Figure 7. T. marneffei quadruple laccase mutant conidia induced significantly more proinflammatory cytokine production by THP-1 macrophages. A) Tumor necrosis factor α (TNF-α, B) interleukin-6 (IL-6), and C) interleukin-1β (IL-1β) levels in THP-1 cells co-cultured with conidia from T. marneffei wild-type, quadruple lac gene disruptant, the single gene disruptants, ΔlacA, ΔlacB, ΔlacC, ΔpbrB, and the pbrB complemented quadruple mutant. Measurements of TNF-α, IL-6, and IL-1β were achieved using supernatants pooled from 3 sets of experiments and expressed as mean ± SEM. The * denotes a p < 0.05 in TNF-α, IL-6 and IL-1β production between wild-type and quadruple lac gene disruptant conidia.
Discussion
Laccase-encoding genes are widely distributed in the fungal kingdom, especially among the ascomycete and basidiomycete species. Fungal laccases function in various metabolic processes including nutrient acquisition, growth and development, and protection against life-threatening stressors.Citation28,37,38 The presence of paralogous laccase-encoding genes, which presumably originated from gene duplication events, may be to fulfill a variety of targeted functions during the life cycle and to respond to environmental conditions. Multiplicity of laccase genes allows broader substrate recognition which confers an advantage in the competition for space and nutrients.Citation39,40 Expression of the various laccases ranges from constitutive to inducible triggered by exogenous regulators such as pH, H2O2, temperature, a variety of aromatic compounds related to lignin or its derivatives, metal ions (e.g. copper and iron) and concentrations of carbon (e.g., glucose and sucrose) and nitrogen sources.Citation14,16,28 The mechanism by which laccases scavenge H2O2 is not clearly understood; however, the catalyzed product of this enzyme, melanin, is described as a scavenger of oxidative radicals. Melanin acts as a free radical trap and stabilizes harmful reactive oxygen species.Citation37,41 The expression of laccase acting indirectly to scavenge intracellular H2O2 and protect cells from lipid oxidative damage has been demonstrated in the heterologous expression system. Trametes sp. 5930 lac gene expressed in Pichia pastoris responds to exogenous H2O2 and the latter enhances transcriptional gene products of the glutathione-dependent antioxidative system of P. pastoris.
T. marneffei genome possesses a family of laccase-encoding gene based on sequence similarity. The genetic redundancy of lac genes may be to provide the robustness of biological functions. According to the previous report, LacA and LacC are classified into ascomycete laccase clade I together with uncharacterized T. marneffei laccases (PMAA100410, PMAA050860 and PMAA008350).Citation32 PbrB laccase is in ascomycete laccase clade II involving in conidial pigment synthesis. LacB is quite similar to ferroxidases than laccase clades but LacB is not grouped into ascomycete laccases nor ferroxidases. Similarity search of lacB yielded 78% sequence homology to C. neoformans lac1. LacB might be ferroxidase-laccase enzyme. Analysis of subcellular localizations by ProtComp 9.0 (http://linux1.softberry.com/berry.html) suggested that LacA and LacC are membrane bound or secreted enzymes while LacB localizes at plasma membrane. The transcripts of lacA and lacC were presented in hyphal (28°C) and yeast (37°C) cells and induction of lacA and lacB expression was evident during H2O2-mediated oxidative stress. Since the localization of fungal laccases associate to physiological functions,Citation28 the lacA and lacC expressions are possibly to support growth. The low level of lacA and lacC transcripts could be from the use of brain heart infusion (an enrichment media) as a culture medium. In contrast, extracellular of ABTS catalytic activity depletes in mutants with lacA or lacC deletions (see Fig. S1) suggesting their co-function to oxidize extracellular substance. When fungal cells encountered with exogenous H2O2, transcripts of lacA and lacB were increased. LacA is expected to be secreted while LacB is at plasma membrane. These data suggest that LacA and LacB may be involved in extracellular and/or intracellular oxidative stress defense. The presence of PbrB is found in asexual development structures and this laccase participates in DHN melanin production during asexual development.Citation32 The melanin product provides fitness and a nonspecific protection against various stressors.Citation26-29 The different expression patterns suggest the T. marneffei lac gene redundancy with divergent function. Glucose-repressible expression has been demonstrated in fungi such as Cryptococcus neoformans,Citation17 Cerrena unicolorCitation33 and Trametes versicolor.Citation34 Especially in C. neoformans, a virulence factor lac1 expression is extremely sensitive to glucose. Similarly, T. marneffei laccase activity increased in both acidic and H2O2-treated conditions, whereas it decreased in the presence of high glucose (5%).
Generation of T. marneffei lac mutants via target gene deletion reveals the distinct roles of laccases but their functions involve in the fitness of the airborne infectious propagules. Deletion of lac genes expressed at 37°C and during oxidative stress did not affect stress resistance, growth or development. However when coupled with a pbrB gene deletion, encoding a laccase required for DHN-melanin production in conidia, conidiation in this quadruple mutant (ΔlacA ΔlacB ΔlacC ΔpbrB) was delayed. This delay of conidiation occurs only in quadruple mutant suggesting their necessary functions during asexual development. Since lacA and lacC express at 28°C whereas lacB respond to oxidative stress, it is possible that these genes participate in conidiation by manipulate stressors occurred during melanin synthesis, melanin deposition and/or involve in morphogenesis of conidial cell wall. Even though laccase activities of single, double and triple lac deletions were lower than wild-type significantly (p < 0.05), these strains were not stress sensitive unlike the quadruple mutant. Since conidia were used to test stress susceptibility and pbrB is expressed during conidiation, the data reflect lacA, lacB and/or lacC expression during asexual development and their functions contribute to conidial fitness. These explanations need further study about conidial cell wall architecture of quadruple mutant. Moreover, not only was conidiation affected but an increasing sensitivity to stress conditions such as H2O2-mediated oxidative stress, SDS-mediated cell wall stress, and antifungal agents (e.g. clotrimazole, itraconazole, and fluconazole) was also evident. These findings support the hypothesis that laccases are required in conidia to tolerate various kinds of stressors.
In summary, the ΔlacA ΔlacB ΔlacC ΔpbrB mutant is stress sensitive due to the cumulative loss of laccase activities in vegetative cells that can in part be masked by the conidial laccase encoded by pbrB. Loss of pbrB in the background of the 3 lac mutants uncovers the stress resistance phenotype, demonstrating partial redundancy due to overlapping spatial and temporal expression patterns of these 4 laccase-encoding genes in T. marneffei.
T. marneffei laccases also play a role in the host-pathogen interaction. Laccases have been associated with virulence in many fungal pathogens and the varied activities of the enzyme have been well documented in C. neoformans.Citation42-45 Laccase itself directly protects C. neoformans from the antifungal activity of macrophages by functioning as an iron scavenger during infection, which is a distinctly different role from the key role in melanin biosynthesis.Citation38,42 As a defense against host immune cells, laccase is believed to oxidize iron, which decreases the production of hydroxyl radicals in alveolar macrophages.Citation42,46 Since host effector cells first interact with inhaled resting conidia, we have begun to examine the roles of laccases in the pathobiology of T. marneffei. To investigate the different activities of laccases in T. marneffei we compared the wild-type to single and compound lac gene mutants. Laccase activity was significantly reduced in the quadruple lac gene deletion strain compared to the wild-type and the single lac gene deletion mutants. Despite deletion of 4 lac genes in the quadruple mutant, residual activity on the 2,6 DMP substrate used in the assay was still evident. This may be bona fide laccase activity from one of the other more diverged laccase-like genes in but may equally be the result of other enzymes such as peroxidases.Citation47 The comparisons of the phagocytic and killing activities of THP-1 macrophage cells against conidia from the T. marneffei quadruple lac gene deletion strain and the wild-type strain revealed clear differences in these processes. The phagocytic index was significantly higher in the quadruple mutant as early as 15 min after engaging THP-1 cells and the phagocytosis percentages notably different by 60 min. Moreover, we determined that there were significant differences in killing between the quadruple deletion strain and the other strains by 4 and 8 hours of co-incubation, with the quadruple deletion strain showing increased susceptibility to the fungicidal responses of the THP-1 cells. Hence, it seems that laccases protect the fungal cells from the cytotoxic interactions with THP-1 cells. This result is consistent with observations that showed that infection of mice with a laccase-deficient strain of C. neoformans resulted in significantly lower pulmonary fungal burden in comparison to mice infected with wild-type yeast cells.Citation43 In summary, our data support a role for T. marneffei laccases in the protection of conidia against phagocytic cells early after infection.
Interestingly, the data showed that a single deletion of a lac gene was not sufficient to alter the characteristics assessed in our assays. The T. marneffei quadruple lac gene deletion strain was more sensitive to oxidative stress (H2O2), cell wall stress (SDS), and antifungal agents compared to the wild-type, whereas strains with single, double and triple lac gene deletions behaved similar to the wild-type. The quadruple mutant was also more susceptible to antifungal activity of THP-1 cells. Similarly, a study of LAC genes in C. neoformans revealed that deletion of both lacA and lacB was required to alter susceptibility to H2O2 or nitric oxide.Citation44 In addition, the deletion of both laccases reduced the survival of C. neoformans in primary macrophages.
A recent study in C. neoformans isolates from patients found a significant positive correlation between laccase activity with ex vivo survival in cerebrospinal fluid (CSF) macrophages co-cultures and the in vivo rate of fungal clearance.Citation48 Interestingly, higher laccase activity in C. neoformans enhanced survival ex vivo and was correlated with increased resistance to clearance following antifungal treatment in patients. Our data is in accord with these findings, as the wild-type T. marneffei conidia were significantly more resistant to phagocytosis and killing that the quadruple lac mutant, which suggests that laccase may play a prominent role in vivo.
The comparison of the inflammatory responses to the laccase quadruple mutant compared to the wild-type revealed significant differences in pro-inflammatory cytokine responses by the THP-1 cells. Consistent with this finding, the levels of TNF-α in mice infected with a C. neoformans laccase-deficient strain was significantly increased compared with TNF-α levels in mice infected with wild-type C. neoformans.Citation45 Similarly, albino (alb1, conidial polyketide synthase deficient) A. fumigatus conidia induced significantly more IL-6, TNF-α and IL-10 in PBMC compared to melanized wild-type conidia.Citation49 In principal, increased levels of pro-inflammatory cytokines like TNF-α, IL-6 and IFN-γ have been related to an efficient response to A. fumigatus infection, through induction of potent antifungal cellular responses to clear the pathogen.Citation50 A less robust host pro-inflammatory response, as seen with the T. marneffei wild-type conidia in comparison to that produced with the quadruple disruptant, may obstruct the effectiveness of the antifungal defense mechanisms, especially in an immunocompromised host. To our knowledge, this is the first study showing that conidial laccases of T. marneffei modulate the pro-inflammatory cytokine response.
Laccase is a critical enzyme in melanin biosynthesis and we have previously shown that pbrB mutants have a defect in conidial pigmentation.Citation32 This phenotype was not exacerbated by the quadruple mutants suggesting that the pbrB gene is specific for conidial pigmentation in T. marneffei and the other genes are not redundant for this role. As only the quadruple mutant displayed significantly increased susceptibility to innate host defenses, it is clear that each of these genes and their specific expression patterns play an overlapping but coordinated role in T. marneffei. Similarly, in A. fumigatus, mutation of the abr2 gene encoding a conidial laccase resulted in poor conidial pigmentation, but this deletion did not alter virulence or impact the susceptibility of the mutant to ROS or diamide compared to the wild-type.Citation51-53
In conclusion, these studies reveal that T. marneffei has a number of laccase-encoding genes that produce laccases with both specific and redundant activities during normal growth condition and under stress conditions. Low level of lacA and lacC transcripts were present in both hyphal (28°C) and yeast (37°C) vegetative cell types. Expression of lacA and lacB is upregulated in response to oxidative stress at 37°C during yeast cell growth. Deletion of lacA, lacB, lacC, and pbrB genes not only altered DHN-melanin biosynthesis during asexual development, but delayed conidiation as well. The ΔlacA ΔlacB ΔlacC ΔpbrB mutants were sensitive to various stressors including H2O2, SDS, and antifungal agents (e.g., clotrimazole, itraconazole, and fluconazole). The results indicate that laccases play a role in protecting T. marneffei conidia against phagocytic cells such as THP-1. In addition, this study identified a laccase-dependent effect during the innate phase of initial engagement of the immune system, suggesting that laccase contribute to virulence by promoting T. marneffei resistance to macrophage killing, through a reduction in phagocytosis and intracellular death as well as modulating the cytokine milieu. Further studies are required in order to elucidate how laccases are capable of protecting T. marneffei against these host responses. Analysis of the role of this group of enzymes may clarify the pathogenicity mechanisms used by this fungal pathogen and to validate laccase and/or melanization as a target for combating the disease.
Materials and methods
Fungal strains and media. Experiments were conducted using T. marneffei strain F4 (CBS no. 119456), wild-type G681 (ΔpkuA::pyrG+) and lac deletion strains (ΔlacA::pyrG+, ΔlacB::pyrG+, ΔlacC::MT1612 pyrG+, ΔpbrB::pyrG+32, ΔlacA ΔlacB::pyrG+, ΔlacC ΔlacB::pyrG+, ΔlacA ΔlacB::pyrG+ ΔlacC, ΔlacA ΔlacB ΔlacC ΔpbrB::pyrG+ and ΔlacA ΔlacB ΔlacC ΔpbrB::pbrB pyrG+ (pbrB complemented strain). G681 is a derivative of the type strain FRR2161.Citation54 All transformants and G681 strain were generated from G526 (Δpku pyrG−) stain which kuA is deleted to inhibit non-homologous DNA end joining repair. All transformants were grown at 28°C on Aspergillus nidulans synthetic medium (ANM).Citation55 After incubation for 10 to 14 days on ANM, the conidia were harvested by suspension in PBS pH 7.2 containing 0.01% Tween 80. The conidial suspension was then filtered through sterile glass wool (Corning) where fungal hyphae were trapped on the surface while conidia pass through the glass wool. The eluate containing mycelium-free conidia was centrifuged for 15 minutes at 4,000 g and then the pellet was washed twice in PBS pH 7.2. The conidial suspension, at a concentration of 107 conidia/ml was used for the experiments.
Measurement of laccase activity. Screening of laccase activity was done on plates containing minimal medium (MM) agar pH 5.5 (0.27% (w/v) glucose, 10 mM MgSO4, 29.4 mM KH2PO4, 13 mM glycine, vitamin B1 3 µM and 20 mM 2,2′-azino-bis (3-ethylbenzthiazoline-6-sulfonic acid) (ABTS, Fluka, Germany) spread on top of agar). Conidial suspension (10Citation6) was pipetted onto MM-ABTS agar plates and incubated at 28°C or 37°C for 1 week. Rhus vernificera laccase (Sigma-Aldrich, UK) 10 unit was used as a positive control. Presence of Green color indicates laccase activity. To determine cytoplasmic laccase activity, T. marneffei grown in BHI (Difco, USA) broth at 37°C for 3 days were washed with cold PBS, collected by centrifugation and resuspended with cold yeast nitrogen base (YNB) broth pH 7 containing 0.1% w/v glucose. Turbidity of the cell suspension was measured at 600 nm and adjusted to a value of 10. The yeast cell suspension (1 ml) was pipetted into individual treatment tubes with 19 ml YNB broth containing 0.1% w/v glucose and prepared in triplicate. Treatment conditions included pH 7, pH 5 adjusted by 1M HCl, pH 5 with 5% glucose, 1 mM H2O2 pH 5 and 1 mM H2O2 pH 5 with 5% glucose. After incubation at 37°C in a shaking incubator (150 rpm) for 1 hour, cells were washed with cold PBS and cytoplasmic protein was extracted by bead beating. The extract was examined by SDS-PAGE and proteins quantified by Bradford dye-binding assay (Biorad, USA). Protein extract (300 µg) was mixed with the laccase substrate (10 mM 2,6-dimethoxyphenol (DMP) (Sigma-Aldrich, UK) in 0.1 M Na2HPO4/citric acid buffer at pH 5) and oxidized product displaying a yellow to brown color was measured at 468 nm.Citation56,57 Mean of absorbance, standard error of the mean and p-value were calculated by online t test calculator (www.graphpad.com).
Expression of lac genes
To examine the expression of the laccase-encoding genes, T. marneffei F4 cells were grown in BHI at 28°C and 37°C for 3 days, harvested and washed with cold PBS before extracting RNA. Yeast cells from the 37°C culture were treated with 1 mM H2O2 at pH 5. At 0, 30 and 60 minutes of incubation, treated cells were washed with cold PBS before performing RNA extraction. RNA was extracted from each sample using Nucleospin Extract II kit. The extracted RNA samples were checked for gDNA contamination by PCR before performing cDNA synthesis using 200 ng (28°C and 0 min at 37°C samples) or 100 ng (H2O2 treatment for 30 minutes) of RNA (Omniscript RT kit). As a control for RT-PCR, cDNA of 18s RNA transcript was amplified from 50 ng RNA using Pm1/Pm2 primers.Citation58 RT-PCR was performed in duplicate. Primers used to amplify lac transcripts are shown in Table S1.
Generation of transformants
The lac genes containing the 5′ and 3′ untranslated regions were amplified from genomic DNA of the T. marneffei G681 strain (ΔpkuA::pyrG+) using primers listed in Table S1. PCR-amplified lac fragments were ligated into pGEM-T Easy (Promega) to generate the constructs pASlac19 (lacA), pASlac42 (lacB) and pASlac7 (lacC). To generate the deletion constructs, pASlac19 was digested with EcoRV/XhoI to remove lacA sequences from +20 to +1,840 (relative to the ATG) and the non-coding flanking regions ligated to a SmaI/XhoI fragment containing the pyrG selectable marker cassette (pAB4342).Citation59 For lacB, pASlac42 was digested with EcoRV/ClaI to remove lacB sequences from −449 to +1,746 and the non-coding flanking regions ligated to a SmaI/ClaI fragment containing the pyrG selectable marker cassette. For lacC, pASlac7 was digested with HindIII/NruI to remove lacC sequences from −1,095 to +3,049 and the non-coding flanking regions ligated to a HindIII/EcoRV fragment containing glufosinate resistant cassette (pMT1612).Citation60 The deletion constructs were linearized by digestion with NotI and purified before transformation. DNA-mediated transformation of T. marneffei G526 strain (ΔpkuA pyrG−) was performed as described previously.Citation59 Transformants of lacA and lacB deletions were selected on medium without uracil and genomic DNA samples were extracted from 2 isolated colonies to check the replacement of target gene by transformation cassette. Deletion of lacC produced ΔlacC::MT1612 pyrG− uracil auxotrophic strain. To remove this auxotrophy, the ΔlacC::MT1612 pyrG− strain was transformed with the T. marneffei pyrG targeting vector gene (pLS7413)Citation61 to generate ΔlacC::MT1612 pyrG+ transformants. PCR and southern blot analysis of genomic DNA from each strain was performed to confirm homologous recombination and target gene loss. Amplification of T. marneffei fetC is an internal PCR control. Primers used to amplify each lac are described in Table S1.
Stress susceptibility test
To prepare conidial suspension, conidia were harvested from ANM plates incubated at 28°C for 2 weeks. Ten-fold serial dilutions of conidia were pipetted onto BHI (Difco, USA) medium pH 5.4 containing each kind of stressors such as 2.1 mM H2O2 (oxidative stressor), 20 µg/ml SDS (cell wall stressor) and antifungal agents (0.1 µg/ml clotrimazole, 0.04 µg/ml itraconazole, and 40 µg/ml fluconazole). Plates were incubated in 37°C incubator for 1 week before observation of transformant growth.
Growth assay
To assay growth rates, 2 independent colonies of each transformant were used to inoculate solid ANM agar medium at a density such that approximately 10 conidia were seeded per plate. The plates were incubated at 28°C and the diameter of 5 colonies was measured daily over a week. Statistical values (mean and SEM) and significance were calculated using the t test.
Melanin detection
Approximately 10Citation8 conidia of each mutant were inoculated into 200 ml BHI broth and incubated in 37°C shaking incubator for 5 days. Fungal cells were washed with PBS before performing melanin immunolabeling and extraction described in previous work.Citation23 Positive staining and particles left after extraction process indicate that melanin can be produced.
THP-1 infected with T. marneffei
The human monocytic cell line THP-1 (ATCC TIB-202) was cultured in RPMI 1640 medium (Gibco, USA) containing 10% (v/v) heat-inactivated FBS (Gibco). For the induction of cellular differentiation, cells (2 × 10Citation6 per ml) were seeded into 24-well culture plates (Costar, Corning, NY) in 1 ml of RPMI 1640 medium with 10% (v/v) FBS and 100 ng/ml phorbol myristate acetate (PMA) (Sigma, St. Louis, Mo) for 72 hours. After incubation, non-attached cells were removed by aspiration and the adherent cells were washed with RPMI 1640 3 times. THP-1 cells in RPMI 1640 without PMA were used as control (undifferentiated) cells. The conidia from wild-type or mutant strains suspended in RPMI 1640 medium with 10% (v/v) FBS were added to the THP-1 monolayers.
Phagocytosis assay
The phagocytosis assay was initiated by adding 4 × 10Citation6 conidia to 2 × 10Citation6 macrophage cells in each well (MOI = 2). THP-1 cells were allowed to interact with conidia for 15, 30, 60 and 120 minutes. At each time interval, supernatants were discarded, and the wells were washed gently 3 times with PBS pH 7.2 to remove unbound conidia. THP-1 were removed from the wells after treatment with 0.25% trypsin-EDTA (Gibco) for 5 min at 37°C and then washed twice to remove trypsin-EDTA. The cells were fixed by adding 0.5% paraformaldehyde in PBS (Sigma-Aldrich Gmbh, Germany). The phagocytosis was assessed by light microscopy, and the percentage of phagocytosis was the total number of macrophage cells from a 100 cell count that internalized fungal conidia. The phagocytic index was determined by calculating the average number of intracellular conidia per macrophage as follows:
Measurement of conidial survival in THP-1
THP-1 cells were infected with wild-type, mutant or complemented strains of T. marneffei conidia at an MOI=2 for 2 hours. Unbound conidia were removed by washing the wells 3 times with PBS/0.05% Tween 20 (PBS-T). Extracellular conidia were killed with 50 μg/ml of nystatin as described.Citation62 THP-1 cells were then supplemented with fresh media for an additional 2, 4 and 8 hours at 37°C prior to lysis by the addition of 1.0% Triton X-100 followed by serial dilutions of the released conidia and plating onto PDA medium and incubating at 25°C (3 replicate plates/well). The colony forming unit (CFU) of wild-type T. marneffei from cell lysates after 2 hours of phagocytosis was used to establish a baseline value (CFU control) for comparison with the CFUs at subsequent time intervals. The percentage of killing in CFU was calculated as follow:
Induction of cytokines in human monocyte cell line THP-1
To assess cytokine production, THP-1 cells were infected with wild-type,mutant or complemented strains of T. marneffei conidia at an MOI = 5 and incubated for 2, 8 or 24 hours at 37°C. After incubation, the supernatants were collected and kept at −20°C until the cytokine assays were performed. All of the experiments were repeated at least 3 times and each set was done in duplicate. TNF-α, IL-6 and IL-1β were measured by commercial ELISA kits (BioLegend, San Diego, CA), according to the manufacturer's instructions. The concentrations of cytokines in the experimental samples were calculated according to the optical densities at 450 nm obtained from wells containing cytokine standard. The measurable concentrations of TNF-α and IL-6 ranged from 7.8 to 500 pg/ml, while IL-1β ranged from 2.0 to 125 pg/ml.
Statistical analysis
All data are expressed as mean ± standard error of the mean (SEM) of the number of determinations carried out in triplicate for the percentage of phagocytosis, phagocytic index and percentage of killed CFU. Variables were tested for normality and then the different groups were compared using the One-Way ANOVA, where P < 0.05 was considered as statistically significant between the groups.
Differences in the cytokine production between wild-type and laccase mutants were analyzed by One-Way ANOVA. The level of significance was set at P < 0.05.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KVIR_S_1193275.zip
Download Zip (572.5 KB)Funding
This study was financially supported from the National Research University Project under Thailand's Office of the Higher Education Commission (SY), the Royal Golden Jubilee PhD Research Assistant Fellowship of the Thailand Research Fund (AS), the Research Fund of Faculty of Medicine at Chiang Mai University (AS) and the National Health and Medical Research Council of Australia (AA). JDN is supported in part by NIH AI52733.
References
- Drouhet E. Penicilliosis due to Penicillium marneffei: a new emerging systemic mycosis in AIDS patients travelling or living in Southeast Asia. Review of 44 cases reported in HIV infected patients during the last 5 years compared to 44 cases of non AIDS patients reported over 20 years. J Mycol Méd 1993; 4:195-224.
- Duong TA. Infection due to Penicillium marneffei, an emerging pathogen: review of 155 reported cases. Clin Infect Dis 1996; 23:125-130.
- Wong SY, Wong KF. Penicillium marneffei infection in AIDS. Patholog Res Int 2011; 2011:764293; PMID:21331327; http://dx.doi.org/10.4061/2011/764293
- Supparatpinyo K, Perriens J, Nelson KE, Sirisanthana T. A controlled trial of itraconazole to prevent relapse of Penicillium marneffei infection in patients infected with the human immunodeficiency virus. N Engl J Med 1998; 339:1739-1743; PMID:9845708; http://dx.doi.org/10.1056/NEJM199812103392403
- Kawila R, Chaiwarith R, Supparatpinyo K. Clinical and laboratory characteristics of penicilliosis marneffei among patients with and without HIV infection in Northern Thailand: a retrospective study. BMC Infect Dis 2013; 13:464; PMID:24094273; http://dx.doi.org/10.1186/1471-2334-13-464
- Sil A, Andrianopoulos A. Thermally dimorphic human fungal pathogens-polyphyletic pathogens with a convergent pathogenicity trait. Cold Spring Harb Perspect Med 2014; 5:a019794; PMID:25384771; http://dx.doi.org/10.1101/cshperspect.a019794
- Cogliati M, Roverselli A, Boelaert JR, Taramelli D, Lombardi L, Viviani MA. Development of an in vitro macrophage system to assess Penicillium marneffei growth and susceptibility to nitric oxide. Infect Immun 1997; 65:279-284; PMID:8975924
- Rongrungruang Y, Levitz SM. Interactions of Penicillium marneffei with human leukocytes in vitro. Infect Immun 1999; 67:4732-4736; PMID:10456924
- Roilides E, Lyman CA, Sein T, Petraitiene R, Walsh TJ. Macrophage colony-stimulating factor enhances phagocytosis and oxidative burst of mononuclear phagocytes against Penicillium marneffei conidia. FEMS Immunol Med Microbiol 2003; 36:19-26; PMID:12727361; http://dx.doi.org/10.1016/S0928-8244(03)00035
- Nyberg K, Johansson U, Johansson A, Camner P. Phagolysosomal pH in alveolar macrophage. Environ Health Perspect 1992; 97:149-152. PMID:1327733
- Becker KL, Ifrim DC, Quintin J, Netea MG, van de Veerdonk FL. Antifungal innate immunity: recognition and inflammatory networks. Semin Immunopathol 2015; 37(2):107-116; PMID:25527294; http://dx.doi.org/10.1007/s00281-014-0467-z
- Kudeken N, Kawakami K, Saito A. Mechanisms of the in vitro fungicidal effects of human neutrophils against Penicillium marneffei induced by granulocyte-macrophage colony-stimulating factor (GM-CSF). Clin Exp Immunol 2000; 119:472-478; PMID:10691919; http://dx.doi.org/10.1046/j.1365-2249.2000.01158
- Vanittanakom N, Cooper CR JR, Fisher MC, Sirisanthana T. Penicillium marneffei infection and recent advances in the epidemiology and molecular biology aspects. Clin Microbiol Rev 2006; 19:95-110; PMID:16418525
- Piscitelli A, Giardina P, Lettera V, Pezzella C, Sannia G, Faraco V. Induction and transcriptional regulation of laccases in fungi. Curr Genomics 2011; 12:104-112; PMID:21966248; http://dx.doi.org/10.2174/138920211795564331
- Zhu X, Williamson PR. Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res 2004; 5:1-10; PMID:15381117; http://dx.doi.org/ 10.1016/j.femsyr.2004.04.004
- Shraddha , Shekher R, Sehgal S, Kamthania M, Kumar A. Laccase: Microbial sources, production, purification, and potential biotechnological applications. Enzyme Res 2011; 2011:1-11; PMID:21755038; http://dx.doi.org/10.4061/2011/217861
- Waterman SR, Hacham M, Panepinto J, Hu G, Shin S, Williamson PR. Cell wall targeting of laccase of Cryptococcus neoformans during infection of mice. Infect Immun 2007; 75:714-722; PMID:17101662; http://dx.doi.org/10.1128/IAI.01351-06
- Hamilton AJ, Jeavons L, Youngchim S, Vanittanakom N, Hay RJ. Sialic acid-dependent recognition of laminin by Penicillium marneffei conidia. Infect Immun 1998; 66:6024-6026; PMID:9826390
- Hamilton AJ, Jeavons L, Youngchim S, Vanittanakom N. Recognition of fibronectin by Penicillium marneffei conidia via a sialic acid-dependent process and its relationship to the interaction between conidia and laminin. Infect Immun 1999; 67:5200-5205; PMID:10496896
- Sternberg S. The emerging fungal threat. Science 1994; 266:1632-1634; PMID:7702654; http://dx.doi.org/10.1126/science.7702654
- Borneman AR, Hynes MJ, Andrianopoulos A. The abaA homologue of Penicillium marneffei participates in two developmental programmes: conidiation and dimorphic growth. Mol Microbiol 2000; 38:1034-1047; PMID:11123677; http://dx.doi.org/ 10.1046/j.1365-2958.2000.02202
- Boyce KJ, Schreider L, Andrianopoulos A. In vivo yeast cell morphogenesis is regulated by a p21-activated kinase in the human pathogen Penicillium marneffei. PLoS Pathog 2009; 5:e1000678; PMID:19956672; http://dx.doi.org/ 10.1371/journal.ppat.1000678
- Youngchim S, Hay RJ, Hamilton AJ. Melanization of Penicillium marneffei in vitro and in vivo. Microbiol 2005; 151:291-299; PMID:15632446; http://dx.doi.org/ 10.1099/mic.0.27433-0
- Liu H, Wei L, Guo T, Tan W. Detection of DOPA-melanin in the dimorphic fungal pathogen Penicillium marneffei and its effect on macrophage phagocytosis in vitro. PLoS One 2014; 9:e92610; PMID:24647795; http://dx.doi.org/10.1371/journal.pone.0092610
- Cánovas D, Andrianopoulos A. Developmental regulation of the glyoxylate cycle in the human pathogen Penicillium marneffei. Mol Microbiol 2006; 62:1725-1738; PMID:17427290
- Thurston CF. The structure and function of fungal laccases. Microbiol 1994; 140:19-26. http://dx.doi.org/10.1099/13500872-140-1-19
- Nagai M, Kawata M, Watanabe H, Ogawa M, Saito K, Takesawa T, Kanda K, Sato T. Important role of fungal intracellular laccase for melanin synthesis: purification and characterization of an intracellular laccase from Lentinula edodes fruit bodies. Microbiol 2003; 149:2455-2462; PMID:12949171; http://dx.doi.org/10.1099/mic.0.26414-0
- Baldrian P. Fungal laccases-occurrence and properties. FEMS Microbiol Rev 2006; 30:215-242; PMID:16472305
- Williamson PR. Laccase and melanin in the pathogenesis of Cryptococcus neoformans. Front Biosci 1997; 2:e99-107; PMID:9342305
- Gómez, BL, Nosanchuk, JD, Díez S, Youngchim S, Aisen P, Cano LE, Restrepo A, Casadevall A, Hamilton AJ. Detection of melanin-like pigments in the dimorphic fungal pathogen Paracoccidioides brasiliensis in vitro and during infection. Infect Immun 2001; 69:5760-5767; PMID:11500453; http://dx.doi.1i.org/10.1128/IAI.69.9.5760-5767.2001
- Nosanchuk JD, Gomez BL, Youngchim S, Díez S, Aisen P, Zancopé-Oliveira RM, Restrepo A, Casadevall A, Hamilton AJ. Histoplasma capsulatum synthesizes melanin-like pigments in vitro and during mammalian infection. Infect Immun 2002; 70:5124-5131; PMID:12183562; http://dx.doi.org/10.1128/IAI.70.9.5124-5131
- Sapmak A, Boyce KJ, Andrianopoulos A, Vanittanakom N. The pbrB gene encodes a laccase required for DHN-melanin synthesis in conidia of Talaromyces (Penicillium) marneffei. PLoS One 2015; 10(4):e0122728; PMID:25866870; http://dx.doi.org/10.1371/journal.pone.0122728
- Antecka A, Bizukojc M, Ledakowicz S. The kinetic model of laccase biosynthesis by Cerrena unicolor. Chem Process Eng 2009; 30:403-416.
- Tavares APM, Coelho MAZ, Coutinho JAP, Xavier AMRB. Laccase improvement in submerged cultivation: induced production and kinetic modeling. J Chem Technol Biot 2005; 80:669-676. http://dx.doi.org/10.1002/jctb.1246
- Kim D, Kwak E, Choi HT. Increase of yeast survival under oxidative stress by the expression of the laccase gene from Coprinellus congregatus. J Microbiol 2006; 44:617-621; PMID:17205039
- Yang Y, Fan F, Zhuo R, Gong Y, Wan X, Jiang M, Zhang X. Expression of laccase gene from white rot fungus in Pichia pastoris can enhance the resistance of yeast to H2O2-mediated oxidative stress by stimulating the glutathione-based antioxidative system. Appl Environ Microbiol 2012; 78:1-14; PMID:22706050; http://dx.doi.org/10.1128/AEM.00218-12
- Eisenman HC, Casadevall A. Synthesis and assembly of fungal melanin. Appl Microbiol Biotechnol 2012; 93:931-940; PMID:22173481; http://dx.doi.org/10.1007/s00253-011-3777-2
- Jacobson ES. Pathogenic roles for fungal melanins. Clin Microbiol Rev 2000; 13:708-717; PMID:11023965; http://dx.doi.org/10.1128/CMR.13.4.708-717.2000
- Castilho FJD, Torres RA, Barbosa AM, Dekker RFH, Garcia JE. On the diversity of the laccase gene: a phylogenetic perspective from Botryosphaeria rhodina (Ascomycota: Fungi) and other related taxa. Biochem Genet 2009; 47:80-91; PMID:19160039; http://dx.doi.org/10.1007/s10528-008-9208-0
- Giardina P, Faraco V, Pezzella C, Piscitelli A, Vanhulle S, Sannia G. Laccases: a never-ending story. Cell Mol Life Sci 2010; 67:369-385; PMID:19844659; http://dx.doi. org/10.1007/s00018-009-0169-1
- Liu GY, Nizet V. Color me bad: microbial pigments as virulence factors. Trends Microbiol 2009; 17:406-413; PMID:19726196; http://dx.doi.org/10.1016/j.mpmed.2009.06.006
- Liu L, Tewari RP, Williamson PR. Laccase protects Cryptococcus neoformans from antifungal activity of alveolar macrophages. Infect Immun 1999; 67:6034-6039; PMID:10531264
- Mednick AJ, Nosanchuk JD, Casadevall A. Melanization of Cryptococcus neoformans affects lung inflammatory responses during cryptococcal infection. Infect Immun 2005; 73:2012-2019; PMID:15784542; http://dx.doi.org/10.1128/IAI.73.4.2012-2019.2005
- Missall TA, Moran JM, Corbett JA, Lodge JK. Distinct stress responses of two functional laccases in Cryptococcus neoformans are revealed in the absence of the thiol-specific antioxidant Tsa1. Eukaryot Cell 2005; 4:202-208; PMID:15643075; http://dx. doi.org/10.1128/EC.4.1.202-208.2005
- Qiu Y, Davis MJ, Dayrit JK, Hadd Z, Meister DL, Osterholzer JJ, Williamson PR, Olszewski MA. Immune modulation mediated by cryptococcal laccase promotes pulmonary growth and brain dissemination of virulent Cryptococcus neoformans in mice. PLoS One 2012; 7:1-14; PMID:23110112; http://dx.doi.org/10.1371/journal.pone.0047853
- Zhu X, Gibbons J, Garcia-Rivera J, Casadevall A, Williamson PR. Laccase of Cryptococcus neoformans is a cell wall-associated virulence factor. Infect Immun 2001; 69:5589-5596; PMID:11500433; http://dx.doi.org/10.1128/IAI.69.9.5589-5596.2001
- Scherer M, Fischer R. Purification and characterisation of laccase II of 482 Aspergillus nidulans. Arch Microbiol 1998; 170:78-84; PMID:9683643
- Sabiiti W, Robertson E, Beale MA, Johnston SA, Brouwer AE, Loyse A, Jarvis JN, Gilbert AS, Fisher MC, Harrison TS, et al. Efficient phagocytosis and laccase activity affect the outcome of HIV-associated cryptococcosis. J Clin Invest 2014; 124:2000-2008; PMID:2474314; http://dx.doi.org/10.1172/JCI72950
- Chai LY, Netea MG, Sugui J, Vonk AG, van de Sande WW, Warris A, Kwon1-Chung KJ, Kullberg BJ. Aspergillus fumigatus conidial melanin modulates host cytokine response. Immunobiol 2010; 215:915-920; PMID:19939494; http://dx.doi.org/10.1016/j.imbio.2009.10.002
- Stevens DA. Th1/Th2 in aspergillosis. Med Mycol 2006; 44:s229-s235. http://dx.doi.org/10.1080/13693780600760773
- Tsai H-F, Chang YC, Washburn RG, Wheeler MH, Kwon-Chung KJ. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J Bacteriol 1998; 180:3031-3038; PMID:9620950
- Krappmann S, Sasse C, Braus GH. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryot Cell 2006; 5:212-215; PMID:16400185; http://dx. doi.org/10.1128/EC.5.1.212-215.2006
- Sugareva V, Hart A, Brock M, Hübner K, Rohde M, Heinekamp T, Brakhage AA. Characterisation of the laccase-encoding gene abr2 of the dihydroxynaphthalene-like melanin gene cluster of Aspergillus fumigatus. Arch Microbiol 2006; 186:345-355; PMID:16988817; http://dx.doi.org/10.1007/s00203-006-0144-2
- Bugeja HE, Boyce KJ, Weerasinghe H, Beard S, Jeziorowski A, Pasricha S, Payne M, Schreider L, Andrianopoulos A. Tools for high efficiency genetic manipulation of the human pathogen Penicillium marneffei. Fungal Genet Biol 2012; 49:772-778; PMID:22921264. http://dx.doi.org/10.1016/j.fgb.2012.08.003
- Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta 1996; 113:51-56; PMID:5940632
- Baldrian P. Purification and characterization of laccase from the white-rot fungus Daedalea quercina and decolorization of synthetic dyes by the enzyme. Appl Microbiol Biotechnol 2004; 63:560-563; PMID:14504838; http://dx.doi.org/10.1007/s00253-003-1434-0
- Madzak MC, Mimmi E, Caminade A, Brault A, Baumberger S, Briozzo P, Mougin C, Jolivalt C. Shifting the optimal pH of activity for a laccase from the fungus Trametes versicolor by structure-based mutagenesis. Protein Eng Des Sel 2006; 19:77-84; PMID:16368720; http://dx.doi.org/10.1093/protein/gzj004
- Vanittanakom N, Vanittanakom P, Hay RJ. Rapid identification of Penicillium marneffei by PCR-based detection of specific sequences on the rRNA gene. J Clin Microbiol 2002; 40:1739-1742; PMID:11980953; http://dx.doi.org/10.1128/JCM.40.5.1739-1742.2002
- Borneman AR, Hynes MJ, Andrianopoulos A. An STE12 homolog from the asexual, dimorphic fungus Penicillium marneffei complements the defect in sexual development of an Aspergillus nidulans steA mutant. Genetics 2001; 157:1003-1014; PMID:11238390
- Boyce KJ, Bugeja HE, Weerasinghe H, Payne M, Schreider L, Park C, Woodward T, Andrianopoulos A. Strategies for the molecular genetic manipulation and visualization of the human fungal pathogen Penicillium marneffei. Fungal Genet Rep 2012; 59:1-12.
- Boyce KJ, Schreider L, Kirszenblat L, Andrianopoulos A. The two-component histidine kinases DrkA and SlnA are required for in vivo growth in the human pathogen Penicillium marneffei. Mol Microbiol 2011; 82:1164-1184; PMID:22059885. http://dx.doi.org/10.1111/j.1365-2958.2011.07878.x
- Wasylnka JA, Moore MM. Aspergillus fumigatus conidia survive and germinate in acidic organelles of A549 epithelial cells. J Cell Sci 2002; 116:179-1587; PMID:12640041; http://dx.doi.org/10.1242/jcs.00329
