In The Lancet Infectious Diseases, Hong Du and coworkers reported that mcr-1 colistin resistance gene is emerging in Enterobacteriaceae with carbapenem resistance in Jiangsu Province, China.Citation1 Although it posed clinical concern that expedited spread of the mcr-1-harbouring plasmid into multidrug-resistant bacteria might give super-bugs with pan-drug resistance,Citation1 the full genome sequence of the mcr-1-harbouring plasmid from the Enterobacterium with the multi-drug resistance remained unclear.
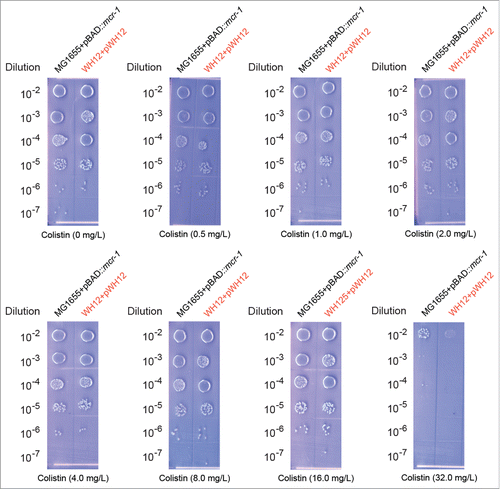
We thereby screened the Jiangsu E. coli isolates with multi-drug resistance collected in our stock for the presence of the mcr-1 colistin resistance gene (). Consequently, the E. coli strain (designated as WH12) was particularly noted in that it exhibits appreciable resistance to 13 antibiotics including ampicillin, cefotaxime, gentamycin, etc. Driven by the fact that the E. coli strain WH12 (MIC of colistin is 32 mg/L, ) was positive for the mcr-1 gene, we successfully isolated the corresponding mcr-1-harbouring plasmid referred to pWH12. The next-generation Illumina MiSeq sequencing was applied to decode the full genome of pWH12. An array of 350-bp paired-end reads produced here, were assembled with GS De Novo Assembler into a long contig, giving the complete genome of the mcr-1-harbourig plasmid pWH12 ().
Figure 1. Genomic evidence that the plasmid pWH12 is a variant of the paradigm mcr-1-harbouring pHNSHP45 plasmid. A.) Genetic organization and structure of the mcr-1-carring plasmid pWH12 from the swine lung microbiota. The genome map was generated using the CGView server (http://stothard.afns.ualberta.ca/cgview_server/index.html). Circles from inside to outside denote the GC screw, GC content and the open reading frames (ORF) in both DNA strands. The server of RAST (Rapid Annotation using Subsystem Technology) (http://www.nmpdr.org/FIG/wiki/view.cgi/FIG/RapidAnnotationServer) was applied for the annotation of plasmid genome, and the mcr-1 gene was highlighted in red. B.) Genomic comparison of the pWH12 with pHNSHP45, a prototype of the mcr-1-harbouring plasmid. Arrows represent putative genes in the linear comparison of plasmid genomes. The DNA fragments/genes that are either missing or extra-inserted in the pWH12 plasmid are highlighted in green arrows labeled with green letters. The mcr-1 gene was indicated in red. In particular, the pHNSHP45Citation3 is the only one mcr-1-carring plasmid with known genome sequence prior to our study. Unlike the other mcr-1-positive plasmid pKH457-3-BECitation2 that is from the cattle isolate in Belgium and has the plasmid signature of IncP, both pHNSHP45Citation3 and pWH12 are hosted by E. coli isolated from swine in China, and located on the Incl2 backbone. Genome comparison was performed using the BLAST tool.
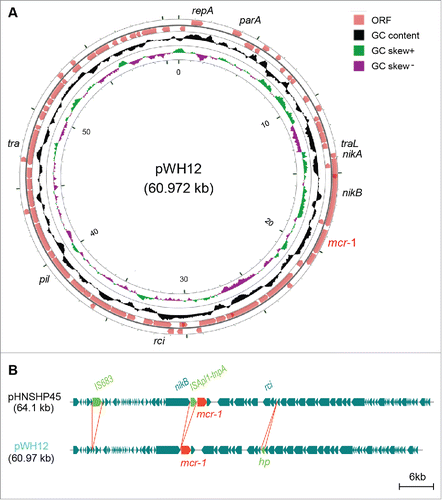
Unlike the pKH457-3-BE, a IncP-type, mcr-1-positive plasmid from the cattle isolate in Belgium in that it is the only one with known genome sequence (77.798 kb) beyond China,Citation2 the pWH12 was determined to be a IncI2-type plasmid whose genome is 60.972 kb in length with the average GC content of 42.54% (). Genome annotation suggested that the plasmid pWH12 encode around 90 putative open reading frames (ORFs), including the elements of the type IV secretion system (). Apart from 3 gaps, the pWH12 plasmid can be aligned well with the paradigmatic mcr-1-harbouring plasmid pHNSHP45Citation3 at the level of 99% similarity (). The difference between pWH12 and pHNSHP45 included the deletions of 2 mobile element-like sequences and one extra-insert of a hypothetical protein (hp) (). In particular, both the insertion sequence of IS683 (that is originally located in pHNSHP45 at the position of nucleotide acids from 2823 to 5536) and the tnpA transposase gene encoded by an insertion transposon-like sequence ISApl1 upstream of the mcr-1 are missing in the pWH12 plasmid (). It clearly suggested that pWH12 is a recent variant of the mcr-1-harbouring pHNSHP45 plasmid. Although the strain WH12 exhibited appreciable resistance to 13 antimicrobial drugs, we failed to note that any other antibiotic resistance genes than the mcr-1 are present in the plasmid pWH12 (). It raised 2 possibilities of the either chromosome-encoded resistance mechanism or coexistence of some unknown plasmids producing other drug-resistance. In fact, the discovery of pWH12 as a variant of the mcr-1-positive plasmid with the backbone of pHNSHP45 suggests its dynamic evolutionary activity under the environmental selective pressures, e.g., massive use of colistin as a veterinary medicine.
Given the fact that the mcr-1-positive strain WH12 in this study was isolated from Jiangsu Province, 2012, and the prototypical type of mcr-1-harbouring plasmid, pHNSHP45 is sampled in the Southern China, from 2011-2014, it is in rational to anticipate the variants of the mcr-1-carrying pHNSHP45 plasmid are circulating in China. This hypothesis was validated by our recent observation that unexpected diversity is present in the mcr-1-harbouring plasmid reservoirs of human gut microbiota from the diarrhea patients.Citation4 In light that Enterobacterial strains reported by Hong Du et al.Citation1 are collected in Jiangsu Province from 2015 to 2016, expedited dissemination of the mcr-1 gene might be transferred by the variants of the pHNSHP45. Dissemination/prevalence of the mcr-1 gene in food (milk and meat), animals, and human beings (even sputum) inevitably prompted us to believe the involvement of food-chain transmission paths. Given that the polymyxin is an ultimate line of refuge therapy against fatal infections by multidrug-resistant Gram-negative pathogens,Citation5,6 it is required to reconsider veterinary/clinical practices and strengthen the need for active surveillance in aiming to restrict the global dissemination of the mcr-1 gene as well as its transmission into inter/intra-species producing other drug resistance like NDM-9.Citation7
Taken together, our finding represents first genomic evidence that a variant of pHNSHP45 is emerging in the colistin-resistant E. coli that also produce other multi-drug resistance.Citation8
Figure 2. Determination of the minimal inhibitory concentration (MIC) of colistin conferred by the plasmid-borne MCR-1. The clinical E. coli strain referrs to WH12 that carrys a mcr-1-positive plasmid pWH12 (highlighted in red), whereas the positive control is the engineered E. coli strain, i.e., MG1655 with the arabinose-inducible expression of the mcr-1 gene. To determine the MIC of the antibiotic colistin, the mig-log phase cultures (OD600 = 0.7) in serial dilution were spotted on LBA plates supplemented with colistin at varied level (0, 0.5, 1.0, 2.0, 4.0, 8.0, 16.0 and 32.0 mg/L) and 0.2% arabinose. The LBA plates were maintained overnight at 37°C. The expression vector pBAD24 is used for functional assay of the mcr-1 gene in E. coli.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
Y.F. designed this project; Y.F., R.G., Q.W., P.L., and Z.L. performed experiments and analyzed the data; Y.F. contributed reagents and tools; Y.F. wrote this manuscript.
Funding
This work was supported by Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholars (Grant No.: LR15H190001), the National Natural Science Foundation of China (Grant No.: 31570027), and the start-up package from Zhejiang University (Y.F.). Dr. Feng is a recipient of the “Young 1000 Talents” Award.
References
- Du H, Chen L, Tang YW, Kreiswirth BN. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis 2016; 16(3): 287-288; PMID:26842776; http://dx.doi.org/10.1016/S1473-3099(16)00056-6
- Surbi Malhotra-Kumar BBX, Anupam J Das, Christine Lammens, Patrick Butaye, Herman Goossens. Colistin resistance gene mcr-1 harboured on a multidrug resistant plasmid. Lancet Infect Dis 2016; 16(3): 283-284; PMID:26774247; http://dx.doi.org/10.1016/S1473-3099(16)00012-8
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16(2): 161-168; http://dx.doi.org/10.1016/S1473-3099(15)00424-7
- Ye H, Li Y, Li Z, Gao R, Zhang H, Wen R, Gao GF, Hu Q, Feng Y. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. mBio 2016; 7(2). pii: e00177-16; PMID:27048797; http://dx.doi.org/10.1128/mBio.00177-16
- Nation RL, Li J, Cars O, Couet W, Dudley MN, Kaye KS, Mouton JW, Paterson DL, Tam VH, Theuretzbacher U, et al. Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect Dis 2015; 15: 225-34; PMID:25459221; http://dx.doi.org/10.1016/S1473-3099(14)70850-3
- Paterson DL, Harris PN. Colistin resistance: a major breach in our last line of defence. Lancet Infect Dis 2016; 16(2): 132-133; PMID:26603171; http://dx.doi.org/10.1016/S1473-3099(15)00463-6
- Yao X, Doi Y, Zeng L, Lv L, Liu JH. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis 2016; 16(3): 288-289; PMID:26842777; http://dx.doi.org/10.1016/S1473-3099(16)00057-8
- Gao R, Li Y, Lin J, Tan C, Feng Y. Unexpected complexity of multidrug resistance in the mcr-1-harbouring Escherichia coli. Sci China Life Sci 2016 May 13; http://dx.doi.org/10.1007/s11427-016-5070-1
