Campylobacter spp. is Gram-negative microaerophilic foodborne pathogen, annually accounting for approximately 96 million cases of foodborne illnesses worldwide according to a report from the World Health Organization (WHO).Citation1 Clinical symptoms developed by human campylobacteriosis includes severe abdominal pains, diarrhea, and fever.Citation2 In addition, Campylobacter is the major cause of Guillain-Barré Syndrome, a serious neurological disorder resulting in muscular paralysis.Citation3 Although C. jejuni is an important public health concern, the mechanisms for C. jejuni infection have not yet been clearly understood.Citation4 Particularly, it remains overall unknown how this fastidious pathogenic bacterium overcomes unfavorable environmental conditions and is transmitted to humans. As a natural host for C. jejuni, poultry is known as the major source of transmitting C. jejuni to humans.Citation5 However, Campylobacter is also isolated from a wide range of domestic animals and wildlifeCitation5,6 and even from environmental samples, such as soil, compost, and water.Citation7,8 This suggests that Campylobacter may possess unique survival mechanisms to sustain its viability under harsh environmental conditions.
Biofilm formation is an important survival mechanism for Campylobacter.Citation9 Bacterial cells in biofilms are physiologically different from planktonic cells, often exhibiting increased resistance to environmental stress and antimicrobials.Citation10 Biofilm development in C. jejuni is affected by many factors, including: 1) microbial factors, such as bacterial motility,Citation11,12 bacterial surface polysaccharides,Citation13,14 and cellular morphology,Citation15 2) environmental factors, such as growth temperature and osmolality.Citation16 In addition, biofilm formation is enhanced in mixed culture with other bacteria, such as Enterococcus faecalis, Staphylococcus simulans, and Pseudomonas aeruginosa,Citation17,18 and microbial commensalism with Pseudomonas spp extends the survival of C. jejuni under aerobic conditions.Citation19
Oxidative stress is associated with the unavoidable generation of toxic reactive oxygen species (ROS) during aerobiosis, and oxidative stress resistance significantly affects the survival of C. jejuni under oxygen-rich conditions.Citation20 C. jejuni possesses oxidative stress defense genes, such as alkyl hydroperoxide reductase (ahpC), catalase (katA), and superoxide dismutase (sodB), and the expression of these genes are modulated by PerR, CosR, and Fur.Citation20 Recently, it has been reported that the biofilm formation of C. jejuni is enhanced under aerobic conditions.Citation21 In our previous study, we demonstrated that oxidative stress played an important role in the biofilm development of C. jejuni under microaerobic conditions.Citation22 The mutation of oxidative stress defense genes, particularly ahpC, significantly enhances biofilm formation under microaerobic conditions, suggesting that oxidative stress is associated with biofilm formation in C. jejuni.Citation22 In this study, we investigated if oxidative stress defense would affect the biofilm formation of C. jejuni under aerobic conditions.
C. jejuni strain NCTC 11168 was routinely grown in Mueller-Hinton (MH) media at 42°C under microaerobic conditions (5% O2, 10% CO2, and 85% N2). Biofilm assay was performed as described in our previous report.Citation22 Occasionally, in this study, the biofilm assay was performed with different materials, such as tissue culture 96-well plate (Corning, #3595), polystyrene plastic 96-well plate (Corning #3370), glass 96-well plate (Cayman, US), and stainless steel small containers (diameter, 50 mm). Briefly, overnight culture of C. jejuni was collected and resuspended in MH broth to an optical density at 600 nm (OD600) of 0.1. Bacterial suspension was incubated at 42°C with shaking (200 rpm) under microaerobic condition for 5 h. After mixing the bacterial suspension with the same volume of fresh MH broth, 200 μl of the culture suspension was distributed to each well of 96-well tissue culture, polystyrene, and glass plates, and 1 ml of the culture suspension was added to a stainless steel container. After incubation at 42°C under microaerobic and aerobic (i.e., normal atmosphere) conditions without agitation, the biofilm samples were washed with PBS (pH 7.4) twice and incubated 20 min at room temperature to dry. Biofilms in the 96-well plates were stained by adding 50 µl (250 µl for the stainless container) of 1% crystal violet solution to each well and incubated at room temperature for 15 min. After removing the unstained crystal violet solution, the stained biofilm samples were washed 3 times with PBS (pH 7.4). Stain was eluted from the biofilms in a 96-well plate with 100 µl (500 µl for the stainless container) elution buffer (10% acetic acid and 30% methanol) and transferred to a new 96-well plate. The eluted crystal violet from the biofilms was measured at 595 nm with a spectrophotometer (Varioskan, Thermo Scientific, US). Each assay was performed with triplicate samples and repeated at least 3 times.
The total ROS levels were measured according to our previous study.Citation22 The total ROS level in biofilms was determined with 5-(6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA, Life Technologies, #C6827), a general oxidative stress indicator. Biofilms were prepared in the same way as described above in 96-well plates, washed twice with PBS, and exposed with 100 μl PBS containing 10 μM CM-H2DCFDA for 30 min at room temperature. Fluorescence was measured with a multi-well plate reader (Varioskan, Thermo Scientific, US) at the excitation and emission of 495 nm and 527 nm, respectively. The ROS levels were normalized to the total protein concentrations that were determined with Bradford protein assay (Bio-Rad, US).
The transcriptional levels of oxidative stress defense genes in biofilms were analyzed with qRT-PCR as described previously.Citation23 Briefly, biofilms were collected and washed twice with PBS (pH 7.4) and resuspended in RNAprotect Bacteria Reagent (Qiagen, # 76506) for RNA protection from RNase. Total RNA was isolated with RNeasy Mini Kit (Qiagen, # 74104) according to the manufacturer's instructions. Residual DNA in the extracted total RNA was removed with RNase-free DNase (Thermo Scientific, #AM2222). cDNA was synthesized using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, #4368814) and quantification of cDNA was carried out using Quanti Tect SYBR Green PCR kit (Qiagen, # 204143). qRT-PCR was performed on 7900HT Fast Real Time PCR System (Applied Biosystems, US), and the results were analyzed by using the 2-ΔΔCt method.Citation24 The transcriptional levels of 3 oxidative stress defense genes, ahpC, sodB and katA, were analyzed with normalization to the expression level of cjr01 encoding a 16S rRNA.Citation23 The sequences of primers for ahpC, sodB, katA, and cjr01 are presented in .
Table 1 The sequence of primers used in this study.
Confocal microscopic analysis of biofilms was performed as described in our previous study.Citation22 Briefly, biofilms were formed in MH broth under aerobic conditions with/without an antioxidant (1 nM N-acetyl cysteine; NAC) at 42°C for 12 h and 24 h. The biofilms were fixed with 4% paraformaldehyde after washing twice with PBS. The biofilms were stained with SYTO9 and propidium iodine (PI) for 20 min at room temperature and then washed twice with PBS. Confocal microscopic analysis was carried out with an inverted confocal microscope (IX-81, Olympus, Japan) and Volocity 3D Image Analysis Software (PerkinElmer Inc., US).
We measured the levels of biofilm formation on the surface of various abiotic materials under microaerobic and aerobic conditions. C. jejuni showed augmented levels of biofilm formation on plastic, tissue-culture treated plate, stainless steel, and glass under aerobic conditions compared to microaerobic conditions (). Reuter et al. first reported aerobic growth enhances the biofilm formation of C. jejuni NCTC 11168 in borosilicate glass tubes.Citation21 In this study, we demonstrated C. jejuni effectively develops biofilms on various surfaces, such as plastic, stainless steel, and glass, under aerobic conditions. Interestingly, C. jejuni also developed more biofilms on stainless steel under aerobic conditions compared to microaerobic conditions (). In addition, the versatility of C. jejuni in the development of biofilms on different surfaces would enable this bacterial pathogen to adapt to and survive in various environmental niches under oxygen-rich conditions. Our previous study demonstrated that the increased accumulation of ROS in the ahpC mutant significantly augmented biofilm formation in C. jejuni, and antioxidant treatment reduced the biofilm level of the ahpC mutant to that of the wild type.Citation22 Based on our previous findings, we hypothesized that oxidative stress would affect biofilm formation under aerobic conditions as well. The generation of ROS is an inevitable process of aerobiosis.Citation25 Cross-membrane transfer of oxygen occurs so freely that the intracellular concentration of oxygen is equivalent to the oxygen level of immediate extracellular environment; thus, microaerophilic microorganisms reduce oxidative stress by living in habitats with low oxygen levels.Citation25 Since C. jejuni is a microaerophile and requires only low oxygen concentrations (3∼15%) for optimal growth, aerobic culture conditions will impose more serious oxidative stress on C. jejuni than microaerobic conditions. We first compared the levels of ROS accumulation in C. jejuni biofilms between aerobic and microaerobic conditions. The ROS levels were substantially higher under aerobic conditions than microaerobic conditions in C. jejuni (). The addition of an antioxidant (i.e., 1 nM N-acetyl cysteine) in culture media significantly reduced the levels of total ROS (), and antioxidant treatment also reduced biofilm formation (). The results strongly suggest that the ROS accumulation plays a critical role in the biofilm development of C. jejuni under aerobic conditions.
Figure 1. Biofilm formation and oxidative stress response in C. jejuni under aerobic conditions. (A) The levels of biofilm formation of C. jejuni on different materials were compared between microaerobic and aerobic conditions. C. jejuni produced more biofilms under aerobic (normal atmospheric) conditions than microaerobic (5% O2, 10% CO2) conditions. The biofilm formation of C. jejuni also significantly increased on plastic, glass, and stainless steel. (B) Total ROS accumulation in C. jejuni biofilms under different culture conditions. The accumulated ROS levels were notably increased under aerobic conditions compare to microaerobic conditions. Also, the addition of 1 nM NAC, an antioxidant, significantly reduced accumulated ROS levels in both culture conditions. (C) Effect of antioxidant treatment on biofilm formation under aerobic and microaerobic conditions. Antioxidant treatment effectively reduced biofilm formation in C. jejuni. (D) Transcriptional levels of oxidative stress defense genes in biofilms under aerobic and microaerobic conditions. The expression levels of 3 oxidative stress defense genes increased after exposure to aerobic conditions. The relative transcriptional levels were determined by normalization to cjr01 encoding 16S rRNA. The results show the means and standard deviations of triplicate samples in 3 different tubes in a single experiment. The assay with triplicate samples was repeated at least 3 times on different days, and similar results were obtained in the experiments. The biofilm assay data were analyzed with the one-way ANOVA and Student's t-test. The total ROS accumulation levels were analyzed with Student's t-test. Statistical analysis of qRT-PCR carried out using 2-way ANOVA. All the data were analyzed with GraphPad Prism 6 (GraphPad Software INC., US). *: P < 0.05, **: P < 0.01, ***: P < 0.001.
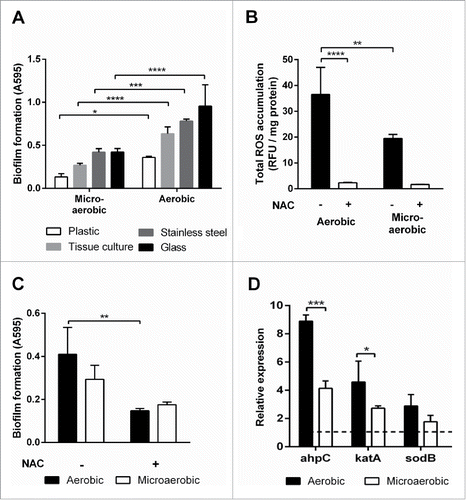
Based on the findings that oxidative stress stimulates biofilm formation under aerobic conditions (), we hypothesized that the expression levels of oxidative stress defense genes will be altered in biofilms under aerobic conditions. To confirm this, qRT-PCR was employed to quantify the transcriptional levels of ahpC, sodB, and katA, which are the sole gene in C. jejuni encoding alkyl hydroperoxide reductase, superoxide dismutase, and catalase, respectively. Interestingly, the transcriptional levels of the antioxidant genes were markedly increased under aerobic conditions compared to microaerobic conditions (). The ahpC gene exhibited higher transcriptional levels than katA and sodB in biofilms in both microaerobic and aerobic conditions (). The results also support our previous findings that ahpC plays a critical role the development of biofilms in C. jejuni.Citation22 Since C. jejuni cells residing in biofilms encounter increased oxidative stress under aerobic conditions (), the function of oxidative stress defense genes would be more critical under aerobic conditions than microaerobic conditions to detoxify increased ROS stress.
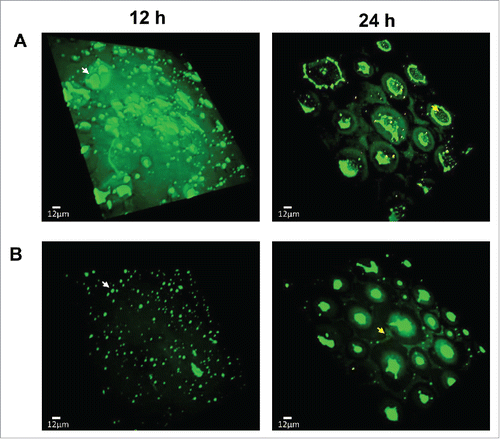
The development of biofilms consists of multiple stages, including the attachment of free floating bacteria to surface, formation of microcolonies, development of mature structures (e.g., mushroom-like structures and channels), and dispersal of sessile cells to free floating cells.Citation26 Biofilms grown under aerobic conditions were observed with confocal microscopy in the presence and absence of an antioxidant (i.e., N-acetylcysteine). Interestingly, C. jejuni easily developed mature biofilms under aerobic conditions (). The dispersal of bacterial cells from biofilms, which is the last event of biofilm development, was observed in 24 h (). Dead cells (i.e., red cells after the Live/Dead staining) were not observed notably in biofilms (24 h in ), and rapid formation of biofilms was noticed even at the early stage of development (12 h in ). Thus, the dispersal of biofilms is likely to be caused by maturation, not by cell death. Interestingly, antioxidant treatment substantially reduced the formation of microcolonies and the dispersal of bacterial cells was not observed in the presence of an antioxidant, suggesting that antioxidant treatment delayed the development of biofilms under aerobic conditions (). The results confirmed that oxidative stress stimulates C. jejuni development of biofilms under aerobic conditions.
Figure 2. Confocal microscopy of C. jejuni biofilms under aerobic conditions in the absence (A) and presence (B) of an antioxidant. Biofilms were stained with SYTO9 and propidium iodine (Life Technologies) for live (i.e., green) and dead (i.e., red) cells, respectively. N-acetylcysteine was used at 1 nM for the biofilm samples in panel B. C. jejuni developed mature biofilms under aerobic conditions. An antioxidant treatment delayed biofilm formation under aerobic conditions in C. jejuni. Microcolonies were observed in 12 h, and mature biofilms were developed in 24 h. Exemplary microcolonies and dispersed cells are indicated with white and yellow arrows, respectively. The biofilms were detected with an Olympus IX-81 motorized microscope base system. The experiment was repeated 3 times, and similar results were observed in all the experiments.
In this study, we demonstrated that aerobic conditions enhance biofilm formation in C. jejuni via oxidative stress. In addition to environmental factors that are known to be involved in biofilm formation, such as temperature, osmolality, and pH,Citation16,27 oxygen is also a key player in biofilm formation in some bacterial species. It has been reported that oxygen induces biofilm formation in Shewanella putrefaciensCitation28 and oxygen depletion induces the detachment of Shewanella oneidensis biofilms.Citation29 The intestinal tracts provide C. jejuni with optimal growth conditions, such as low oxygen concentrations and high nutrients, but after C. jejuni is excreted from host animals, this bacterium will unavoidably encounter harsh oxygen stress in normal environmental conditions.Citation20 Oxygen concentrations in the environment, such as water and air, will pose increased oxidative stress to microaerophilic C. jejuni, and the increased oxidative stress would stimulate biofilm formation. Our recent study also demonstrated that aerobic conditions promoted the development of a viable-but-non-culturable (VBNC) state in C. jejuni.Citation30 Based on our previous report and findings in this study, aerobic conditions may stimulate sessile C. jejuni cells to develop biofilms and planktonic cells to enter the VBNC state. Either the formation of biofilms or the development of VBNC stage would significantly affect the survival of this microaerophilic pathogen under harsh oxygen-rich conditions. Importantly, oxidative stress appears to be a critical factor facilitating these physiological changes in C. jejuni under aerobic conditions. Although the elucidation of molecular details still awaits future studies, the findings in this study exhibited that oxidative stress plays an important role in the stimulation of biofilm development in C. jejuni under aerobic conditions.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Drs. Lynn McMullen and Michael Gänzle (University of Alberta) for sharing their laboratory facilities.
Funding
This study is supported by NSERC Discovery Grant (401843-2012-RGPIN) to BJ.
References
- Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, Lake RJ, Praet N, Bellinger DC, de Silva NR, Gargouri N, et al. World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med 2015; 12:e1001923-e; PMID:26633896; http://dx.doi.org/10.1371/journal.pmed.1001923
- Snelling WJ, Matsuda M, Moore JE, Dooley JS. Campylobacter jejuni. Lett Appl Microbiol 2005; 41:297-302; PMID:16162134; http://dx.doi.org/10.1111/j.1472-765X.2005.01788.x
- Hughes RA, Cornblath DR. Guillain-Barré syndrome. Lancet 2005; 366:1653-66; PMID:16271648; http://dx.doi.org/10.1016/S0140-6736(05)67665-9
- Acheson D, Allos BM. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Diseases 2001; 32:1201-6; PMID:NOT_FOUND; http://dx.doi.org/10.1086/319760
- Hermans D, Pasmans F, Messens W, Martel A, Van Immerseel F, Rasschaert G, Heyndrickx M, Van Deun K, Haesebrouck F. Poultry as a host for the zoonotic pathogen Campylobacter jejuni. Vector Borne Zoonotic Dis 2012; 12:89-98; PMID:22133236; http://dx.doi.org/10.1089/vbz.2011.0676
- Jokinen C, Edge TA, Ho S, Koning W, Laing C, Mauro W, Medeiros D, Miller J, Robertson W, Taboada E, et al. Molecular subtypes of Campylobacter spp., Salmonella enterica, and Escherichia coli O157:H7 isolated from faecal and surface water samples in the Oldman River watershed, Alberta, Canada. Water Res 2011; 45:1247-57; PMID:20971491; http://dx.doi.org/10.1016/j.watres.2010.10.001
- Trimble LM, Alali WQ, Gibson KE, Ricke SC, Crandall P, Jaroni D, Berrang M, Habteselassie MY. Prevalence and concentration of Salmonella and Campylobacter in the processing environment of small-scale pastured broiler farms. Poult Sci 2013; 92:3060-6; PMID:24135612; http://dx.doi.org/10.3382/ps.2013-03114
- Levesque S, St-Pierre K, Frost E, Arbeit RD, Michaud S. Use of amplified-fragment length polymorphism to study the ecology of Campylobacter jejuni in environmental water and to predict multilocus sequence typing clonal complexes. Appl Environ Microbiol 2012; 78:2470-3; PMID:22267674; http://dx.doi.org/10.1128/AEM.06527-11
- Murphy C, Carroll C, Jordan KN. Environmental survival mechanisms of the foodborne pathogen Campylobacter jejuni. J Appl Microbiol 2006; 100:623-32; PMID:16553716; http://dx.doi.org/10.1111/j.1365-2672.2006.02903.x
- Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol 2008; 6:199-210; PMID:18264116; http://dx.doi.org/10.1038/nrmicro1838
- Joshua GW, Guthrie-Irons C, Karlyshev AV, Wren BW. Biofilm formation in Campylobacter jejuni. Microbiology 2006; 152:387-96; PMID:16436427; http://dx.doi.org/10.1099/mic.0.28358-0
- Kalmokoff M, Lanthier P, Tremblay TL, Foss M, Lau PC, Sanders G, Austin J, Kelly J, Szymanski CM.. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J Bacteriol 2006; 188:4312-20; PMID:16740937; http://dx.doi.org/10.1128/JB.01975-05
- Naito M, Frirdich E, Fields JA, Pryjma M, Li J, Cameron A, Gilbert M, Thompson SA, Gaynor EC. Effects of sequential Campylobacter jejuni 81-176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J Bacteriol 2010; 192:2182-92; PMID:20139192; http://dx.doi.org/10.1128/JB.01222-09
- Jowiya W, Brunner K, Abouelhadid S, Hussain HA, Nair SP, Sadiq S, Williams LK, Trantham EK, Stephenson H, Wren BW, et al. Pancreatic amylase is an environmental signal for regulation of biofilm formation and host interaction in Campylobacter jejuni. Infect Immun 2015; 83:4884-95; PMID:26438798
- Frirdich E, Biboy J, Adams C, Lee J, Ellermeier J, Gielda LD, Dirita VJ, Girardin SE, Vollmer W, Gaynor EC. Peptidoglycan-modifying enzyme Pgp1 is required for helical cell shape and pathogenicity traits in Campylobacter jejuni. PLoS Pathog 2012; 8:e1002602; PMID:22457624; http://dx.doi.org/10.1371/journal.ppat.1002602
- Reeser RJ, Medler RT, Billington SJ, Jost BH, Joens LA. Characterization of Campylobacter jejuni biofilms under defined growth conditions. Appl Environ Microbiol 2007; 73:1908-13; PMID:17259368; http://dx.doi.org/10.1128/AEM.00740-06
- Teh KH, Flint S, French N. Biofilm formation by Campylobacter jejuni in controlled mixed-microbial populations. Int J Food Microbiol 2010; 143:118-24; PMID:20805009; http://dx.doi.org/10.1016/j.ijfoodmicro.2010.07.037
- Ica T, Caner V, Istanbullu O, Nguyen HD, Ahmed B, Call DR, Beyenal H. Characterization of mono-and mixed-culture Campylobacter jejuni biofilms. Appl Environ Microbiol 2012; 78:1033-8; PMID:22179238; http://dx.doi.org/10.1128/AEM.07364-11
- Hilbert F, Scherwitzel M, Paulsen P, Szostak MP. Survival of Campylobacter jejuni under conditions of atmospheric oxygen tension with the support of Pseudomonas spp. Appl Environ Microbiol 2010; 76:5911-7; PMID:20639377; http://dx.doi.org/10.1128/AEM.01532-10
- Kim J-C, Oh E, Kim J, Jeon B. Regulation of oxidative stress resistance in Campylobacter jejuni, a microaerophilic foodborne pathogen. Front Microbiol 2015; 6:751; PMID:26284041
- Reuter M, Mallett A, Pearson BM, van Vliet AH. Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl Environ Microbiol 2010; 76:2122-8; PMID:20139307; http://dx.doi.org/10.1128/AEM.01878-09
- Oh E, Jeon B. Role of alkyl hydroperoxide reductase (AhpC) in the biofilm formation of Campylobacter jejuni. PloS One 2014; 9:e87312; PMID:24498070; http://dx.doi.org/10.1371/journal.pone.0087312
- Hwang S, Kim M, Ryu S, Jeon B. Regulation of oxidative stress response by CosR, an essential response regulator in Campylobacter jejuni. PloS One 2011; 6:e22300; PMID:21811584; http://dx.doi.org/10.1371/journal.pone.0022300
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/10.1006/meth.2001.1262
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 2008; 77:755-76; PMID:18173371; http://dx.doi.org/10.1146/annurev.biochem.77.061606.161055
- Monroe D. Looking for chinks in the armor of bacterial biofilms. PLoS Biol 2007; 5:e307; PMID:18001153; http://dx.doi.org/10.1371/journal.pbio.0050307
- Hostacka A, Ciznar I, Stefkovicova M. Temperature and pH affect the production of bacterial biofilm. Folia Microbiol 2010; 55:75-8; PMID:20336508; http://dx.doi.org/10.1007/s12223-010-0012-y
- Wu C, Cheng YY, Yin H, Song XN, Li WW, Zhou XX, Zhao LP, Tian LJ, Han JC, Yu HQ. Oxygen promotes biofilm formation of Shewanella putrefaciens CN32 through a diguanylate cyclase and an adhesin. Sci Rep 2013; 3:1945; PMID:23736081
- Thormann KM, Saville RM, Shukla S, Spormann AM. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J Bacteriol 2005; 187:1014-21; PMID:15659679; http://dx.doi.org/10.1128/JB.187.3.1014-1021.2005
- Oh E, McMullen L, Jeon B. Impact of oxidative stress defense on bacterial survival and morphological change in Campylobacter jejuni under aerobic conditions. Front Microbiol 2015; 6:295; PMID:25914692
- Hwang S, Zhang Q, Ryu S, Jeon B. Transcriptional regulation of the CmeABC multidrug efflux pump and the KatA catalase by CosR in Campylobacter jejuni. J Bacteriol 2012; 194:6883-91; PMID:23065977; http://dx.doi.org/10.1128/JB.01636-12
