ABSTRACT
The commensal fungal pathogen Candida albicans is a leading cause of lethal systemic infections in immunocompromised patients. One of the main mechanisms of host immune evasion and virulence by this pathogen is the switch from yeast form to hyphal growth morphologies. Micro RNAs (miRNAs), a small regulatory non-coding RNA, has been identified as an important part of the immune response to a wide variety of pathogens. In general, miRNAs act by modulating the intensity of inflammatory responses. miRNAs act by base-paring binding to specific sequences of target mRNAs, generally causing their silencing through mRNA degradation or translational repression. To study the impact of C. albicans cell morphology upon host miRNA expression, we investigated the differential modulation of 9 different immune response-related miRNAs in primary murine bone marrow-derived macrophages (BMDMs) exposed to either yeasts or hyphal forms of Candida albicans. Here, we show that the different growth morphologies induce distinct miRNA expression patterns in BMDMs. Interestingly, our data suggest that the C-Type lectin receptor Dectin-1 is a major PRR that orchestrates miR155 upregulation in a Syk-dependent manner. Our results suggest that PRR-mediating signaling events are key drivers of miRNA-mediated gene regulation during fungal pathogenesis.
Introduction
Candida albicans is the most prevalent cause of serious mycosis in humans, responsible for a wide variety of diseases, including invasive systemic infections as well as mucosal and skin infections. Although mucosal infections can also occur in healthy individuals, C. albicans is a leading cause of lethal invasive systemic infections in immunocompromised patients.Citation1 C. albicans is a pleomorphic fungus able to grow and colonize the host in yeast, pseudo hypha and hypha morphologies. These morphologies are tightly regulated by environmental and host stimuli, such as temperature, pH, CO2 levels, and presence of serum, among many other factors.Citation2 The ability to switch to a hyphal morphology is suspected to contribute to the evasion from immune surveillance and thus has been linked to virulence.Citation3,4 Although similar in principle, it is widely accepted that each morphology has a distinct cell wall composition with differences in the relative abundance of glucan, mannan and chitin, which plays an important role in host interactions.Citation5
Macrophages are pivotal modulators of the early innate immune response during C. albicans infections, since these effector cells can recognize specific components of the fungal cell wall and thus initiate an appropriate host response to the pathogen.Citation6 To identify different fungal cell wall components, macrophages carry a plethora of different surface receptors, also known as Pattern Recognition Receptors (PRRs), that specifically recognize pathogen-associated molecular patterns (PAMPs).Citation7,8 In the context of the immune response to C. albicans, relevant PRRs include the Toll-like receptors (TLRs) 2 and 4,Citation9,10 the C-Type Lectin Receptor Dectin-1Citation11 and others, as reviewed elsewhere.Citation12 Upon recognition of exposed cell wall β-glucans, Dectin-1 modulates host gene expression via intracellular signaling pathways. The activation of Dectin-1 induces 2 different pathways, a Syk-dependent pathway and a Syk-independent pathway, through Raf-1.Citation13 Of note, Syk-signaling in phagocytes also operates via a phagosomal pathway.Citation14
In recent years, microRNAs (miRNAs) have emerged as important regulators of host immune responses. Of note, certain miRNAs are induced upon activation of TLRs. For instance, both miR146 and miR155 are induced by lipopolysaccharide (LPS) activation of TLR4.Citation15 Nonetheless, miRNAs show different expression behaviors in mouse macrophages depending on the stimuli.Citation16 In RAW264.7 macrophages, LPS increases miR155 expression but reduces that of miR125b.Citation17 miR132 is activated in human monocytes co-cultured with Aspergillus fumigatus hyphae, but not by conidia or bacterial LPS.Citation18 miRNAs also play a pivotal role in modulating immune responses, although the identification of target genes involved in these processes has been challenging. Nevertheless, many miRNAs such as miR146,Citation19,20 miR125b,Citation21miR221,Citation17,22 miR132,Citation23,24 miR9,Citation25 miR145,Citation26 miR223,Citation27 and miR155 Citation15,28,29 have been related to host response to pathogens and regulation of the inflammatory process.
Recently, the regulation of host miRNAs in response to C. albicans has been reported.Citation30 However, to the best of our knowledge, there is no published report about the effects of different pathogen growth morphologies on the regulation of miRNAs. In this work, we initially investigated the expression profiles of select immune-relevant miRNAs in BMDMs stimulated with different C. albicans morphologies. We also analyzed the pathway of activation of miR155 in BMDMs during interaction with C. albicans hyphae, due to the importance of this miRNA in immune responses. Ten miRNAs were selected for analyses in mouse BMDMs, miR9, miR125b, miR132, miR145, miR146a, miR155, miR199, miR221, miR223 and miR455. Here, we show that C. albicans morphologies impact the expression of immune-related miRNAs. Interestingly, the responses of BMDMs from mice lacking TLR2, TLR4, both TLR2/4 and Dectin-1 indicate that different C. albicans morphologies trigger different expression patterns for miR155. Notably, while TLR2 and TLR4 have a minor role, Dectin-1 seems to be a main activator of miR155 expression through a Syk-dependent pathway.
Results
In this work, we investigated the expression profiles of select immune-relevant miRNAs in BMDMs stimulated with different morphologies of C. albicans growth. After one hour incubation with macrophages, more than 50% of yeast C. albicans cells had already filamented, and after 2 hours more than 50% of the macrophages were killed by the newly formed hyphae (data not shown).Citation31 Therefore, in order to avoid any bias due to cells transitioning between morphologies, only killed C. albicans cells were used. So, an initial screening was designed to evaluate the impact of C. albicans cell wall ligands specific for each cell morphology on murine primary macrophages' miRNA expression. BALB/c BMDMs were isolated and incubated with either heat-killed (HK-) yeasts or hyphae. A multiplicity of infection (MOI) of 2 was used for 8 and 16 hours for these interactions followed by RNA isolation at these intervals. The experiments were performed as 3 independent experiments, with each performed on different days with cells from different mice.
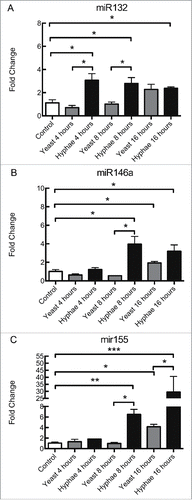
Initially, the expression levels of miR125b, miR132, miR145, miR146a, miR155 and miR455 in BMDMs were tested as detailed in the methods. Both miR125b and miR132 were significantly upregulated in macrophages exposed to HK-hyphae for 8 or 16 hours, but not after incubation with yeast forms (, Fig. S1B and S1C). Although at 16 hours macrophages exposed to yeasts showed a trend toward upregulation of miR125b, this result was not statistically significant (Fig. S1B). Also, the accumulation of both miR125b and miR132 were significantly higher in macrophages exposed to hyphae than to yeast form after 8 hours, whereas only the upregulation of miR132 remained higher at 16 hours (, Fig. S1B and S1C). miR146a and miR155 were upregulated in macrophages exposed to both HK-yeasts and hyphae at 8 hours, but only the expression of miR146a was increased at 16 hours (, Fig. S1D and Fig. S1A). After interaction for 8 hours, the accumulation of miR146a was significantly higher in BMDMs exposed to HK-hyphae than for those exposed to yeasts, but the variations were not statistically different at 16 hours (Fig. S1D). miR455 was also upregulated in BMDMs stimulated with HK-hyphae for 8 hours, but not at 16 hours or when stimulated with HK-yeast cells (, and Fig. S1E). The expression of miR145 did not significantly vary during any of the conditions employed (data not shown). We also quantified expression levels of miR9, miR199, miR221 and miR223. After 8 hours incubation, miR9 was upregulated in BMDMs exposed to both HK-yeasts and hyphae, but when exposed to hyphae, BMDMs expressed more miR9 than when exposed to yeasts (Fig. S2A). miR199 was upregulated in BMDMs after 8 hours exposed to HK-killed hyphae and yeasts, however HK-hyphae induced response was significantly higher than the response induced by UV-killed yeasts (Fig. S2B). miR221 and miR223 were significantly higher expressed in BMDMs exposed to HK-hyphae for 8 hours compared to the unstimulated control, but yeast forms did not alter their expression compared to these controls ( and Fig. S2C and S2D).
Table 1. Differential expression patterns of 9 selected miRNAs in mouse BMDMs with or without exposure to C. albicans heat-killed yeasts or hyphae.
Heat killing of fungal pathogens alters the fungal cell wall architecture by strongly exposing glucans, which are otherwise covered by mannans that form the outer-most layer of the fungal cell wall.Citation7 To avoid this bias, we performed experiments using C57Bl/6 BMDMs and C. albicans cells killed by ultraviolet radiation (UV). We observed a higher aggregation propensity of the fungi caused by UV-killing compared to heat-killing, which led us to increase the MOI to 5. Interactions were then performed for 4, 8 and 16 hours. To confirm there were responses to UV-killed cells, levels of TNFα mRNA were also measured by qRT-PCR at each time point. The levels of TNFα were significantly higher at 4 hours after incubation with both UV-killed yeasts and hyphae relative to unchallenged BMDMs, although the hyphae induced higher levels of this cytokine. At 8 hours, the levels of TNFα mRNA returned to basal levels. At 16 hours, the levels of TNFα were again slightly increased when compared to the unstimulated control (S3 Fig.).
As observed in the interaction with heat-killed C. albicans cells ( and Fig. S1C-D), BMDMs increased their expression of miR132 and miR146a in response to UV-killed hyphae at 8 and 16 hours of incubation, while only miR132 was upregulated in this condition at 4 hours (). UV-killed hyphae induced statistically different responses from that of UV-killed yeast for miR132 at 4 and 8 hours whereas miR146a expression levels varied significantly at 8 and 16 hours (). By contrast, miR155 was only upregulated after incubation with UV-killed hyphae at 8 and 16 hours, but not at 4 hours (). UV-killed yeasts produced a response in miR155 expression in BMDMs only at 16 hours, and this response was significantly different from the response to UV-killed hyphae at the same time interval (). C57/Bl6 BMDMs exposed to heat-killed C. albicans showed a similar pattern of expression of miR132, although there were some important differences in miR146a and miR155 expression (S4A-C Fig.). For instance, there are differences in miR146a and miR155 expression in response to HK-yeasts at 8 hours, and in miR155 expression in response to HK-hyphae at 16 hours between the 2 strains (S1A and D Fig. and S4 B and C Fig.). In addition, incubation with yeasts killed with different methodologies (HK- or UV-) resulted in different miR155 expression patterns in C57Bl/6 BMDMs ( and S4C Fig.). This suggests that both the different methods of killing the fungal cells and different mouse strains influence the variations in the expression of these miRNAs.
Figure 1. Expression levels of miRNAs in BMDMs exposed to UV-killed C. albicans cells. Expression levels of miR132 (A), miR146a (B), and miR155 (C) in C57Bl/6 BMDMs incubated with C. albicans UV-killed yeasts or hyphae at a MOI of 5. Expression levels were measured by qRT-PCR. Results represent the mean of fold change from 3 independent pooled experiments ± SD. Asterisks indicate p-value; (*) p < 0.05, (**) p < 0.01, (***) p < 0.001, (****) p < 0.0001.
These findings led us to focus on the miR155 response given its status as a typical multifunctional miRNA, with a pivotal role in various physiological and pathological processes. miR155 also has a critical role in the modulation of innate-cell mediated as well as adaptive immune responses. To unravel a possible signaling mechanism leading to the accumulation of miR155, we analyzed the influence of some of the main PRRs mediating fungal recognition by macrophages. Hence, BMDMs from C57Bl/6 mice lacking TLR2, TLR4, both TLR2/4 or Dectin-1 genes were employed. In these experiments, BMDMs were incubated for 8 hours with UV-killed hyphae with a MOI of 5.
As shown in , the absence of either TLR2 or TLR4 alone failed to alter the accumulation of miR155 in BMDMs under these conditions. However, the deletion of both TLR2 and TLR4 resulted in an upregulation that was significantly higher than in wild type (WT) BMDMs or in BMDMs lacking each of these receptors alone. Remarkably, the genetic deletion of Dectin-1 gene completely abolished the accumulation of miR155 (). The levels of miR155 in unstimulated control BMDMs did not vary over time (data not shown).
Figure 2. Dectin-1 is responsible for regulating miR155 expression in BMDMs exposed to C. albicans hypha. Expression levels of miR155 in (A) BMDMs obtained from different strains of C57Bl/6 mice, incubated for 8 hours with C. albicans UV-killed hyphae or in (B) Wild-type BMDMs incubated with either β-glucan or C. albicans UV-killed hyphae. (A) BMDMs from Wild-type mice and mice deficient in the PRRs TLR2, TLR4, both TLR2 and TLR4, or Dectin 1 (Wild-type, ΔTLR2, ΔTLR4, ΔTLR2/4 and ΔDectin1) were incubated for 8 hours with UV-killed C. albicans hyphae at a MOI of 5. (B) BMDMs from Wild-type C57Bl/6 mice were incubated for 8 hours with either β-glucan (depleted Zymosan) at 100 µg/mL or UV-killed C. albicans hyphae at a MOI of 5. Expression levels were measured by qRT-PCR. Results represent the mean of fold change from 3 independent pooled experiments ± SD. Asterisks indicate p-value; (*) p < 0.05, (**) p < 0.01, (***) p < 0.001, (****) p < 0.0001.
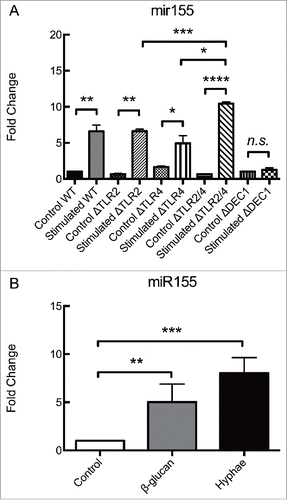
To verify if interactions with Dectin-1 was indeed stimulating the expression of miR155 in BMDMs from C57Bl/6 mice, the BMDMs were incubated with a specific agonist of Dectin-1. BMDMs were incubated with 100μg/mL of depleted Zymosan, a commercial reagent rich in β-glucans that activates Dectin-1 without activating other PRRs. After 8 hours, the BMDMs showed an upregulation of miR155 similar to that measured in BMDMs challenged with UV-killed hyphae at a MOI of 5 for the similar time interval (). So, the stimulation of Dectin-1 in BMDMs by either commercial β-glucans or β-glucans in the cell walls of C. albicans results in the enhanced expression of miR155.
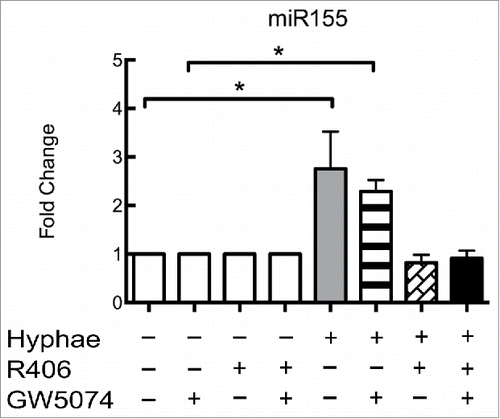
The activation of Dectin-1 can induce 2 different response pathways, a Syk-dependent signaling pathway and a Raf-1-dependent pathway. Both pathways have been related to C. albicans responses in macrophages,Citation32,33 as well as in other phagocytes or antigen-presenting cells. To determine which pathway is more important for regulating the expression of miR155, we performed an interaction assay using inhibitors specific for each of these pathways. BMDMs from WT C57BL/6 mice were incubated with GW5074, R406 or both for one hour prior to performing the 8 hours incubation with UV-killed hyphae (MOI of 5). While BMDMs incubated with the Raf-1 inhibitor GW5074 challenged with hyphae displayed the same expression levels as the BMDMs subjected to hyphae alone (). BMDMs treated with the specific Syk inhibitor R406 alone or in combination with GW5074 no longer increased miR155 expression (). Inhibitor treatment alone had no effect on unstimulated BMDMs ().
Figure 3. Dectin-1 activation of miR155 occurs via a Syk-dependent pathway. Effect of Dectin-1 pathway inhibitors on the expression of miR155 by different C57Bl/6 BMDMs with or without exposure to C. albicans UV-killed hyphae at an MOI of 5 for 8 hours. BMDMs with or without exposure to Dectin-1 inhibitors GW5074 (inhibits Raf-1), R406 (Syk inhibitor) or both compounds. Expression levels were measured by qRT-PCR. Results represent the mean of fold change from 3 independent pooled experiments ± SD. Asterisks indicate p-value; (*) p < 0.05, (**) p < 0.01, (***) p < 0.001, (****) p < 0.0001.
To verify the effects of miR155 over its targets, 5 targets were analyzed after induction with C. albicans hyphae for 8 hours. The chosen targets were MyD88,Citation34,35 SHIP1,Citation28,36 SOCS1,Citation27 IKKεCitation37,38 and C/EBPβ.Citation39,40 It was observed that the differences in expression of MyD88 (R2 = 0.980, p < 0.0001), SHIP1 (R2 = 0.767, p = 0.002), SOCS1 (R2 = 0.988, p < 0.0001) and IKKε (R2 = 0.996, p < 0.0001) were indeed correlated to the differences in miR155 expression (Fig. S4 and Supplementary text). Differences of expression in C/EBPβ, on the other hand, showed very little correlation compared to miR155 (R2 = 0.072 and p = 0.484) (Fig. S5 and Supplementary text). It is feasible that miR155 is causing the down regulation in its targets, considering it was already validated experimentally that miR155 targets directly these mRNAs.
Discussion
Here, we show that miR9, miR125b, miR132, miR146a, miR155, miR199, miR221, miR223 and miR455 can be differentially induced upon incubation of BMDMs with different morphologies of heat-killed C. albicans. We also show that Dectin-1 in murine BMDMs is critical for induction of mir155 by hyphae in a Syk-dependent pathway.
In our experiments, macrophages were challenged with C. albicans for several hours in RPMI1640 containing serum, at 37˚C and with high carbon dioxide rates, all of which strongly and rapidly induce the filamentation program in this fungus. Filamentation gene programs are fully activated within 30 minutes of cultivation under these conditions, even when yeast cell morphology persists.Citation31 We () and others have found Citation30 that miRNA expression levels are only altered after significantly longer durations of interaction of macrophages with morphotypes of C. albicans. Therefore, we opted to use killed C. albicans cells, due to the difficulties intrinsic to ex vivo assays analyzing specific C. albicans cell morphologies. Although C. albicans cell walls that went through heat-killing (and to a smaller degree, UV-killing) will not have the same ligand exposure as live cells, it is still possible to use this model for collecting valuable data concerning this interaction.Citation30,41–44
The main criterion for selecting these miRNAs was the documented importance of their targets in the regulation of innate immunity. The results described by our work show that hyphal morphologies distinctly regulate the accumulation of most of these miRNAs, suggesting that differential activation of PRRs by hyphae and yeasts may play a role in their regulation and in downstream host immune responses. This is the first report demonstrating the pronounced impact of C. albicans hyphae in miRNA expression in primary BMDMs. Previous work using BMDMs incubated with heat-killed C. albicans yeasts showed that only miR155, miR146, miR125a and miR455 were upregulated compared to unchallenged BMDMs.Citation30 Here, we show that miR155 and miR146a are indeed induced by heat-killed yeasts along with miR9, which was not differentially regulated in these conditions previously. However, in our hands, miR455 is only upregulated following hyphal challenge. Recently, miR221 was shown to be upregulated in human lung epithelium in response to C. albicans infection.Citation45 Of note, BMDMs interacting with hyphae also increased their levels of other miRNAs, including 125b, miR132, miR199, and miR223. While these miRNAs have been associated with responses to other pathogens,Citation18,46–48 this is the first report demonstrating their specific induction by C. albicans.
Upon activation of a host effector cell by a microbe, miRNAs are induced and commonly act as regulators, modulating the host response. Even minute changes in mature miRNA expression can tip the balance in transcriptional programs due to the regulatory role of these molecules. Most miRNAs affect their mRNA targets directly. These targets are often components of key steps of PRR pathways, transcriptional factors or cytokines. For instance, miR9, miR199, miR221 and miR223 exert a regulatory activity on inflammation.Citation48–51 miR146 impairs pro-inflammatory cytokines in dendritic cells by targeting the mRNAs of the TLR pathway proteins TRAF6 and IRAK1.Citation19,52 The expression of these miRNAs in response to C. albicans could possibly be explained as a negative feedback loop upon activation of PRR's signaling cascades. Their main function would be avoiding an exacerbated inflammatory response and a return to baseline levels, thus maintaining homeostasis and, therefore, exerting a protective function.
miR155 is an extremely interesting miRNA to study in this context. It is induced in macrophages by a wide variety of microorganisms, including viruses, bacteria and other fungi.Citation18,29,53–55 miR155 can dampen the inflammatory response to LPS, possibly acting as a negative feedback loop.Citation56 However, miR155 also has a pro-inflammatory effect, which probably occurs via repression of negative regulators.Citation28,57,58 It also enhances TNFα transcription, mRNA stability and translation and consequently inflammation.Citation17,59,60 Mice overexpressing miR155 have an increased propensity for developing haematopoietic cancer and chronic inflammation derived from myeloproliferative disorders, whereas mice deficient in this miRNA show decreased immune response.Citation28,40,61,62 miR155 showed to have an indirect correlation with some of its targets in our model. miR155 seems to have an anti-inflammatory role by targeting MyD88 and IKKε, while it seems to have the opposite role by targeting SHIP1 and SOCS1. This shows that miR155 has itself many functions in this process, probably being part of a very complex regulation mechanism. However, the immune response elicited by C. albicans in macrophages is a very complex network of signaling pathways, and it's extremely difficult to isolate the effects of miR155 in the targets. Further experiments using miR155 knockouts or loss and gain of function techniques such as antagomirs (miRNA antagonists) are still necessary to validate these effects of miR155 over the expression of its targets.
Thus, miR155 seems to have conflicting and paradoxical roles in inflammation depending on the context. One can speculate that the hyphal morphology, being associated with increased virulence and infection, may induce increased miR155 expression and, consequently, enhancing the inflammatory response by macrophages. This corresponds with our findings. Activation of Dectin-1 has a fundamental role in the response to C. albicans, inducing many pathways that result in an inflammatory outcome. In fact, Dectin-1 signaling is necessary to discriminate between yeasts and hyphae of C. albicans, and its deficiency results in enhanced fungal dissemination and susceptibility to infection in mice.Citation63 The distinct morphologies of C. albicans may play different roles during the infection. The hyphal form has been linked to virulence because of its capacity to mask its cell wall's β-glucan under a layer of mannans, dampening the recognition of the fungus by Dectin-1.Citation3 Although UV-killed and HK-Candida albicans cells (both yeasts and hyphae) show increased exposure of β-glucans in the cell wall when compared to live cells,Citation3 it is known that these levels of exposure vary in in vivo infections due to organ-tropism.Citation64 Also, drugs such as Caspofungin that affect cell wall integrity increase β-glucan exposure, thereby enhancing Dectin-1 activation.Citation64,65
One example of a pathway induced by Dectin-1 is the Syk pathway, which controls specific gene expression in response to the pathogen via multiple transcription factors including NFAT, JNK, p38, AP-1 and NFκB.Citation66 From those, NFκB and AP-1 have already been related to the induction of miR155 expression in response to different stimuli.Citation15,28,29,67 In this context, miR155 would have a pro-inflammatory role, resulting in enhanced inflammation when macrophages are interacting with hyphae. However, whether this increased expression of miR155 and the corresponding augmented inflammatory response will provide protection to C. albicans is still unknown, and should be further investigated.
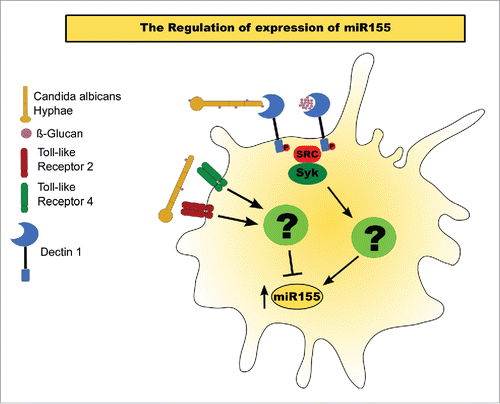
Based on our results, we propose a model of how the interaction of C. albicans hyphae with BMDM's PRRs triggers a miR155 response (). Increasing exposure of β-glucan in the C. albicans' cell wall during and after Dectin-1-mediated phagocytosis induces the expression of miR155. Our findings suggest that miR155 upregulation is controlled by the Syk signaling pathway, whereas the specific pathway through which Syk regulates this miRNA response remains unknown. However, TLR2 or TLR4 activation by C. albicans seems to reduce the accumulation of mature miR155, albeit through an unknown mechanism. With the activation of either receptor, the upregulation of this miRNA appears to be attenuated, whereas the lack of both receptors enhances miR155 when compared to the WT control. However, whether this phenomenon happens via a direct effect on mature miR155, by regulation over its transcription, or through interference with other pathways that induce miR155, such as Dectin-1 signaling, requires further study.
Figure 4. Model of miR155 activation in BMDMs exposed to C. albicans hyphae. miR155 activation by C. albicans. Exposure of β-glucans in the fungal cell wall interact with Dectin 1, inducing a Syk-dependent pathway that results in the upregulation of mature miR155. Activation of either TLR 2 or 4 inhibits the accumulation of the mature miRNA.
Taken together, our data show that miRNA responses can vary depending of the fungal cell morphology the innate immune cells, such as macrophages, are encountering. This points to the existence of enormously complex, but also highly dynamic, mechanisms intimately involved in controlling the host immune response to fungal pathogens. Further studies examining the differential modes of regulation by the 2 forms of C. albicans, as well as by pseudo hyphal cells, would augment our understanding to this process. Deciphering the complexity underlying miRNA regulation in microbial pathogenesis by genome-wide approaches, including in vivo validation, will therefore be one important avenue to pursue in order to more fully understand the mechanisms of immune response to C. albicans and other important pathogens.
Material and methods
Fungal strains (Candida albicans)
C. albicans strain SC5314 Citation68 yeast cells were kept frozen until used. Prior to interaction assays, cells were thawed and plated on YPD agar and cultivated for 2 d at 30˚C. A single colony was then picked and inoculated in liquid YPD overnight at 30˚C and 150rpm, then the yeast cells were washed in PBS and counted. Hyphal cells were obtained by incubating 5x10Citation6 yeast cells in 10mL RPMI medium supplemented with 10% fetal bovine serum for 3 hours at 37˚C and 150rpm. Cultures were monitored microscopically to ensure that more than 95% of cells were in hyphal morphology. Heat-killed cells were obtained by incubating yeast or hyphal cells at 65˚C for 3 hours in a water bath. UV-killed cells were obtained by exposing the cells in a Petri dish to UV light (Phillips Ultra-violet lamp 15W) inside a cell hood for 30 minutes. Aliquots were taken from both heat-killed and UV-killed cells and incubated in YPD plates to verify killing. The hyphal inoculum corresponds to the number of yeast-like cells that had undergone yeast-to-hyphae transition and thus showed filamentous growth morphologies.
Bone marrow-derived macrophages (BMDM)
Animal housing and all experimental procedures were approved by the Animal Ethics Committee of the University of Brasilia (UnB DOC 52657/2011), and by the Veterinärmedizinische Universität Wien (Medical University of Vienna; BMBWK-66.009/0057-II/10b/2010). Bone marrows from BALB/c and C57BL/6 mice were used for these experiments. BALB/c mice were obtained from the University of Brasilia's animal facility. C57BL/6 mice were obtained from the University of Veterinary Medicine of Vienna's animal facility. BMDMs were obtained as previously described.Citation69 Briefly, mice were euthanized, and femurs and tibias were extracted. Then, the bone marrow was flushed and ∼6×10Citation6 obtained cells were kept for 8 d in 2 Petri dishes containing RPMI medium supplemented with 20% heat-inactivated fetal bovine serum and 30% of culture supernatant from L929 fibroblasts (ATCC CCL-1). Adherent cells were then collected, counted in a cell counter (Luna) and cultured in fresh RPMI medium supplemented with 10% FBS and 5% of L929 culture supernatant. The cells were then left to adhere for 24 hours prior to the interaction experiments.
Host-pathogen interaction experiments
For interactions experiments, 106 BMDMs were cultivated overnight in a 6-well plate, as detailed above. BMDMs were stimulated with C. albicans yeasts or hyphae at a MOI from 2 to 5. For incubation with β-glucan, Zymosan depleted (Invivogen, tlrl-dzn) in a final concentration of 100 μg/mL was used. In the interaction experiments performed with Dectin-1 pathway inhibitors, BMDMs were incubated with or without 10 µM GW5074 (Sigma Aldrich, G6416), 5 µM R406 (Santa Cruz Biotech, CAS 841290-81), or both inhibitors for one hour prior to the initiation of fungal interactions.
Quantitative RT-PCR
RNA was isolated using MirVana kit (Invitrogen, AM1560) according to the manufacturer's specifications. Samples were quantified by nanodrop (Thermo Scientific). cDNAs from miRNAs were generated using 10ng of RNA from each sample according to the Taqman protocol (Life Technologies). Quantitative RT PCR for miRNA was performed using Taqman probes according to the manufacturer. U6 snRNA was used as endogenous control.
Statistical analysis
The expression level of transcripts was calculated using the ΔΔCt formula.Citation70 Differential expression was calculated using the values of ΔCt of the samples and testing with Two-way ANOVA with Tukey's posttest performed in GraphPad Prism version 5.00 for Mac, GraphPad Software, San Diego California USA, www.graphpad.com. A p-value of 0.05 or lower defined statistical significance. The results were represented using the relative fold change expression compared to the control, where the control values were considered 1.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KVIR_S_1200215.zip
Download Zip (493.4 KB)Acknowledgments
We thank the diligent work of Prof. Dr. Cynthia Maria Kyaw and Prof. Dr. Joshua Nosanchuk in reviewing and proofreading this manuscript.
Funding
This study was supported by research funding from Project PRONEX number 193.000.571/2009 from FAP-DF/CNPq. KK was supported in part by a grant from the European Commission (FP7-FUNGITECT).
References
- Moran G, Coleman D, Sullivan D. An introduction to the medically important Candida species. Candida Candidiasis; 2nd Ed 2012.
- Whiteway M, Oberholzer U. Candida morphogenesis and host-pathogen interactions. Curr Opin Microbiol 2004; 7:350-7; PMID:15358253; http://dx.doi.org/10.1016/j.mib.2004.06.005
- Bain JM, Louw J, Lewis LE, Okai B, Walls CA, Ballou ER, Walker LA, Reid D, Munro CA, Brown AJP, et al. Candida albicans Hypha Formation and Mannan Masking of β-Glucan Inhibit Macrophage Phagosome Maturation. mBio 2014; 5:e01874-14; PMID:25467440; http://dx.doi.org/10.1128/mBio.01874-14
- Saville SP, Lazzell AL, Bryant AP, Fretzen A, Monreal A, Solberg EO, Monteagudo C, Lopez-Ribot JL, Milne GT. Inhibition of filamentation can be used to treat disseminated candidiasis. Antimicrob Agents Chemother 2006; 50:3312-6; PMID:17005810; http://dx.doi.org/10.1128/AAC.00628-06
- Staniszewska M, Bondaryk M, Rabczenko D, Smolenska-Sym G, Kurzatkowski W. Cell wall carbohydrates content of pathogenic Candida albicans strain morphological forms. Med Dosw Mikrobiol 2013; 65:119-28; PMID:24180139
- Jouault T, Sarazin A, Martinez-Esparza M, Fradin C, Sendid B, Poulain D. Host responses to a versatile commensal: PAMPs and PRRs interplay leading to tolerance or infection by Candida albicans. Cell Microbiol 2009; 11:1007-15; PMID:19388906; http://dx.doi.org/10.1111/j.1462-5822.2009.01318.x
- Gow NAR, Netea MG, Munro CA, Ferwerda G, Bates S, Mora‐Montes HM, Walker L, Jansen T, Jacobs L, Tsoni V, et al. Immune Recognition of Candida albicans β‐glucan by Dectin‐1. J Infect Dis 2007; 196:1565-71; PMID:18008237; http://dx.doi.org/10.1086/523110
- van der Graaf CA, Netea MG, Verschueren I, van der Meer JW, Kullberg BJ. Differential cytokine production and Toll-like receptor signaling pathways by Candida albicans blastoconidia and hyphae. Infect Immun 2005; 73:7458-64; PMID:16239547; http://dx.doi.org/10.1128/IAI.73.11.7458-7464.2005
- Blasi E, Mucci A, Neglia R, Pezzini F, Colombari B, Radzioch D, Cossarizza A, Lugli E, Volpini G, Del Giudice G, et al. Biological importance of the two Toll-like receptors, TLR2 and TLR4, in macrophage response to infection with Candida albicans. FEMS Immunol Med Microbiol 2005; 44:69-79; PMID:15849871; http://dx.doi.org/10.1016/j.femsim.2004.12.005
- Gasparoto TH, Tessarolli V, Garlet TP, Torres SA, Garlet GP, da Silva JS, Campanelli AP. Absence of functional TLR4 impairs response of macrophages after Candida albicans infection. Med Mycol 2010; 48:1009-17; PMID:20465519; http://dx.doi.org/10.3109/13693786.2010.481292
- Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, et al. Dectin-1 is required for β-glucan recognition and control of fungal infection. Nat Immunol 2007; 8:31-8; PMID:17159984; http://dx.doi.org/10.1038/ni1408
- Bourgeois C, Kuchler K. Fungal pathogens-a sweet and sour treat for toll-like receptors. Front Cell Infect Microbiol 2012; 2:142; PMID:23189270; http://dx.doi.org/10.3389/fcimb.2012.00142
- Gringhuis SI, den Dunnen J, Litjens M, van der Vlist M, Wevers B, Bruijns SC, Geijtenbeek TB. Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-kappaB activation through Raf-1 and Syk. Nat Immunol 2009; 10:203-13; PMID:19122653; http://dx.doi.org/10.1038/ni.1692
- Bourgeois C, Majer O, Frohner IE, Lesiak-Markowicz I, Hildering KS, Glaser W, Stockinger S, Decker T, Akira S, Muller M, et al. Conventional dendritic cells mount a type I IFN response against Candida spp. requiring novel phagosomal TLR7-mediated IFN-beta signaling. J Immunol 2011; 186:3104-12; PMID:21282509; http://dx.doi.org/10.4049/jimmunol.1002599
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U A 2006; 103:12481-6; http://dx.doi.org/10.1073/pnas.0605298103
- Schulte LN, Westermann AJ, Vogel J. Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res 2013; 41:542-53; PMID:23143100; http://dx.doi.org/10.1093/nar/gks1030
- Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 2007; 179:5082-9; PMID:17911593; http://dx.doi.org/10.4049/jimmunol.179.8.5082
- Das Gupta M, Fliesser M, Springer J, Breitschopf T, Schlossnagel H, Schmitt A-L, Kurzai O, Hünniger K, Einsele H, Löffler J. Aspergillus fumigatus induces microRNA-132 in human monocytes and dendritic cells. Int J Med Microbiol 2014; 304:592-6; PMID:24841251; http://dx.doi.org/10.1016/j.ijmm.2014.04.005
- Park H, Huang X, Lu C, Cairo MS, Zhou X. miR-146a and miR-146b Regulate Human Dendritic Cell Apoptosis and Cytokine Production by Targeting of TRAF6 and IRAK1. J Biol Chem [Internet] 2014; 290(5):2831-41; Available from: http://www.ncbi.nlm.nih.gov/pubmed/25505246
- Cheng HS, Sivachandran N, Lau A, Boudreau E, Zhao JL, Baltimore D, Delgado-Olguin P, Cybulsky MI, Fish JE. MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol Med 2013; 5:949-66; PMID:23733368; http://dx.doi.org/10.1002/emmm.201202318
- Guo Z, Gu Y, Wang C, Zhang J, Shan S, Gu X, Wang K, Han Y, Ren T. Enforced expression of miR-125b attenuates LPS-induced acute lung injury. Immunol Lett 2014; 162:18-26; PMID:25004393; http://dx.doi.org/10.1016/j.imlet.2014.06.008
- El Gazzar M, McCall CE. MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. J Biol Chem 2010; 285:20940-51; PMID:20435889; http://dx.doi.org/10.1074/jbc.M110.115063
- Zhang L, Huang D, Wang Q, Shen D, Wang Y, Chen B, Zhang J, Gai L. MiR-132 inhibits expression of SIRT1 and induces pro-inflammatory processes of vascular endothelial inflammation through blockade of the SREBP-1c metabolic pathway. Cardiovasc Drugs Ther 2014; 28:303-11; PMID:24924687; http://dx.doi.org/10.1007/s10557-014-6533-x
- Kong H, Yin F, He F, Omran A, Li L, Wu T, Wang Y, Peng J. The Effect of miR-132, miR-146a, and miR-155 on MRP8/TLR4-Induced Astrocyte-Related Inflammation. J Mol Neurosci MN 2015; 57:28-37; PMID:25957996; http://dx.doi.org/10.1007/s12031-015-0574-x
- Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci 2009; 106:5282-5287; PMID:19289835; http://dx.doi.org/10.1073/pnas.0810909106
- Starczynowski DT, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, Muranyi A, Hirst M, Hogge D, Marra M, Wells RA, et al. Identification of miR-145 and miR-146a as mediators of the 5q– syndrome phenotype. Nat Med 2010; 16:49-58; PMID:19898489; http://dx.doi.org/10.1038/nm.2054
- Chen Q, Wang H, Liu Y, Song Y, Lai L, Han Q, Cao X, Wang Q. Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1beta production in macrophages by targeting STAT3. PloS One 2012; 7:e42971; PMID:22937006; http://dx.doi.org/10.1371/journal.pone.0042971
- O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U A 2009; 106:7113-8; http://dx.doi.org/10.1073/pnas.0902636106
- Koch M, Mollenkopf HJ, Klemm U, Meyer TF. Induction of microRNA-155 is TLR- and type IV secretion system-dependent in macrophages and inhibits DNA-damage induced apoptosis. Proc Natl Acad Sci U A 2012; 109:E1153-62; http://dx.doi.org/10.1073/pnas.1116125109
- Monk CE, Hutvagner G, Arthur JSC. Regulation of miRNA Transcription in Macrophages in Response to Candida albicans. PLoS ONE 2010; 5:e13669; PMID:21060679; http://dx.doi.org/10.1371/journal.pone.0013669
- Hnisz D, Bardet AF, Nobile CJ, Petryshyn A, Glaser W, Schöck U, Stark A, Kuchler K. A Histone Deacetylase Adjusts Transcription Kinetics at Coding Sequences during Candida albicans Morphogenesis. PLoS Genet 2012; 8:e1003118; PMID:23236295; http://dx.doi.org/10.1371/journal.pgen.1003118
- Galan-Diez M, Arana DM, Serrano-Gomez D, Kremer L, Casasnovas JM, Ortega M, Cuesta-Dominguez A, Corbi AL, Pla J, Fernandez-Ruiz E. Candida albicans beta-glucan exposure is controlled by the fungal CEK1-mediated mitogen-activated protein kinase pathway that modulates immune responses triggered through dectin-1. Infect Immun 2010; 78:1426-36; PMID:20100861; http://dx.doi.org/10.1128/IAI.00989-09
- Gazi U, Rosas M, Singh S, Heinsbroek S, Haq I, Johnson S, Brown GD, Williams DL, Taylor PR, Martinez-Pomares L. Fungal recognition enhances mannose receptor shedding through dectin-1 engagement. J Biol Chem 2011; 286:7822-9; PMID:21205820; http://dx.doi.org/10.1074/jbc.M110.185025
- Tang B, Xiao B, Liu Z, Li N, Zhu E-D, Li B-S, Xie Q-H, Zhuang Y, Zou Q-M, Mao X-H. Identification of MyD88 as a novel target of miR-155, involved in negative regulation of Helicobacter pylori-induced inflammation. FEBS Lett 2010; 584:1481-6; PMID:20219467; http://dx.doi.org/10.1016/j.febslet.2010.02.063
- Huang RS, Hu GQ, Lin B, Lin ZY, Sun CC. MicroRNA-155 silencing enhances inflammatory response and lipid uptake in oxidized low-density lipoprotein-stimulated human THP-1 macrophages. J Investig Med 2010; 58:961-7; PMID:21030878; http://dx.doi.org/10.2310/JIM.0b013e3181ff46d7
- Cremer TJ, Ravneberg DH, Clay CD, Piper-Hunter MG, Marsh CB, Elton TS, Gunn JS, Amer A, Kanneganti T-D, Schlesinger LS, et al. MiR-155 Induction by F. novicida but Not the Virulent F. tularensis Results in SHIP Down-Regulation and Enhanced Pro-Inflammatory Cytokine Response. PLoS ONE [Internet] 2009; 4(12):e8508 [cited 2016 Mar 4]; 4. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2794384/; PMID:AMBIGUOUS
- Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi J-TA, Braich R, Manoharan M, Soutschek J, Ohler U, et al. A viral microRNA functions as an ortholog of cellular miR-155. Nature 2007; 450:1096-9; PMID:18075594; http://dx.doi.org/10.1038/nature05992
- Xiao B, Liu Z, Li B-S, Tang B, Li W, Guo G, Shi Y, Wang F, Wu Y, Tong W-D, et al. Induction of microRNA-155 during Helicobacter pylori Infection and Its Negative Regulatory Role in the Inflammatory Response. J Infect Dis 2009; 200:916-25; PMID:19650740; http://dx.doi.org/10.1086/605443
- Worm J, Stenvang J, Petri A, Frederiksen KS, Obad S, Elmén J, Hedtjärn M, Straarup EM, Hansen JB, Kauppinen S. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF. Nucleic Acids Res 2009; 37:5784-92; PMID:19596814; http://dx.doi.org/10.1093/nar/gkp577
- Costinean S, Sandhu SK, Pedersen IM, Tili E, Trotta R, Perrotti D, Ciarlariello D, Neviani P, Harb J, Kauffman LR, et al. Src homology 2 domain–containing inositol-5-phosphatase and CCAAT enhancer-binding protein β are targeted by miR-155 in B cells of Eμ-MiR-155 transgenic mice. Blood 2009; 114:1374-82; PMID:19520806; http://dx.doi.org/10.1182/blood-2009-05-220814
- Takahara K, Tokieda S, Nagaoka K, Inaba K. Efficient capture of Candida albicans and zymosan by SIGNR1 augments TLR2-dependent TNF-α production. Int Immunol 2012; 24:89-96; PMID:22207132; http://dx.doi.org/10.1093/intimm/dxr103
- Xu R, Sun H-F, Williams DW, Jones AV, Al-Hussaini A, Song B, Wei X-Q, Xu R, Sun H-F, Williams DW, et al. IL-34 Suppresses Candida albicans Induced TNFα Production in M1 Macrophages by Downregulating Expression of Dectin-1 and TLR2, IL-34 Suppresses Candida albicans Induced TNFα Production in M1 Macrophages by Downregulating Expression of Dectin-1 and TLR2. J Immunol Res J Immunol Res 2015; 2015; 2015:e328146
- Blanco-Menéndez N, Fresno C del, Fernandes S, Calvo E, Conde-Garrosa R, Kerr WG, Sancho D. SHIP-1 Couples to the Dectin-1 hemITAM and Selectively Modulates Reactive Oxygen Species Production in Dendritic Cells in Response to Candida albicans. J Immunol 2015; 195:4466-78; http://dx.doi.org/10.4049/jimmunol.1402874
- Ganesan S, Rathinam VAK, Bossaller L, Army K, Kaiser WJ, Mocarski ES, Dillon CP, Green DR, Mayadas TN, Levitz SM, et al. Caspase-8 Modulates Dectin-1 and Complement Receptor 3–Driven IL-1β Production in Response to β-Glucans and the Fungal Pathogen, Candida albicans. J Immunol 2014; 193:2519-30; PMID:25063877; http://dx.doi.org/10.4049/jimmunol.1400276
- Muhammad SA, Fatima N, Syed N-H, Wu X, Yang XF, Chen JY. MicroRNA Expression Profiling of Human Respiratory Epithelium Affected by Invasive Candida Infection. PLoS ONE 2015; 10:e0136454; PMID:26313489; http://dx.doi.org/10.1371/journal.pone.0136454
- Cai P, Piao X, Liu S, Hou N, Wang H, Chen Q. MicroRNA-gene expression network in murine liver during Schistosoma japonicum infection. PloS One 2013; 8:e67037; PMID:23825609; http://dx.doi.org/10.1371/journal.pone.0067037
- Dorhoi A, Iannaccone M, Farinacci M, Faé KC, Schreiber J, Moura-Alves P, Nouailles G, Mollenkopf H-J, Oberbeck-Müller D, Jörg S, et al. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J Clin Invest 2013; 123:4836-48; PMID:24084739; http://dx.doi.org/10.1172/JCI67604
- Gong AY, Hu G, Zhou R, Liu J, Feng Y, Soukup GA, Chen XM. MicroRNA-221 controls expression of intercellular adhesion molecule-1 in epithelial cells in response to Cryptosporidium parvum infection. Int J Parasitol 2011; 41:397-403; PMID:21236259; http://dx.doi.org/10.1016/j.ijpara.2010.11.011
- Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I, Leiser A, Schwartz PE, Rutherford T, Mor G. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene 2008; 27:4712-23; PMID:18408758; http://dx.doi.org/10.1038/onc.2008.112
- Ehrenborg E. MicroRNA-9 regulates the expression of peroxisome proliferator-activated receptor δ in human monocytes during the inflammatory response. Int J Mol Med [Internet] 2013; 31(5):1003-10 [cited 2015 Oct 22]; Available from:http://www.spandidos-publications.com/10.3892/ijmm.2013.1311; PMID:23525285
- Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S, et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation 2012; 125:2892-903; PMID:22580331; http://dx.doi.org/10.1161/CIRCULATIONAHA.111.087817
- Saba R, Gushue S, Huzarewich RL, Manguiat K, Medina S, Robertson C, Booth SA. MicroRNA 146a (miR-146a) is over-expressed during prion disease and modulates the innate immune response and the microglial activation state. PloS One 2012; 7:e30832; PMID:22363497; http://dx.doi.org/10.1371/journal.pone.0030832
- Bandyopadhyay S, Long ME, Allen LA. Differential expression of microRNAs in Francisella tularensis-infected human macrophages: miR-155-dependent downregulation of MyD88 inhibits the inflammatory response. PloS One 2014; 9:e109525; PMID:25295729; http://dx.doi.org/10.1371/journal.pone.0109525
- Kumar R, Halder P, Sahu SK, Kumar M, Kumari M, Jana K, Ghosh Z, Sharma P, Kundu M, Basu J. Identification of a novel role of ESAT-6-dependent miR-155 induction during infection of macrophages with Mycobacterium tuberculosis. Cell Microbiol 2012; 14:1620-31; PMID:22712528; http://dx.doi.org/10.1111/j.1462-5822.2012.01827.x
- Pareek S, Roy S, Kumari B, Jain P, Banerjee A, Vrati S. MiR-155 induction in microglial cells suppresses Japanese encephalitis virus replication and negatively modulates innate immune responses. J Neuroinflammation 2014; 11:97; PMID:24885259; http://dx.doi.org/10.1186/1742-2094-11-97
- Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci U A 2009; 106:2735-40; http://dx.doi.org/10.1073/pnas.0811073106
- Contreras J, Rao DS. MicroRNAs in inflammation and immune responses. Leukemia 2012; 26:404-13; PMID:22182919; http://dx.doi.org/10.1038/leu.2011.356
- Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, Margioris AN, Tsichlis PN, Tsatsanis C. The Kinase Akt1 Controls Macrophage Response to Lipopolysaccharide by Regulating MicroRNAs. Immunity 2009; 31:220-31; PMID:19699171; http://dx.doi.org/10.1016/j.immuni.2009.06.024
- Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P, Szabo G. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem 2011; 286:1436-44; PMID:21062749; http://dx.doi.org/10.1074/jbc.M110.145870
- Rajaram MV, Ni B, Morris JD, Brooks MN, Carlson TK, Bakthavachalu B, Schoenberg DR, Torrelles JB, Schlesinger LS. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc Natl Acad Sci U A 2011; 108:17408-13; http://dx.doi.org/10.1073/pnas.1112660108
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eμ-miR155 transgenic mice. Proc Natl Acad Sci 2006; 103:7024-9; PMID:16641092; http://dx.doi.org/10.1073/pnas.0602266103
- O’Connell RM, Kahn D, Gibson WSJ, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 Promotes Autoimmune Inflammation by Enhancing Inflammatory T Cell Development. Immunity 2010; 33:607-19; http://dx.doi.org/10.1016/j.immuni.2010.09.009
- Cheng SC, van de Veerdonk FL, Lenardon M, Stoffels M, Plantinga T, Smeekens S, Rizzetto L, Mukaremera L, Preechasuth K, Cavalieri D, et al. The dectin-1/inflammasome pathway is responsible for the induction of protective T-helper 17 responses that discriminate between yeasts and hyphae of Candida albicans. J Leukoc Biol 2011; 90:357-66; PMID:21531876; http://dx.doi.org/10.1189/jlb.1210702
- Wheeler RT, Kombe D, Agarwala SD, Fink GR. Dynamic, morphotype-specific Candida albicans beta-glucan exposure during infection and drug treatment. PLoS Pathog 2008; 4:e1000227; PMID:19057660; http://dx.doi.org/10.1371/journal.ppat.1000227
- Mora-Montes HM, Netea MG, Ferwerda G, Lenardon MD, Brown GD, Mistry AR, Kullberg BJ, O’Callaghan CA, Sheth CC, Odds FC, et al. Recognition and blocking of innate immunity cells by Candida albicans chitin. Infect Immun 2011; 79:1961-70; PMID:21357722; http://dx.doi.org/10.1128/IAI.01282-10
- Rizzetto L, Buschow SI, Beltrame L, Figdor CG, Schierer S, Schuler G, Cavalieri D. The Modular Nature of Dendritic Cell Responses to Commensal and Pathogenic Fungi. PLoS ONE 2012; 7:e42430; PMID:22879980; http://dx.doi.org/10.1371/journal.pone.0042430
- Imaizumi T, Tanaka H, Tajima A, Yokono Y, Matsumiya T, Yoshida H, Tsuruga K, Aizawa-Yashiro T, Hayakari R, Inoue I, et al. IFN-gamma and TNF-alpha synergistically induce microRNA-155 which regulates TAB2/IP-10 expression in human mesangial cells. Am J Nephrol 2010; 32:462-8; PMID:20948191; http://dx.doi.org/10.1159/000321365
- Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5’-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 1984; 198:179-82; PMID:6394964; http://dx.doi.org/10.1007/BF00328721
- Bourgeois C, Majer O, Frohner I, Kuchler K. In vitro systems for studying the interaction of fungal pathogens with primary cells from the mammalian innate immune system. Methods Mol Biol Clifton NJ 2009; 470:125-39; http://dx.doi.org/10.1007/978-1-59745-204-5_11
- Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/10.1006/meth.2001.1262
