ABSTRACT
To cope with hyperosmotic stress encountered in the environments and in the host, the pathogenic as well as non-pathogenic microbes use diverse transport systems to obtain osmoprotectants. To study the role of Shigella sonnei ProU system in response to hyperosmotic stress and virulence, we constructed deletion and complementation strains of proV and used an RNAi approach to silence the whole ProU operon. We compared the response between wild type and the mutants to the hyperosmotic pressure in vitro, and assessed virulence properties of the mutants using gentamicin protection assay as well as Galleria mellonella moth larvae model. In response to osmotic stress by either NaCl or KCl, S. sonnei highly up-regulates transcription of proVWX genes. Supplementation of betaine greatly elevates the growth of the wild type S. sonnei but not the proV mutants in M9 medium containing 0.2 M NaCl or 0.2 M KCl. The proV mutants are also defective in intracellular growth compared with the wild type. The moth larvae model of G. mellonella shows that either deletion of proV gene or knockdown of proVWX transcripts by RNAi significantly attenuates virulence. ProU system in S. sonnei is required to cope with osmotic stress for survival and multiplication in vitro, and for infection.
Introduction
Shigella is a facultative intracellular Gram-negative pathogen, known as the etiologic agent of bacillary dysentery since the 1890s. Although Shigella was defined a genus with 4 species S. dysenteriae, S. flexneri, S. boydii and S. sonnei in the 1950s,Citation1 it has become clear that they are pathogenic lineages of Escherichia coli of multiple origins.Citation2 The primary transmission is the fecal-oral route, so it is life threatening in developing countries because of poor sanitation. Shigella strains are among the most prevalent causative agents of moderate-to-severe diarrhea, and especially affect children under 5 y old in developing countries.Citation3 The widespread of multiple antibiotic resistant strains, has made Shigella treatment increasingly difficult and there is urgent need for vaccine development.Citation4 Shigella is highly invasive to the colon and the rectum and it has the ability to proliferate in the cell cytoplasm and trigger the host pro-inflammatory response. It causes variable clinical manifestations ranging from short term illness, typically watery diarrhea, to a long lasting one manifesting with fever, bloody diarrhea with intestinal cramps and mucopurulent feces.Citation5
Keeping a stable osmotic balance between the cell cytoplasm and the outer environment is an important challenge to all cell types, especially the unicellular organisms. For bacteria, the high surface area to total volume ratios makes them vulnerable when they encounter osmotic stress; bacteria could tolerate the osmolarity changes in the environment through either solutes efflux or water movement across the cytoplasmic membrane.Citation6 The external osmolarity changes are translated by the microorganisms to an adaptation process; this happens by a rapid K+ ion influx through specific transporters, and at the same time, microorganisms produce counter ions like glutamate.Citation7 However, high intracellular concentration of K+ and glutamate only support microbial adaptation to moderately high osmolarity. At very high osmolarity, further accumulation of K+ and glutamate becomes impossible for growth, and therefore, bacteria exploit less deleterious compounds called osmoprotectants, like polyoles (trehalose), amino acids (proline), and methyl-amines (glycine betaine).Citation7 Osmoprotectants can accumulate intracellularly via cellular uptake or synthesis from their precursors. Glycine betaine is one of the most important osmoprotectants for bacteria.Citation7
For pathogenic bacteria, osmoregulation is a very important factor for establishing infection. For example, Staphylococcus aureus has a PutP proline transport system that helps in host tissue colonization.Citation8-10 Other studies have found a link between osmotic stress and the expression of virulence genes in Pseudomonas aeruginosa Citation11,12 and between the transport of compatible solutes and pathogen colonization in Listeria monocytogenes.Citation13,14 In Salmonella enterica serovar typhimurium and E. coli K-12, osmoprotectants are mainly accumulated through the ProP and ProU transport systems.Citation15-19
ProP is a member of the major facilitator superfamily of permeases, and it is known as a symporter.Citation20 ProP is of low affinity for proline and glycine betaine, it has Km of ≈0.1 mM for both.Citation20 ProU system efficiently scavenges glycine betaine,Citation21 Km of ≈1 µM,Citation16 as well as proline betaine for bacteria to cope extreme osmotic stress.Citation22,23 It is composed of 3 proteins, i.e., ProV, ProW, and ProX, which are encoded by an operon proVWX,Citation18 of which ProV belongs to the ATP-binding cassette (ABC) superfamily.Citation24,25 ProX is a periplasmic soluble substrate-binding proteinCitation19,26,27 which could bind and deliver glycine betaine to the inner membrane protein ProW, whereas ProV hydrolyses ATP providing energy for transporting substrates against the concentration gradient.Citation24,25
In 2005, Lucchini and his colleagues analyzed genomic expression of S. flexneri during infection by DNA microarray.Citation28 proVWX genes were found highly up-regulated in both epithelial HeLa and macrophage-like U937 cells; in particular, proV was upregulated by 57-fold in U937 cells.Citation28 Besides, previous studies found that the level of ProU transcription in E. coli is induced upon exposure to hyperosmotic stress.Citation29 These findings suggested that Shigella faced extreme osmotic stress in the host cell cytosol and up-regulation of ProU was necessary for Shigella to cope with this hostile cellular niche, allowing Shigella intracellular survival and growth to establish infection. Additionally, the orthologous ProXVWZ system in Mycobacterium tuberculosis has been shown to actively transport glycine betaine into macrophages, which contributed to early steps in colonization of the cellular niche.Citation30 Hence, we have investigated the impact of the ProU system on osmotic stress response and pathogenesis of S. sonnei. Our data have shown that the ProU transport system is important for S. sonnei to cope with hyperosmotic stress in the host cell cytoplasm, and for rapid intracellular proliferation to establish infection.
Results
proV deletion and complementation
In order to study the function of ProU we decided to construct a proV deletion mutant since proV is the most highly upregulated gene inside host cells and the first gene in the proVWX operon.Citation28 ProV deletion was constructed in wild type S. sonnei strain (20071599) Citation31 using the phage λ Red recombination system.Citation32 All proV constructs for mutagenesis as well as complementation were confirmed by PCR and agarose gel electrophoresis (Fig. S1). All PCR products were further confirmed by DNA sequencing using primers c and d (Table S1).
ProU is required to cope with hyperosmotic stress in vitro
Because the ProU system is known to be required for E. coli to cope with extreme osmotic stress,Citation22,23 we tested whether this was also the case for S. sonnei. We first measured growth of the wild type, ΔproV and the complemented ΔproV/pProV strains in M9 medium supplemented with 0.3 M NaCl or 0.3 M KCl by measuring the optical density (OD600nm) each hour untill the late stationary phase (10 hours). While the wild type S. sonnei grew well, the growth of ΔproV strain was impaired in the presence of 0.3 M of NaCl or KCl (). All the tested strains were able to grow properly in M9 medium without any supplements (Fig. S2). Overexpression of ProV in trans not only reversed the growth defect of the ΔproV strain, but also made the strain grow faster than the wild type. This suggested that deletion of proV is solely responsible for the slow growth phenotype of the mutant and also suggested that excess ProV made the system more effective and that ProU may also transport osmoprotectants other than betaine, such as K+, available in the medium to facilitate bacterial growth (). We further tested the growth of wild type and the ΔproV strains in M9 medium supplemented with 0.2 M NaCl or 0.2 M KCl. Under these milder conditions, the ΔproV strain grew equally well as the wild type strain up to 6 hours. However, the growth of the ΔproV strain ceased at 6 hours and remained flat to 10 hours. In contrast, the wild type grew steadily, reaching OD600nm = 0.7 and 0.8 in M9 supplemented with 0.2 M NaCl and 0.2 M KCl, respectively (). Thus, in the presence of 0.2 M NaCl, growth of ΔproV was more than 1.5-fold reduced (OD600nm 0.4 vs. 0.7) compared to the wild type at the stationary phase 10 hours (). In the presence of 0.2 M KCl, there was a 2-fold reduction of ΔproV growth (OD600nm 0.4 vs. 0.8) compared to the wild type at the stationary phase ().
Figure 1. S. sonnei growth under various osmotic conditions. Growth curves of wild type (triangle), proV mutant (gray square), MirproV (RNAi proV strain) (gray trigonal) and ΔproV /pProV strains (circle) in M9 medium supplemented with 0.3 M NaCl (A), 0.3 M KCl (B), 0.2 M NaCl (C, E) and 0.2 M KCl (D, F) in the presence (open symbols) or absence (closed symbols) of 500 µM betaine (C, D, E, F). The results are means of 3 successive wells ± standard deviation (n = 3).
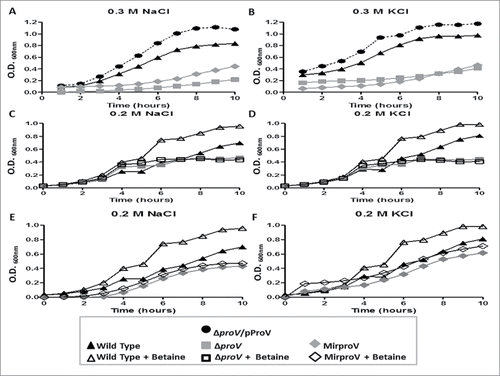
Betaine is a well-known compatible solute that is a preferable osmoprotectant for many organisms.Citation33 We therefore tested the ability of both wild type and the ΔproV mutant in utilizing betaine under hyperosmotic conditions. In M9 medium containing 0.2 M NaCl or 0.2 M KCl, supplementation of betaine (500 µM) significantly elevated the growth of the wild type (, S3). In contrast, the growth suppression of the ΔproV mutant persisted in the presence of 500 µM betaine (, S3). Taken together, these data demonstrated that a functional ProU system is required to transport betaine for S. sonnei, and that betaine can correct the growth defects of the wild type induced by high concentrations of NaCl and KCl. In contrast, the growth of the ΔproV mutant remained severely impaired under high osmotic stress, and could not be corrected by addition of betaine in the medium ().
proV paralogues are able to compensate the loss of proV
As shown in , the ΔproV strain grew equally well as the wild type strain in the first 6 hours albeit its growth was ceased thereafter. This result suggested that the ProU system was functional at least in the first 6 hours when proV is removed. Given the fact that proV encodes an ATPase and all ATPase of the ABC superfamily share highly conserved sequence and structure,Citation34 we reasoned that ATPase from other transporting systems may compensate for the loss of proV. Using ProV as query sequence we identified putative 25 ATPases (Table S3), belonging to other transport systems in the S. sonnei SSO46 genome (http://www.mgc.ac.cn/ShiBASE/Search.htm). We cloned 3 of them in random, oppF, glnQ and malK, which are responsible for transporting oligopeptides, glutamate, and maltose, respectively. Over-expressing each of them was able to complement the ΔproV strain for better growth in LB broth supplemented with 0.2 M NaCl although OppF appeared less effective than GlnQ and MalK (Fig. S5). These data support our hypothesis that removal of proV only rendered ProU system partially inactive. Since the proV::Kan intermediate strain used for producing ΔproV strain potentially had polar effect on expression of proWX, we performed complementation test for growth in L-broth. As seen in , expression of proV in trans fully complemented the ΔproV strain but not the proV::Kan strain for growth in L-broth (Fig. S7). Thus, ProU system alone is required for S. sonnei growth under the hyper osmotic condition used. To fully assess the ProU function and regulation, it was necessary to delete the whole operon. However, despite repeated efforts we were unable to obtain a ΔproVWX, or ΔproX or ΔproW strain with intact virulence plasmid, which would allow formation of small and smooth red colonies on Congo red agar. All strains harboring these deletions formed large pale and rough colonies (data not shown). Thus, S. sonnei must rearrange its genome when completely loosing ProU. Hence, we searched for an alternative approach for knockdown of the ProU system.
Silencing the proVWX operon using RNAi
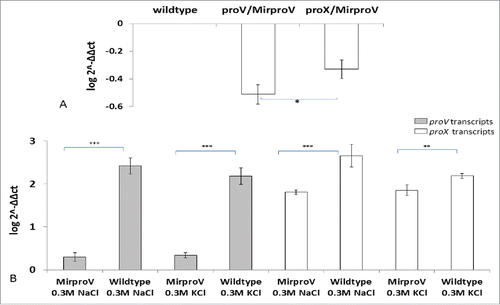
Tchurikov and his colleaguesCitation35 described an RNAi methodology to knockdown gene expression in E. coli. According to their work Citation35 Mirlon (which expresses the inverted sequence of the gene to be knocked down, lon), is the most potent in silencing the target genes. We adopted described Mirlon approach to knockdown ProU system in S. sonnei, hence termed MirproV RNA.Citation35 We tested the impact of MirproV on the expression of proV and proX, which are the first and last genes, respectively, in the ProU operon, by qRT-PCR using primers (k and l; m and n respectively, Table S1). The house-keeping gene, cysG, was used as an internal control using primers (o and p, Table S1).Citation36 shows the results of qRT-PCR after normalization with transcripts of cysG and using transcripts from wild type strain as calibrator. The transcripts of both proV and proX were significantly reduced as a result of MirproV RNA expression, which indicated that the RNAi construct in MirproV was successful in ProU attenuation.
Figure 2. qRT-PCR analysis for both proV and proX transcripts in both wild type and MirproV. (A) qRT-PCR analysis of wild type and MirproV strains growth in M9 medium without supplements. Levels of proV and proX transcripts in wild type were set as calibrator (one) and levels of transcripts in MirproV strain were calculated using the 2−ΔΔct method; significant drops of proV and proX transcripts were observed in MirproV strain. Levels of transcription between proV and proX also significantly differ in the MirproV strain (*p< 0.05). (B) qRT-PCR analysis for the expression of proV (gray columns) and proX (open columns) genes in both wild type and MirproV grown in hyperosmotic media. The differences in proV transcripts between wild type and MirproV strains in presence of both 0.3 M NaCl and KCl are highly significant (***p < 0.0001; gray columns); and the differences in proX transcripts between wild type and MirproV in the presence of 0.3 M NaCl or 0.3 M KCl are highly significant (***p = 0.0007, and **p = 0.0025, respectively). All the results are means of 3 successive groups ± standard deviation (n = 3).
Similar to the proV mutant, the MirproV strain grew as well as the wild type in M9 without supplements (Fig. S2) but became intolerant to hyperosmolarity; its growth was severely compromised in M9 medium containing either 0.3 M NaCl or 0.3 M KCl (). In M9 supplemented with 0.2 M NaCl, the MirproV strain had a long lag phase of 4 hours (). This was in contrast to the ΔproV strain, which grew equally well as the wild type strain during this phase of growth under this condition (). Thus, MirproV was more effective in inactivating the ProU system. Moreover, addition of betaine to M9 medium containing 0.2 M NaCl or 0.2 M KCl failed to rescue growth of the MirproV strain (, S3). Thus, expression of MirproV is effective in blocking ProU function and betaine transport.
By linear regression analysis, the significance of inhibition of wild type, proV and MirproV mutants growth by 0.2 M NaCl and 0.2 M KCl supplements was identified (Fig. S3). Furthermore, paired t-test was performed to compare optical density (OD600nm) at the stationary for wild type, ΔproV and MirproV in presence and absence of betaine (Fig. S4). The results showed that betaine supplementation significantly increased the optical density for wild type but not for either mutant strains (Fig. S4).
To gain insight into the transcriptional regulation of ProU and its inactivation by MirproV, we isolated total RNA from wild type and the MirproV strain from M9 cultures containing either 0.3M NaCl or 0.3M KCl and performed qRT-PCR for transcripts of proV and proX. Again the house-keeping gene cysG was used as an internal control for normalization. As expected, the wild type responded to the osmotic stress of NaCl or KCl by massive upregulation of proV and proX transcripts. The MirproV strain also responded, but at a significantly lower level (). These data demonstrated that the ProU system was inducible under high osmotic stress and that MirproV RNA effectively knocked down ProU transcripts which in turn rendered S. sonnei intolerant to hyperosmolarity.
Inactivation of ProU does not disrupt the type 3 secretion system (TTSS)
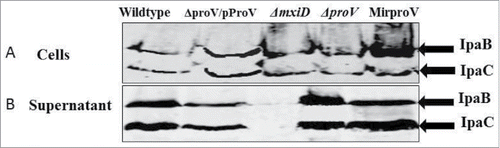
Shigella strains possess a type III secretion system (TTSS), which is essential for cell-invasion, phagosome escape and intracellular replication.Citation37 Therefore, we ought to separate the roles of TTSS and ProU with regard to cell invasion and intracellular growth. We first confirmed that all the mutant strains contained intact TTSS genes (ipaB and mxiD) using primers listed in Table S1. Further, the production and secretion of IpaB and IpaC proteins were investigated by Western blotting using Congo red as an environmental cue in vitro as described previously.Citation38 Because it is known that MxiD is required for type III secretion, we constructed a ΔmxiD mutant as a negative control for Ipa secretion, using the same approach as for ΔproV construction with primers (g and h; Table S1). Similar levels of IpaB and IpaC were detected in the cell lysates and supernatants of the wild type, the ΔproV, the MirproV and the complemented ΔproV/pProV strains whereas Ipa proteins were detected only in cell lysate but not supernatant of the ΔmxiD mutant (). We therefore conclude that removal of proV or knockdown of transcripts did not impair IpaB and IpaC production or secretion – TTSS is functional in the ΔproV and the MirproV strains.
Figure 3. Deletion of proV doesn't disturb the function of TTSS. S. sonnei wild type, the complemented strain ΔproV/PproV, ΔmxiD, ΔproV and MirproV strains were grown to mid-log phase, and TTSS secretion was induced with Congo red. Total proteins from cell lysates (A) and culture supernatants (B) were separated on SDS-PAGE and IpaB and IpaC were detected with anti-IpB and anti-IpaC antibodies.
ProU is required for S. sonnei intracellular growth (ex vivo)
Evidence via DNA microarray analysis indicated that the host cell cytoplasm is a hostile environment which exposes bacteria to hyperosmotic stress.Citation28 We therefore hypothesized that removal of proV or reduction of proVWX transcription by RNAi would adversely influence S. sonnei intracellular growth. To test our hypothesis, the HEK293 cell line, a good model for testing Shigella invasion,Citation39 was used as a host to perform gentamicin protection assay to test intracellular growth of the wild type, the ΔproV and the MirproV strains. As anticipated, we observed a significant drop in intracellular bacterial burden with the ΔproV mutant compared to the wild type 2 hours post infection (). Noticeably, the MirproV strain had a significantly reduced intracellular bacterial burden, compared to the ΔproV mutant. This suggested that suppression of proVWX operon was more effective than mutating proV gene alone because ProV paralogues may compensate the loss of ProV (Fig. S5). As anticipated, expressing ProV in trans restored intracellular growth of the ΔproV mutant, demonstrating that the deletion of proV had no polar effect on the expression of proW and proX downstream; removal of proV was solely responsible for the defective intracellular growth observed with the ΔproV mutant.
Figure 4. Testing the intracellular growth of S. sonnei in vitro using HEK293 cells. (A) Intracellular growth of S. sonnei 2 hours post infection (MOI of 10). Intracellular CFU of the wild type were taken as 100%, and intracellular CFU from strains ΔproV, MirproV and ΔproV/pProV were expressed as percentages to that of wild type. Each value is the mean of 3 independent determinations ± standard deviation. The levels of significance were determined using unpaired t-test, Asterisks (****) indicate p-values< 0.0001, (***) mean p-values = 0.0003. (B) Time course of intracellular growth of S. sonnei (MOI of 10). At indicated time interval post infection, cells were lysed and intracellular CFU were determined by plating on agar. Each value is the mean of triplicates ± standard deviation (n = 3). Doubling time for each strain is calculated by linear regression analysis (Fig. S6). (C, D) Overlay of the flow cytometry analysis of cells were infected with ΔproV (dark gray), wild type (black) or mock-infected (light gray). Both S. sonnei strains were expressing EGFP. Controls are cells mock-infected with saline. Analysis was done 4 hours post infection (C) or overnight (D). Gate A depicts populations of cells emit EGFP signals.
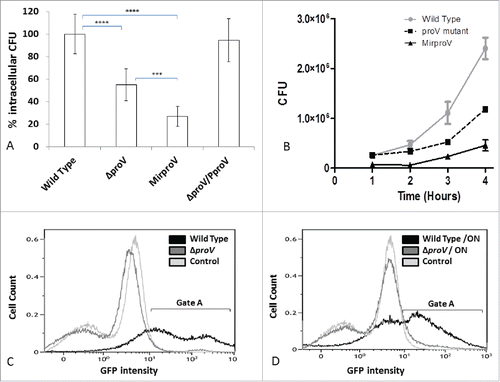
To further test if the ProU system is required for intracellular growth not invasion per se, we performed a time course analysis of intracellular growth after 1, 2, 3, and 4 hours using gentamicin protection assay. At each time point, cells were lysed with Triton X-100, and cell-lysates were plated out on LB agar, and incubated at 37°C overnight and enumerated by colony count. The wild type grew rapidly with a doubling time of approximately 38 min from 1 hour onwards whereas the ΔproV and the MirproV strains had doubling times of 60 and 90 min, respectively (, Fig S7).
To gather further evidence for ProU requirement for intracellular growth we exploited flow cytometry. We transformed all strains with a plasmid that expressed EGFP, and infected HEK293 cells with these green bacteria. As shown in , 38% of the host cells infected with wild type strain emitted green fluorescence 4 hours post infection, indicating that green bacteria were metabolically active inside host cells at this time point. Noticeably, some cells had very strong EGFP signals (fluorescence intensity above 102 units), demonstrating that these cells harbored large numbers of green bacteria as a result of rapid bacterial intracellular growth. In contrast to cells infected with wild type, there were less than 4% of cells infected with the ΔproV mutant emitted green fluorescence (), indicating impaired intracellular growth. We left cell infection overnight and repeated flow cytometry and this showed that 43% of cells infected with wild type emitted EGFP signals, (). It was noticeable that a smaller percentage of cells could emit high fluorescent above 102 scale compared to that of 4 hours post infection (). Presumably, host cells harboring large numbers of green bacteria were dead following the overnight infection. Consistent with this was the notion that more cells were emitting low fluorescence (intensity of 101 to 102 units), suggesting that cells harboring lower bacteria burden survived overnight infection. Noticeably also, there was an increase in green fluorescence in host cells that have been infected with the ΔproV mutant overnight, suggesting limited remaining intracellular growth of this strain (). Taken together, ProU is required for intracellular growth of S. sonnei in host cells.
ProU is requried for virulence in the Galleria mellonella larvae model (in vivo)
The great moth G. mellonella larvae have become a popular in vivo model for assessing bacterial virulence.Citation40,41 We recently adopted this model to assess Shigella virulence and found it comparable with the widely accepted Sereny test.Citation42
Here we exploited this model to compare the virulence of ΔproV and MirproV strains with the wild type S. sonnei. Larvae were challenged with 105 CFU of each strain; 10 larvae per group. Mock-infection was done using the same volume of saline. All groups of larvae were observed daily for 5 d and dead larvae suffered from melanization and loss of motility. As shown in , wild type and the complemented strain (ΔproV/pProV) were able to kill 90% and 80% of larvae, respectively, in 1 day and the remaining larvae died at the third day. Using the same dose of the ΔproV strain, only 40% of larvae died at the first day, and 30% died at the second day while the rest died at the third day. With the MirproV strain, a dose of 105 CFU only caused 20% of death at the first day, 10% at the second day and other 10% at the fourth day, while 60% of the whole population survived to the end of the study (). These data were consistent with those of that MirproV was more effective than proV deletion in inactivating the ProU system.
Figure 5. Testing S. sonnei virulence in vivo using G. mellonella larvae. (A) Fraction survival of G. mellonella larvae model challenged by 105 CFU of wild type S. sonnei strain 20071599 (gray square), the complemented strain (ΔproV/pProV) (black circle), ΔproV (black triangle) and the MirproV (black trigonal), using saline as a control (crosses). The observation lasted for 5 d. The results are means of 3 successive groups (n = 10 larvae). (B) Overlaid histogram of the flow cytometry analysis of hemocytes isolated from G. mellonella larvae mock-infected as a control (light gray), challenged by S. sonnei wild strain 20071599 (dark gray) or by ΔproV (black). Both S. sonnei strains were expressing EGFP. Hemocytes were isolated 4 hours post infection for analysis; gate A depicts hemocytes emit GFP signals.
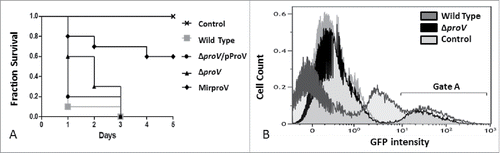
Our recent study has demonstrated that infection of larvae hemocytes is an important mechanism for larvae-killing by S. sonnei.Citation42 Therefore, we infected moth larvae with EGFP-expressing strains (Table S2) and isolated hemocytes from infected and mock-infected larvae for flow cytometry. Larvae were infected with 106 CFU of each strain, and mock-infection was done using saline. Hemocytes were isolated from larvae 4 hours post infection. We used hemocytes from mock-infected larvae to set gates defining EGFP negative and positive populations (). It was apparent that wild type strain colonized more hemocytes giving rise to 2 populations of hemocytes with low and high EGFP intensity, whereas the ΔproV strain colonized less numbers of hemocytes with one population of high-EGFP expressing hemocytes at this time point.
Discussion
In pathogenic bacteria, osmoregulation is a very important mechanism not only for survival during environmental osmotic stress, but also in establishment of infection. Examples of osmoregulatory systems important for virulence include the Staphylococcus aureus PutP,Citation8-10 Pseudomonas aeruginosa PlcH,Citation11,12 and Listeria monocytogenes OpuC.Citation13,14
The ProU transport system is widely distributed in bacteria. Previous studies have firmly established its role in environmental survival under hyperosmolarity in E. coli, but whether it is required in the context of infection remained unknown.Citation7 By studying the orthologous ProXVWZ system in M. tuberculosis, Price and his colleagues have demonstrated the importance of this system in osmotolerance in vitro and during early host cell colonization.Citation30 Here, we have presented compelling evidence for the first time that Shigella ProU system is required for coping with hyperosmolarity both in vitro () and ex vivo (), and for virulence in a moth larvae infection model (). We have also demonstrated that ProU is highly upregulated under hyperosmotic conditions (). This is consistent with the findings for S. flexneri by Lucchini and co-workers Citation28; proV is up-regulated up to 57-fold inside the host cell cytoplasm compared to growth in LB broth. Lucchini suggested that the overall different ionic composition in the host cell cytosol is the cue that triggers this upregulation, because the host cell cytosol should have similar osmolarity as LB broth.Citation28 Supporting evidence for this hypothesis is the notion that transcription of the phoRB regulon and mgtA is also upregulated inside host cells compared to their transcription in LB broth.Citation28 phoRB is required for transporting phosphates and mgtA is required for transporting Mg2+; their upregulation indicates shortages of phosphates and Mg2+ in the host cell cytosol. M. tuberculosis colonizes and modifies phagosome by inhibiting acidification.Citation43 The exact cue for upregulation of the M. tuberculosis ProXVWZ system has not been identified so far.Citation30 Nevertheless, it is clear that both the phagosome and the host cell cytosol impose osmotic stress on invading microbes Citation(30and this study). In both these cellular niches, the microbes use the ProU system to transport glycine betaine to survive and multiply. Glycine betaine is an important free cytoplasmic constituent of eukaryotic cells,Citation44 and it is present at 20 to 60 μM in human serum.Citation45 Hence, it is not surprising that pathogens like M. tuberculosis Citation46 and S. sonnei exploit glycine betaine as an osmoprotectant during growth within the host cells.
ProV encodes an ATPase of the ProU system. Deletion of proV caused significantly slow growth in hyperosmotic media, and complementation by expressing ProV in trans restored wild type rate of growth (). These data reinforce that ProU is an energy dependent transport system, and ΔproV is solely responsible for the slow growth phenotype of the deletion mutant. However, the ΔproV mutant grew well in early phase in the presence of 0.2 M NaCl and 0.2 M KCl (). This result suggested that the ProU system is, at least in part, still functional when proV is removed. By overexpression of 3 paralogues of proV in trans, we have demonstrated that ATPases from other transport systems are able to compensate for the loss of ProV for better growth (Fig. S5). According to complete genome sequence, S. sonnei has intact ProP, a low affinity transporter for proline and glycine betaine. But, the function of ProP appeared insignificant because the growth defect of ΔproV strains cannot be reversed by supplementing betaine (). Furthermore, S. sonnei also has K+ specific transporters, Trk, Kdp, and Kup, according to complete genome sequence. The functions of these systems appeared also insignificant because ΔproV strain had impaired growth in M9 medium, which contained K+ ().
The MirproV strain does not significantly respond to betaine supplementation in M9 medium with 0.2 M NaCl or 0.2 M KCl for better growth (). These data suggest that MirproV could effectively attenuate ProU system. Even though the MirproV strain produced more transcripts of proV and proX in response to 0.3 M NaCl and 0.3 M KCl () compared to its transcripts in M9 medium without any supplements (), these levels of responses were not sufficient to reverse the growth defect (; S3, S4). The effectiveness of MirproV was also demonstrated in gentamicin protection assay; the MirproV strain was more severely defective than the ΔproV mutant in the intracellular growth (). Moreover, the possibility of ‘off-target’ effects by MirproV is low; there are only 4 low score hits when the 42 bp proV sequence is used as query to blast the S. sonnei SSO46 genome (Table S4). Noticeably, mxiJ is one of the hits; mxiJ encodes an essential component of TTSS.Citation47 However, like the ΔproV strain, the MirproV strain produces and secretes Ipa proteins (), suggesting that any ‘off-target’ hits, if they occur, do not significantly change the strain's osmotolerance or virulence. However, we cannot categorically rule out a mild reduction of the MirproV strain in invasion as noted in ; the MirproV strain had reduced intracellular CFU at first hour postinfection.
Despite the fact that we were able to delete proV from a proV::Kan intermediate, which had completely inactivated ProU as expression of ProV in trans did not correct its impaired growth defect (Fig. S7). This proV::Kan strain formed smooth red colonies on Congo red agar, indicating it harbored intact virulence plasmid. However, it was a surprise that we were unable to obtain a strain with intact virulence plasmid when either the whole proVWX operon, or single gene (proW or proX) was removed. Previous studies have shown that S. sonnei could frequently become avirulent by losing its 120-megadalton virulence plasmid.Citation48 Besides, it is well known that both ProU and the TTSS genes are controlled by the same negative regulator: the H-NS protein and this regulatory link provided an additional rational for monitoring the possibility of altered TTSS expression upon ProU attenuation.Citation49,50 So, it was very important to test the stability of this plasmid in wild type and the mutant strains under investigation (ΔproV, MirproV and the complemented ΔproV/pProV). Our data show all the strains possess key plasmid borne virulence genes, ipaB and mxiD. Furthermore, all strains produce and secrete IpaB and IpaC proteins, except ΔmxiD strain that produces but does not secrete Ipa proteins (). Hence, we can conclude that inactivation of the ProU system by either deletion of proV or RNAi approach does not impair genetic stability and function of the virulence plasmid.
Finally, we have once again demonstrated the usefulness of the moth larvae model in assessing bacterial virulence, which enabled us to establish a link between osmotolerance and virulence.
Conclusions
Altogether, we conclude that the ProU system is important for S. sonnei to tolerate hyperosmotic stress in vitro, as well as for survival and proliferation of the bacteria in the stressful intracellular niche. Silencing of the whole proVWX operon is more effective than deleting proV alone because proV paralogues may compensate for the loss of proV. Last but not least, the G. mellonella larvae model is a cost-effective and good model for studying Shigella virulence, and reflects results of more established models such as the Sereny test.
Materials and methods
Bacterial strains and growth conditions
The wild type Shigella strain used in this study was S. sonnei strain 20071599Citation31 and mutants thereof. Bacteria were routinely grown on Congo red TSA plates or in LB broth at 37°C. To obtain EGFP expressing bacteria, strains were transformed with pGEMT-Easy (AmpR) containing EGFP. Afterwards, 100 μg/ml ampicillin was added for selection of plasmid containing strains. All primers and strains used in this study are listed in Tables S1 and S2 respectively.
Genetic engineering
Construction of the mutant bearing in-frame deletion in proV gene was done using phage λ Red recombination system.Citation32 Plasmid, pKD46, which carries the red lambda recombinase genes, was first transformed to strain 20071599. Primers (a and b; Table S1) were used to amplify kanamycin cassette using pKD4 plasmid as a template; the PCR product, Kan-cassette, contained the first and last 51 base pairs of proV coding sequences at the 5′- and 3′-ends, respectively. Introduction of the Kan-cassette into 20071599/pKD46 enabled homologous recombination; the wild type proV gene was replaced with the kan-cassette. Plasmid, pCP20, was then transformed to the proV-Kan mutant for second cross-over to loop out the Kan-cassette, leaving an in-frame deletion scar (102 bp) through the gene. EGFP was cloned using pGEM-T-Easy which was used to express EGFP in wild type and ΔproV strains. Entire proV coding sequence was PCR-amplified from the wild type S. sonnei using primers c and d (Table S1) and cloned into pGEM-T-Easy (ampR), with the 5′-end of the proV coding sequence facing the lacZ promoter. The resultant clone was used to complement the ΔproV mutant.
For RNAi experiments, 2 oligonucleotides were synthesized, which were complementary to each other and 87 bases in length each, they span 45 and 42 base pairs of the promoter and 5′-end of proV coding sequence, respectively (Table S1). The two oligonucleotides were adjusted to the concentration of 100 pmol each, mixed together and heated to 65°C (Tm = 71°C) in a heating block. The heating block was allowed to cool down to 37°C; then incubated at 37°C overnight. The double stranded DNA was cloned into pGEM®-T-Easy Vector and transformed into E. coli DH5α. The resultant clones were analyzed using restriction enzyme SspI and by DNA sequencing to ensure the proV coding sequence 3′-end facing the lacZ promoter, which produced RNA mirrored that of proV gene sense strand (hence called MirproV).
Western blot
Smooth red colonies of S. sonnei tested strains were picked from fresh overnight Congo red TSA plates and were grown into TSB to an OD600nm of 0.7. Bacteria were spun down and pellets were washed once with sterile PBS, and resuspended in 500 µl PBS with 0.01 % Congo red. The bacterial suspensions were incubated at 37°C for 30 min. Cells were again spun down and supernatant was transferred to a fresh micro-tube. The pellets were washed once with PBS then 100 µl of sample buffer was added and boiled at 95°C for 5 min. Total proteins in the supernatant were precipitated using 10% TCA and samples were incubated on ice for 20 min before spinning the protein down at 20000 xg for 20 min at 4°C. The protein pellets were washed with 70% ethanol before re-suspension in 10 µl sample buffer and boiling at 95°C for 5 minCitation51. Both IpaB and IpaC were detected using the monoclonal antibodies H16 (anti-IpaB) and J22 (anti- IpaC), respectively,Citation51 followed by incubation with Alexa Fluor 680 goat anti-mouse IgG (H+L), and the images were visualized using a UV scanner at 700 nm.
In vitro osmotolerance test
M9 medium was prepared, autoclaved and supplemented with filtered 200 µl/100ml of 1 M MgSO4, 0.4g% glucose, 12.5 µg/ml of Nicotinic acid, 45 µg/ml of L-methionine, and 20 µg/ml of L-Tryptophan. Routinely, all strains were grown on M9 agar overnight, smooth colonies were picked and grown in M9 broth 2 hours before the start of the experiment. Then, 96-well plates were set for growth curve; each well contained 200 μl M9 medium with or without supplements: 0.2 and 0.3 M NaCl or 0.2 and 0.3 M KCl in presence and absence of 500 µM betaine. Each culture condition was tested in triplicates for each bacterial strain, and 3 independent experiments were carried out. Afterwards, 107 CFU from each of the tested strains were added to the 96-well plates. Wells without bacterial inocula were used as a blank, while wells without supplements were used to study the growth curve of Shigella strains in M9 medium. The plates were incubated in a shaker incubator at 37°C, and the OD600nm was recorded each 1 hour.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Wild type and MirproV strains were grown in M9 minimal medium to mid-log phase. Total RNA was isolated using RNA isolation kit (Bioline). The house-keeping gene, cycG, was used as an internal control.Citation36 To establish standard curves for each gene primers k and l; m and n and o and p (Table S1) were used to amplify proV, proX and cycG genes, respectively, and serial dilutions (from 100 to 108 molecules/µl) of the genome DNA was used as templates using SYBR-Green qRT-PCR kit on Rotor Gene 6000 (Qiagen). Triplicate RNA samples from triplicate cultures (n = 3) were used to prepare cDNAs, which were quantified by the same PCR procedure. The amplification curves of proV and proX were normalized with that of cycG, and quantification was calculated using the standard curves. Changes in gene expression between wild type (set as calibrator) and the MirproV strains were calculated using the 2−ΔΔct method and proprietary software in the Rotor Gene instrument (version 1.7.34).Citation52 To analyze the impact of high osmotic stress on ProU expression both wild type and MirproV strains were grown in M9 supplemented with 0.3 M NaCl or 0.3 M KCl. The levels of transcripts from wild type and MirproV grown in M9 without salt supplements were used as calibrators. The transcripts of house-keeping gene, cycG, were again used for normalization to calculate the 2−ΔΔct 52.
Gentamicin protection assay
HEK293 (human embryonic kidney stem) cells were seeded and cultured until approximately 80% confluence in 24-well plates and S. sonnei bacteria were added to the cell monolayers at a multiplicity of infection (MOI) of 10. The plates were centrifuged at 700 xg for 10 min at 22°C. The plates were incubated for 40 min at 37°C under 5% CO2 atmosphere to allow bacterial invasion into host cells. Thereafter, cell monolayers were washed twice with PBS, DMEM containing gentamicin (50 µg/ml) was added, and the plates were incubated for required time intervals before terminating infection. Cells were washed 3 times using PBS and lysed with Triton X-100 (0.1 % in H2O) for 10 min. Cell lysates were serially diluted, plated on LB agar, incubated at 37°C overnight and enumerated by colony count.
Infection of Galleria mellonella larvae
Smooth red colonies of S. sonnei strains were selected from Congo red TSA plates, grown for 3 to 5 hours in LB broth to prepare bacterial suspensions. Each group of 10 healthy larvae of approximately similar size were injected with 10 µl (105 CFU) of each bacterial suspension. The mock-infection group received sterile PBS instead of bacteria. All larvae were incubated at 37°CCitation53 and observed for 5 d post infection.Citation42 The experiments have been repeated 3 times for results confirmation and the averages have been used in Kaplan-Meier survival curves.
Flow cytometry
For HEK293 cells, infection was terminated at appropriate time intervals (either 4 hours or overnight); cells were washed with saline twice and then trypsinized for 1 min after which the DMEM medium was added. Cells were spun down and cell pellets were resuspended in saline containing 4% (v/v) paraformaldehyde for fixation.
For the wax moth larvae, 4 hours post-infection with 106 CFU bacteria, hemocytes from 10 larvae were collected by incision between 2 segments near larvae tail to avoid gut interruption. Hemocytes were collected in 1 ml of sterile PBS and the processing was within 10 min to prevent clotting.Citation53 Cells were centrifuged at 500 xg for 10 min at room temperature, and were resuspended in 1 ml PBS containing 4% (v/v) paraformaldehyde for fixation.
Cells were vigorously mixed and then were used for the flow-cytometry. Lasers emitting at 488 nm was set for the detecting signals in FITC channel, which overlap with EGFP excitation. Data acquisition was performed using Kaluza™ software (Beckman Coulter, Inc.). The experiments have been repeated 3 times for results confirmation.
Statistics
The growth curves in are x/y blotting using triplicates for each reading and the error bars are for SD. In , unpaired t test has been used to compare the expression of different genes using 2−ΔΔct method. In , unpaired t test has been used to compare the % intracellular CFU of different Shigella strains; the growth curve of intracellular growth of Shigella strains is x/y blotting using triplicates for each reading and the error bars are for SD.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
Conceived and designed experiments: JY
Performed the experiments: RYM WL
Analyzed the data: RYM JY
Contributed reagents/materials/analysis tools: JY ME ERA
Wrote the paper: RYM JY.
KVIR_S_1227906.doc
Download MS Word (516 KB)Acknowledgments
We are grateful for technical assistance from Clare Harding, Namrta Bhopal, Aliyah Shafiq and Jade Slaven. We also thank Gareth Westrop and Anne-Marie Krachler for critical reading and comments on the manuscript.
Additional information
Funding
References
- Hale TL. Genetic basis of virulence in Shigella species. Microbiol Rev 1991; 55:206-24; PMID:1886518
- Peng J, Zhang X, Yang J, Wang J, Yang E, Bin W, Wei C, Sun M, Jin Q. The use of comparative genomic hybridization to characterize genome dynamics and diversity among the serotypes of Shigella. BMC Genomics 2006; 7:218; PMID:16939645; https://doi.org/10.1186/1471-2164-7-218
- von Seidlein L, Kim DR, Ali M, Lee H, Wang X, Thiem VD, Canh do G, Chaicumpa W, Agtini MD, Hossain A, et al. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med 2006; 3:e353; PMID:16968124; https://doi.org/10.1371/journal.pmed.0030353
- Sansonetti PJ. Slaying the Hydra all at once or head by head? Nat Med 1998; 4:499-500; PMID:9585195; https://doi.org/10.1038/nm0598supp-499
- Sansonetti PJ. Microbes and microbial toxins: paradigms for microbial-mucosal interactions III. Shigellosis: from symptoms to molecular pathogenesis. Am J Physiol Gastrointest Liver Physiol 2001; 280:G319-23; PMID:11171613
- Arakawa T, Timasheff SN. The stabilization of proteins by osmolytes. Biophys J 1985; 47:411-4; PMID:3978211; https://doi.org/10.1016/S0006-3495(85)83932-1
- Lucht JM, Bremer E. Adaptation of Escherichia coli to high osmolarity environments: osmoregulation of the high-affinity glycine betaine transport system proU. FEMS Microbiol Rev 1994; 14:3-20; PMID:8011357; https://doi.org/10.1111/j.1574-6976.1994.tb00067.x
- Schwan WR, Coulter SN, Ng EY, Langhorne MH, Ritchie HD, Brody LL, Westbrock-Wadman S, Bayer AS, Folger KR, Stover CK. Identification and characterization of the PutP proline permease that contributes to in vivo survival of Staphylococcus aureus in animal models. Infect Immun 1998; 66:567-72; PMID:9453610
- Bayer AS, Coulter SN, Stover CK, Schwan WR. Impact of the high-affinity proline permease gene (putP) on the virulence of Staphylococcus aureus in experimental endocarditis. Infect Immun 1999; 67:740-4; PMID:9916085
- Schwan WR, Wetzel KJ, Gomez TS, Stiles MA, Beitlich BD, Grunwald S. Low-proline environments impair growth, proline transport and in vivo survival of Staphylococcus aureus strain-specific putP mutants. Microbiology 2004; 150:1055-61; PMID:15073314; https://doi.org/10.1099/mic.0.26710-0
- D'Souza-Ault MR, Smith LT, Smith GM. Roles of N-acetylglutaminylglutamine amide and glycine betaine in adaptation of Pseudomonas aeruginosa to osmotic stress. Appl Environ Microbiol 1993; 59:473-8; PMID:8434912
- Sage AE, Vasil ML. Osmoprotectant-dependent expression of plcH, encoding the hemolytic phospholipase C, is subject to novel catabolite repression control in Pseudomonas aeruginosa PAO1. J Bacteriol 1997; 179:4874-81; PMID:9244277
- Sleator RD, Wouters J, Gahan CG, Abee T, Hill C. Analysis of the role of OpuC, an osmolyte transport system, in salt tolerance and virulence potential of Listeria monocytogenes. Appl Environ Microbiol 2001; 67:2692-8; PMID:11375182; https://doi.org/10.1128/AEM.67.6.2692-2698.2001
- Wemekamp-Kamphuis HH, Wouters JA, Sleator RD, Gahan CG, Hill C, Abee T. Multiple deletions of the osmolyte transporters BetL, Gbu, and OpuC of Listeria monocytogenes affect virulence and growth at high osmolarity. Appl Environ Microbiol 2002; 68:4710-6; PMID:12324311; https://doi.org/10.1128/AEM.68.10.4710-4716.2002
- Frossard SM, Khan AA, Warrick EC, Gately JM, Hanson AD, Oldham ML, Sanders DA, Csonka LN. Identification of a third osmoprotectant transport system, the osmU system, in Salmonella enterica. J Bacteriol 2012; 194:3861-71; PMID:22609924; https://doi.org/10.1128/JB.00495-12
- Cairney J, Booth IR, Higgins CF. Osmoregulation of gene expression in Salmonella typhimurium: proU encodes an osmotically induced betaine transport system. J Bacteriol 1985; 164:1224-32; PMID:3905768
- Dunlap VJ, Csonka LN. Osmotic regulation of L-proline transport in Salmonella typhimurium. J Bacteriol 1985; 163:296-304; PMID:3924895
- Gowrishankar J. Nucleotide sequence of the osmoregulatory proU operon of Escherichia coli. J Bacteriol 1989; 171:1923-31; PMID:2649479
- May G, Faatz E, Villarejo M, Bremer E. Binding protein dependent transport of glycine betaine and its osmotic regulation in Escherichia coli K12. Mol Gen Genet 1986; 205:225-33; PMID:2949137; https://doi.org/10.1007/BF00430432
- MacMillan SV, Alexander DA, Culham DE, Kunte HJ, Marshall EV, Rochon D, Wood JM. The ion coupling and organic substrate specificities of osmoregulatory transporter ProP in Escherichia coli. Biochim Biophys Acta 1999; 1420:30-44; PMID:10446288; https://doi.org/10.1016/S0005-2736(99)00085-1
- Ko R, Smith LT, Smith GM. Glycine betaine confers enhanced osmotolerance and cryotolerance on Listeria monocytogenes. J Bacteriol 1994; 176:426-31; PMID:8288538
- Gouesbet G, Jebbar M, Talibart R, Bernard T, Blanco C. Pipecolic acid is an osmoprotectant for Escherichia coli taken up by the general osmoporters ProU and ProP. Microbiology 1994; 140 ( Pt 9):2415-22.
- Haardt M, Kempf B, Faatz E, Bremer E. The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli K-12. Mol Gen Genet 1995; 246:783-6; PMID:7898450; https://doi.org/10.1007/BF00290728
- Doige CA, Ames GF. ATP-dependent transport systems in bacteria and humans: relevance to cystic fibrosis and multidrug resistance. Annu Rev Microbiol 1993; 47:291-319; PMID:7504904; https://doi.org/10.1146/annurev.mi.47.100193.001451
- Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol 1992; 8:67-113; PMID:1282354; https://doi.org/10.1146/annurev.cb.08.110192.000435
- Barron A, Jung JU, Villarejo M. Purification and characterization of a glycine betaine binding protein from Escherichia coli. J Biol Chem 1987; 262:11841-6; PMID:3305496
- Higgins CF, Sutherland L, Cairney J, Booth IR. The osmotically regulated proU locus of Salmonella typhimurium encodes a periplasmic betaine-binding protein. J Gen Microbiol 1987; 133:305-10; PMID:3309148
- Lucchini S, Liu H, Jin Q, Hinton JC, Yu J. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect Immun 2005; 73:88-102; PMID:15618144; https://doi.org/10.1128/IAI.73.1.88-102.2005
- Gowrishankar J, Manna D. How is osmotic regulation of transcription of the Escherichia coli proU operon achieved? A review and a model. Genetica 1996; 97:363-78; PMID:9081863; https://doi.org/10.1007/BF00055322
- Price CT, Bukka A, Cynamon M, Graham JE. Glycine betaine uptake by the ProXVWZ ABC transporter contributes to the ability of Mycobacterium tuberculosis to initiate growth in human macrophages. J Bacteriol 2008; 190:3955-61; PMID:18390665; https://doi.org/10.1128/JB.01476-07
- Xu D, Yang X, Wang D, Yu J, Wang Y. Surface display of the HPV L1 capsid protein by the autotransporter Shigella IcsA. J Microbiol 2014; 52:77-82; PMID:24390841; https://doi.org/10.1007/s12275-014-3235-9
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 2000; 97:6640-5; PMID:10829079; https://doi.org/10.1073/pnas.120163297
- Haardt M, Bremer E. Use of phoA and lacZ fusions to study the membrane topology of ProW, a component of the osmoregulated ProU transport system of Escherichia coli. J Bacteriol 1996; 178:5370-81; PMID:8808924
- Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 2008; 72:317-64, table of contents; PMID:18535149; https://doi.org/10.1128/MMBR.00031-07
- Tchurikov NA, Chistyakova LG, Zavilgelsky GB, Manukhov IV, Chernov BK, Golova YB. Gene-specific silencing by expression of parallel complementary RNA in Escherichia coli. J Biol Chem 2000; 275:26523-9; PMID:10849423; https://doi.org/10.1074/jbc.M002833200
- Zhou K, Zhou L, Lim Q, Zou R, Stephanopoulos G, Too HP. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol Biol 2011; 12:18; PMID:21513543; https://doi.org/10.1186/1471-2199-12-18
- Carayol N, Tran Van Nhieu G. Tips and tricks about Shigella invasion of epithelial cells. Curr Opin Microbiol 2013; 16:32-7; PMID:23318141; https://doi.org/10.1016/j.mib.2012.11.010
- Bahrani FK, Sansonetti PJ, Parsot C. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect Immunity 1997; 65:4005-10; PMID:9316999
- Paciello I, Silipo A, Lembo-Fazio L, Curcuru L, Zumsteg A, Noel G, Ciancarella V, Sturiale L, Molinaro A, Bernardini ML. Intracellular Shigella remodels its LPS to dampen the innate immune recognition and evade inflammasome activation. Proc Natl Acad Sci U S A 2013; 110:E4345-54; PMID:24167293; https://doi.org/10.1073/pnas.1303641110
- Leuko S, Raivio TL. Mutations that impact the enteropathogenic Escherichia coli Cpx envelope stress response attenuate virulence in Galleria mellonella. Infect Immun 2012; 80:3077-85; PMID:22710873; https://doi.org/10.1128/IAI.00081-12
- Viegas SC, Mil-Homens D, Fialho AM, Arraiano CM. The virulence of Salmonella enterica Serovar Typhimurium in the insect model Galleria mellonella is impaired by mutations in RNase E and RNase III. Appl Environ Microbiol 2013; 79:6124-33; PMID:23913419; https://doi.org/10.1128/AEM.02044-13
- Mahmoud RY, Stones DH, Li W, Emara M, El-Domany RA, Wang D, Wang Y, Krachler AM, Yu J. The Multivalent Adhesion Molecule SSO1327 plays a key role in Shigella sonnei pathogenesis. Mol Microbiol 2015; 99:658-73; PMID:26481305; https://doi.org/10.1111/mmi.13255
- Wong D, Bach H, Sun J, Hmama Z, Av-Gay Y. Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar-H+-ATPase to inhibit phagosome acidification. Proc Natl Acad Sci U S A 2011; 108:19371-6; PMID:22087003; https://doi.org/10.1073/pnas.1109201108
- Denkert C, Warskulat U, Hensel F, Haussinger D. Osmolyte strategy in human monocytes and macrophages: involvement of p38MAPK in hyperosmotic induction of betaine and myoinositol transporters. Arch Biochem Biophys 1998; 354:172-80; PMID:9633613; https://doi.org/10.1006/abbi.1998.0661
- Melse-Boonstra A, Holm PI, Ueland PM, Olthof M, Clarke R, Verhoef P. Betaine concentration as a determinant of fasting total homocysteine concentrations and the effect of folic acid supplementation on betaine concentrations. Am J Clin Nutr 2005; 81:1378-82; PMID:15941890
- Metris A, George SM, Mulholland F, Carter AT, Baranyi J. Metabolic shift of Escherichia coli under salt stress in the presence of glycine betaine. Appl Environ Microbiol 2014; 80:4745-56; PMID:24858086; https://doi.org/10.1128/AEM.00599-14
- Allaoui A, Sansonetti PJ, Parsot C. MxiJ, a lipoprotein involved in secretion of Shigella Ipa invasins, is homologous to YscJ, a secretion factor of the Yersinia Yop proteins. J Bacteriol 1992; 174:7661-9; PMID:1332940
- Sansonetti PJ, Kopecko DJ, Formal SB. Shigella sonnei plasmids: evidence that a large plasmid is necessary for virulence. Infect Immun 1981; 34:75-83; PMID:6271687
- Dagberg B, Uhlin BE. Regulation of virulence-associated plasmid genes in enteroinvasive Escherichia coli. J Bacteriol 1992; 174:7606-12; PMID:1332938
- Kane KA, Dorman CJ. Rational design of an artificial genetic switch: Co-option of the H-NS-repressed proU operon by the VirB virulence master regulator. J Bacteriol 2011; 193:5950-60; PMID:21873493; https://doi.org/10.1128/JB.05557-11
- Yu J, Edwards-Jones B, Neyrolles O, Kroll JS. Key role for DsbA in cell-to-cell spread of Shigella flexneri, permitting secretion of Ipa proteins into interepithelial protrusions. Infect Immun 2000; 68:6449-56; PMID:11035758; https://doi.org/10.1128/IAI.68.11.6449-6456.2000
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009; 55:611-22; PMID:19246619; https://doi.org/10.1373/clinchem.2008.112797
- Harding CR, Schroeder GN, Collins JW, Frankel G. Use of Galleria mellonella as a model organism to study Legionella pneumophila infection. J Vis Exp 2013; 22:e50964; PMID:24299965
