ABSTRACT
Patients undergoing haematopoietic stem cell transplantation (HSCT) are highly exposed to infectious agents. However, it is not known why certain HSCT recipients rapidly develop severe infections while other, despite similar immunosuppressive conditions, do not. Increasing evidence suggests that such differences may be due, in part, to polymorphisms in immune genes. Thus, the identification of genetic factors influencing susceptibility to infections in HSCT recipients may lead to the development of individualized management strategies. However, studies are challenged by several issues, including the relative small size of existing cohorts, the frequent use of prophylactic or preemptive antimicrobial agents, and the fact that genes responsible for immune functions can be inherited either from the donor or the host. Consequently, the major challenge for today's researchers is to overcome these limitations and find associations that are robust enough to be translated into reliable risk stratification strategies for infectious diseases.
Infectious complications and risk assessment of HSCT recipients
Opportunistic infections caused by bacteria, fungi and viruses represent a major challenge in patients undergoing haematopoietic stem cell transplantation (HSCT). HSCT is characterized by different phases, each associated with a different pattern of immunosuppression. The risk of infection is particularly important for allogeneic HSCT recipients, in which 3 distinct periods are typically described ().Citation1,2 The pre-engraftment period starts with the beginning of the conditioning regimen and extends up to 1 month after transplantation. This phase is characterized by profound neutropenia, which together with the alteration of natural host defense barriers (chemotherapy-induced mucositis, presence of venous catheters) make patients at risk to develop severe bacterial infections (i.e. bacteremia due to Enterobacteria, viridans Streptococci, Enterococci or coagulase-negative Staphylococci). The same factors combined with the extensive use of antibacterial agents subsequently expose patients to the risk of developing invasive fungal infections, in particular candidemia, hepatosplenic candidiasis and early onset invasive aspergillosis (IA).Citation3,4 The pre-engraftment phase is also characterized by T-cell depletion and can be associated with viral infections, such as Herpes-simplex virus (HSV) reactivation.Citation1,2 The post-engraftment period starts with neutrophils counts recovery and extends to ∼100 d after transplantation, at the time when T-cell function starts to recuperate. This phase is characterized by the occurrence of acute graft-versus-host disease (mainly in allogeneic transplant recipients), which requires the administration of immunosuppressive drugs that further contribute to the impairment of T cell functions. This is also the time at risk for Cytomegalovirus (CMV) reactivation, which itself exerts some level of immunosuppression.Citation5,6 During this phase patients are still at risk for invasive fungal infections, i.e., late-onset aspergillosis Citation3,4 and Pneumocystis jirovecii pneumonia.Citation1,2 The late risk period is mainly characterized by the occurrence of chronic graft-vs.-host disease, which further extends the period at risk for infection due to CMV,Citation5,6 Pneumocystis and Aspergillus, and can also be characterized by infection due to Varicella-zoster virus (VZV) and pyogenic bacteria.Citation1,2
Table 1. Risk stratification of allogeneic haematopoietic stem cell transplant recipients.
While these phases are well characterized and the occurrence of corresponding infections quite reproducible at the population level, the pattern of infection at the individual level is difficult to predict. Despite similar underlying conditions, similar age group and similar conditioning regimen, some patients rapidly or repeatedly develop severe infections with one or several pathogens, while other do not. An increasing number of studies have shown that individual susceptibility to infections in immunocompetent patients is influenced by genetic polymorphisms.Citation7-10 By analogy, it has been hypothesized that susceptibility to infections in immunocompromised patients may also result, in part, from polymorphisms in immune genes. However, such investigations in strongly immunocompromised patients raise very specific questions.
First, HSCT represents an iatrogenic situation in which the infectious pattern is different from that observed in the general population, as infections are more frequent, more severe, and often due to different (so-called opportunistic) pathogens. On the one hand, the large number of infections within a limited number of patients may contribute to increase the statistical power to detect genetic associations. On other hand, HSCT patients represent a limited population, making it difficult to gather very large cohorts of patients as it can be done in a much easier way for common diseases. However, the iatrogenic condition may reveal genetic factors influencing hosts and pathogens interactions that have not been encountered during their long co-evolution. Second, because the phases of allogeneic HSCT are typically associated with specific risks, different prophylactic strategies have been developed to prevent the occurrence of frequent infections and/or to limit their consequences, which often differ depending on the center and local epidemiology. The extended use of anti-microbial drugs during these periods (either as a prophylaxis or for the treatment of suspected or proven infections) can dramatically influence the type of infections and pathogens' resistance patterns, thereby inducing major biases and limiting the ability to compare results from studies. Finally, allogeneic transplant recipients represent chimerical individuals in which a part of immunity is inherited from the donor (e.g. white blood cells), while another part comes from the recipient (e.g., epithelial cells, proteins produced in the liver), so that the polymorphisms responsible for susceptibility to infections may originate either from the donor or from the recipient, or both of them.
The correlation between single nucleotide polymorphisms (SNPs) and disease phenotypes can be assessed by performing association studies. Such studies can be conducted in a candidate genes approach, in which polymorphisms are selected based on the hypothesis that they can influence the function of a specific gene or group of genes and subsequent immune response to the pathogen (hypothesis driven approach). They can also be conducted in the genome-wide approach (genome-wide association studies, GWAS, or whole genome/exome sequencing) in which millions of polymorphisms throughout the entire genome are interrogated (hypothesis free approach).Citation11,12 Up to now, to our knowledge, no genetic study exploring the association of polymorphisms within the whole genome with infectious phenotypes has been published in HSCT recipients. This may be due to the complexity and heterogeneity of the HSCT procedures, as well as the relative small number of patients presenting the phenotypes of interest. Even for candidate genes, the majority of association studies published so far are challenged by a small sample size, heterogeneous definition of cases and controls, differences in patients' managements between different centers. Also, studies were restricted by the lack of replication and functional evaluation of associated polymorphisms, a restricted number of studied genes and SNPs, the lack of corrections for covariates and multiple tests. Despite these limitations, some associations seem to be relevant, because they have been replicated by one or several investigators, or are supported by strong functional evidence.
Because the innate immune system is at the interface of host and pathogen, most candidate gene studies focus on innate immune genes. The innate immune system is composed of physical barriers (skin and mucous membranes), cellular elements (monocytes, macrophages, neutrophils, dendritic cells, mast cells, natural killer [NK] cells) as well as soluble factors (cytokines, chemokines and other), which all contribute to contain the spread of infection. At the molecular level, pathogen-associated molecular patterns (PAMPs) are detected by specific receptors (PRRs), including toll-like receptors (TLRs), NOD-like receptors (NODs), c-type lectin receptors (CLRs) and RIG-I-like receptors (RLRs). Microbes can be neutralized by proteins of the complement system, thereby preventing their entrance into the cells, increasing the ability of immune cells to detect the pathogens (opsonization). Effective innate responses are very important for establishing adaptive immune mechanisms, including cytotoxic T-cell responses and B-cell differentiation which are responsible for microbial clearance and maintenance.
Bacterial infections represent an important problem in HSCT recipients, mainly during the pre-engraftment period. Neutropenic patients who develop fever require the immediate administration of broad-spectrum antibiotics, which are usually administered for long periods of time, even when the episode is neither clinically nor microbiologically documented. Thus, the development of bacterial infection is largely determined by environmental factors (e.g. previous administration of antibiotics for the prophylaxis or treatment of neutropenic fever), which may render the role of genetic factors more difficult to uncover. This may explain why only a limited number of studies identified polymorphisms associated with bacterial infections. To our knowledge, polymorphisms from 7 genes have been associated with bacterial infections in HSCT recipients (). Among them, 3 associations are relatively robust.
Table 2. Main pathogens causing infections after haematopoietic stem cell transplantation.
One of the most relevant associations was that of the promoter SNP rs2232582 in the gene encoding lipopolysaccharide-binding protein (LBP) with susceptibility to gram-negative bacterial infections in HSCT recipients.Citation13 The polymorphic, risk allele was present in the recipient. It was discovered in a group of ∼1200 patients and subsequently replicated in a smaller group of 230 patients. LBP binds to lipopolysaccharide (LPS), an essential component of gram-negative bacteria cell walls, and interacts with the surface pattern recognition receptor TLR4 (together with other molecules such as CD14 and MD2) to induce the NF-κB signaling pathways and subsequent production of pro-inflammatory molecules by innate immune cells.Citation14-16 This association was further supported by the fact that patients carrying one or 2 copies of rare allele for rs2232571 SNP (a SNP that is in strong linkage disequilibrium with rs2232582) had higher circulating blood LBP levels before transplantation.Citation17 A haplotypic combination of promoter variants in LBP (containing rs2232571) was associated with severe sepsis in immunocompetent individuals.Citation18
The second relevant association was observed between the 3020insC frameshift mutation (known as SNP13 or L1007finsC) within nucleotide-binding oligomerization domain containing 2/caspase recruitment domain-containing protein 15 (NOD2/CARD15) and the development of sepsis in HSCT recipients. The risk allele was observed in both the donor and the recipient in an initial study of 430 patients,Citation19 and in the donor (but not the recipient) in a subsequent, smaller study of 160 patients.Citation20 NOD2 is a PRR that belongs to NOD-like receptor family. It is the detector of muramyl dipeptide, a component of the cell wall of both gram-positive and gram-negative bacteria. The presence of 3020insC in NOD2/CARD15 is predicted to result in production of a truncated NOD2 protein. In vitro HEK293T cells transfected with plasmids containing 3020insC NOD2/CARD15 mutant showed diminished relative NF-κB activity in response to LPS when compared with wild-type NOD2.Citation21
Another relevant association was found for haplotypes expressing low amounts of mannose-binding lectin (MBL2) and different types of bacterial infections in HSCT patients.Citation22-25 MBL2 is a PRR involved in complement activation and subsequently opsonization and phagocytosis of invading pathogens as well as apoptosis. MBL2 polymorphisms are frequent in the human population and classically classified into groups of haplotypes/diplotypes that are strongly correlated with the serum levels (stratified into high, intermediate and lowCitation26). Although these studies were limited by a relative small number of patients (e.g., < 150) and analyzed different phenotypes (bacterial infections due to gram-positive bacteria only,Citation22 bacterial infections due to either gram-negative and gram-positive bacteriaCitation24,25 or bacterial sepsisCitation23) in different populations (autologousCitation22-24 versus allogeneic HSCTCitation25), they provided relatively consistent results. Overall, low MBL2 expressing haplotypes in autologous and allogeneic HSCT recipient wereCitation23-25 or tended to beCitation22 associated with an increased risk of infection.
Fungal infections are still associated with an important mortality and morbidity in HSCT recipients. The epidemiology of invasive fungal infections is influenced by the use of antifungal prophylaxis (e.g. fluconazole, posaconazole, co-trimoxazole), which often differ according to the center, and influence the ability of investigators to detect genetic associations. To our knowledge, polymorphisms from 18 genes were significantly associated with invasive fungal infections among HSCT recipients, essentially with IA (). Among them, 3 associations are particularly robust, as they were performed in relatively large cohorts of patients, replicated by different groups and/or supported by in vitro experimentsCitation27-29 ( and ).
Figure 1. The most relevant donor and/or recipient risk factors for invasive aspergillosis in HSCT recipients.
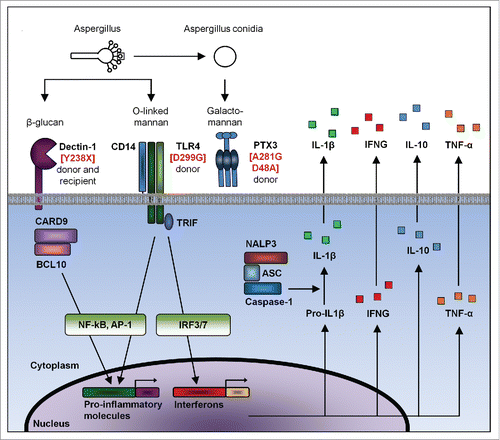
Two nonsynonymous SNPs in TLR4 (D299G and T399I) were associated with the risk of developing IA in HSCT recipients.Citation27 The polymorphism at risk was identified in HSCT donors. The association was replicated in the initial study, and subsequently validated by others.Citation30,31 TLR4 is a PRR essential for recognition of LPS from gram-negative bacteria as well as O-linked mannan from fungi. Functionally those 2 TLR4 polymorphisms were shown to affect lipopolysaccharide recognition by TLR4,Citation32 however their impact on detection of the fungi has been not clarified yet. From these studies and other emerged the concept by which the risk of developing invasive aspergillosis could be predicted before transplantation, by the identification of specific genetic polymorphisms together with other pre-transplant risk factors, such as CMV serostatus, both in HSCT donors and/or the recipients.Citation27
A stop-codon polymorphism (Y238X) in CLEC7A, the gene encoding Dectin-1, was subsequently associated with IA in a cohort of ∼200 HSCT recipients.Citation28 A similar trend was observed in a cohort of ∼180 HSCT recipients.Citation33 In both studies, the risk was conferred by presence of the risk allele in either HSCT donors or recipients. Dectin-1 is a key PRR for β-glucan, an important component of fungal cell walls, and triggers inflammatory responses toward fungal pathogen, as well as their phagocytosis and killing.Citation34-36 Different lines of experiments suggested an effective role for Dectin-1 in cells from both HSCT donors and recipients.Citation28,33 SiRNA inhibition of CLEC7A in lung epithelial cells was associated with a diminished production of inflammatory cytokines after A. fumigatus stimulation, thereby suggesting an antifungal role for Dectin-1 in lung cells from the recipient.Citation28 Human PBMCs and monocytes carrying the Y238X SNP produced lower amounts of inflammatory cytokines upon Aspergillus stimulation compared to WT cells, thereby suggesting that susceptibility to this infection may also depend from impaired Dectin-1 function in donor cells.Citation28 A HSCT model of IA using different combinations of donor and recipient mice (WT and deficient in Dectin-1) further suggested a role for Dectin-1 in both donor and recipient derived cells in the immune response to Aspergillus.Citation28
The most relevant association between IA and genetic variant comes from the study showing the influence of donor haplotype in PTX3 (cluster of 281G and 734A SNPs) with IA in HSCT recipient.Citation29 This association was discovered in a study of 268 HSCT recipients and replicated in another study of 330 recipients.Citation29 The same haplotype was associated with IA in 2 cohorts of solid organ transplant recipients.Citation37,38 PTX3 encodes long pentraxin 3, a soluble PRR that detects galactomannan from the fungi. PTX3 induces many immune processes such as pathogen opsonization, phagocytosis, complement activation, as well as clearance of apoptotic cells.Citation39-41 Functional studies revealed the important role of the donor haplotype at risk in immunity to Aspergillus.Citation29,42-44 The haplotype was associated with reduced PTX3 expression or production as well as reduced anti-Aspergillus phagocytic and killing activity of neutrophils.Citation29 Different mouse models of IA (immunocompetent, neutropenic and HSCT) using PTX3-deficient vs. WT miceCitation42,44 or wild type miceCitation42,43 with/without complementation with soluble PTX3, respectively, further supported the role of long PTX3 in protection from IA. The successful use of soluble PTX3 in animal models suggest that in the future, soluble molecules may be used in the clinical practice to complement defective immune functions in selected patients.Citation38,43,45
Viral infections represent an important problem during HSCT, mainly during the pre- and post-engraftment period. They result from the reactivation of a pre-existing virus in the recipient or from the transmission of a new virus from the donor. As it is the case for bacterial and fungal infections, prophylactic and preemptive strategies have been developed to reduce the burden of viral infections in these high-risk periods, thereby complicating the interpretation of genetic association studies. To our knowledge, polymorphisms in 13 genes showed significant association with the presentation of viral infections in HSCT recipients, essentially with CMV infection (). Yet, only few of them were consistently replicated and/or supported by some level of functional evidence.
Several investigators reported associations between the expression pattern of killer immunoglobulin-like receptors (KIR) genes in allogeneic HSCT donors and the risk of CMV infection in the recipient. KIR are encoded by 15 distinct loci within a ∼200kB region in the leukocyte receptor complex (chromosome 19q13.4) and interact with ligands from HLA class I molecules to regulate the immune function of NK cells.Citation46 Some KIR exert an activating action on NK cells, while other exerts an inhibitory action. Individuals selectively express different combinations of activating and/or inhibitory KIR genes (>50 haplotypes have been described so far), that have been shown to influence susceptibility to infectious diseases.Citation47-49 Four small studies (≤ 230 subjects) were relatively consistent in showing that recipients of donor haplotypes expressing a high number of activating KIRs had a lower risk of CMV infection compared to recipients of those expressing a lower number; however, these studies used different cut-offs (> 1 activating KIR gene versus < 1,Citation50,51 or >5 vs. < 5Citation52,53). Two studies showed that HSCT recipients of donor haplotypes containing the activating KIR2DS2 and KIR2DS4 genes had a reduced risk of CMV infection compared to those receiving other donor haplotypes.Citation53,54
The role of polymorphisms from 7 PRR genes on susceptibility to CMV infections has been investigated among HSCT recipients. Significant associations were observed for polymorphisms in 2 of them, TLR9 and dendritic cell-specific ICAM-3-grabbing non-integrin (DC-SIGN). TLR9 is a PRR recognizing unmethylated CpG motifs within viral DNA. Two TLR9 SNPs in HSCT donors (1174A/G and P545P SNPs, both in strong linkage disequilibrium) were associated with an increased risk of CMV infection in the recipients in 2 independent groups of HSCT patients (138 in the discovery study and 102 in the replication study).Citation55 Furthermore the -1237C/T promoter polymorphism in TLR9 (that is not in linkage disequilibrium with the 1174A/G and the P545P SNPs) was associated with an increased risk of CMV infection in Caucasian HSCT recipients in a single study of ∼220 subjects.Citation56 DC-SIGN is a macrophage and dendritic cell specific PRR recognizing viral glycoproteins. The presence of 2 promoter polymorphisms in DC-SIGN (-139C/T and -939G/A) was associated with and increased risk of CMV infection and/or disease in another study of HSCT recipients (∼130 subjects).Citation57 Immature DCs from individuals carrying 2 copies of -139T and -939G alleles showed decreased expression of DC-SIGN as compared to DCs from individuals carrying 2 copies of -139C and -939A of those polymorphisms.Citation57 Yet, so far, this association was not investigated by others.
A study of 72 HSCT recipients reported associations between polymorphisms in 2 gene influencing T cells functions (CD28 and cytotoxic T-lymphocyte antigen-4, CTLA4) and CMV infection in HSCT recipients.Citation58 CD28 is a co-stimulatory molecules constitutively expressed by naive T cells. CD28 can bind CD80 and CD86 molecules on antigen presenting cells (APC) to initiate signaling pathway important in T-cell activation.Citation59 CTLA4 is a co-inhibitory molecule that is expressed by T-cells upon stimulation and compete with CD28 to bind to CD80 and CD86, thereby playing a key role in the regulation of adaptive immune responses by T lymphocytes.Citation59 Both molecules are recognized as important factors determining the outcome of HSCT.
Conclusions/future standpoints
Increasing evidence from candidate gene studies suggest that polymorphisms in the donor and/or the recipient can influence susceptibility to bacterial, fungal and viral infections after HSCT. The identification of such markers may be useful in the clinical practice, as they may lead to the development of tailored strategies for the management of such patients. For instance, antimicrobial prophylaxes may be administered to patients undergoing a high risk to develop infections, thereby preventing the use of costly and sometimes toxic agents in patients who are at lower risk. However, many centers already use indiscriminate prophylaxis and/or preemptive treatment strategies that have proven to significantly reduce the infectious burden in this population. Alternatively, the use of immunosuppressive drugs after HSCT may be tailored to the individual risk, and/or novel immunomodulators (such as soluble pattern recognition receptors) may be developed to compensate for immune functions that are relatively deficient in selected individuals. However, studies have not translated into personalized management strategies so far. The major limitations include the fact that most studies identified genetic associations of a single polymorphism with a single pathogen. In the clinical practice, patients develop several infections either at the same time or sequentially, and each is influenced by a complex combination of demographic, clinical and genetic factors. The implementation of personalized approaches in the HSCT population would require the integration of these multiple factors within a global risk scoring system. Such approaches are currently limited by the relative small size of existing studies, the small number of available phenotypes as well as the limited number of genes and polymorphisms that have been tested. This goal could probably only be achieved with the implementation of very large cohorts of patients in several centers, with extensive clinical data and samples collection.
Abbreviations
| CCR | = | chemokine (C-C motif) receptor |
| CMV | = | Cytomegalovirus |
| CTLA4 | = | cytotoxic T-lymphocyte-associated protein 4 |
| CXCL | = | CXC-chemokine ligand |
| D | = | donor |
| DC-SIGN | = | dendritic cell-specific ICAM-3-grabbing non-integrin 1 |
| EBV | = | Epstein-Barr virus |
| FCGRIIA | = | Fc fragment of IgG, low affinity IIa receptor |
| FOXP3 | = | forkhead box P3 |
| GVHD | = | graft-versus-host disease |
| HHV-6 | = | human herpes virus 6 |
| HSCT | = | haematopoietic stem cell transplant |
| HSV | = | Herpes simplex virus |
| IFNG | = | interferon gamma |
| HSV | = | Herpes simplex virus |
| IFL3/4 | = | interferon lambda 3/4 |
| IL | = | interleukin |
| IL23R | = | interleukin 23 receptor |
| KIR | = | killer cell immunoglobulin-like receptor |
| LBP | = | lipopolysaccharide binding protein |
| MASP2 | = | mannan-binding lectin serine peptidase 2 |
| MBL | = | mannose banding lectin |
| MCP-1 | = | monocyte chemoattractant protein 1 |
| NOD2/CARD15 | = | nucleotide-binding oligomerization domain containing 2/caspase recruitment domain-containing protein 15 |
| P2X7 | = | purinergic receptor P2X7 |
| PLG | = | plasminogen |
| PTPN22 | = | protein tyrosine phosphatase, non-receptor type 22 |
| PTX3 | = | pentraxin 3 |
| R | = | recipient |
| RAGE | = | advanced glycosylation end product-specific receptor |
| RSV | = | respiratory syncytial virus |
| S100B | = | S100 calcium binding protein B |
| TLR | = | toll-like receptor |
| VEGFA | = | vascular endothelial growth factor A |
| VZV | = | Varicella-zoster virus |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
P.-Y.B. is supported by the Swiss National Foundation (grants number 32003B_127613, and 324730-165954); the Leenaards Foundation; the Santos-Suarez Foundation; and the Loterie Romande. P.-Y. B. is a recipient of Mérieux Research Grant and is a participant in the European Union's Seventh Framework Program (FP7/2007-2013) under grant agreement number HEALTH-2010–260338 (ALLFUN).
References
- Tomblyn M, Chiller T, Einsele H, Gress R, Sepkowitz K, Storek J, Wingard JR, Young JA, Boeckh MJ, Center for International B, et al. Guidelines for preventing infectious complications among hematopoietic cell transplantation recipients: a global perspective. Biol Blood Marrow Transplant 2009; 15:1143-238; PMID:19747629; http://dx.doi.org/10.1016/j.bbmt.2009.06.019
- Kedia S, Acharya PS, Mohammad F, Nguyen H, Asti D, Mehta S, Pant M, Mobarakai N. Infectious complications of hematopoietic stem cell transplantation. J Stem Cell Res Ther 2013; S3:002.
- Marr KA, Carter RA, Crippa F, Wald A, Corey L. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin Infect Dis 2002; 34:909-17; PMID:11880955; http://dx.doi.org/10.1086/339202
- Marr KA. Fungal infections in hematopoietic stem cell transplant recipients. Med Mycol 2008; 46:293-302; PMID:18415836; http://dx.doi.org/10.1080/13693780701885552
- Ljungman P, Hakki M, Boeckh M. Cytomegalovirus in hematopoietic stem cell transplant recipients. Hematol Oncol Clin North Am 2011; 25:151-69; PMID:21236396; http://dx.doi.org/10.1016/j.hoc.2010.11.011
- Ljungman P. CMV infections after hematopoietic stem cell transplantation. Bone Marrow Transplant 2008; 42(Suppl 1):S70-S2; PMID:18724309; http://dx.doi.org/10.1038/bmt.2008.120
- Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nat Rev Genet 2012; 13:175-88; PMID:22310894
- Zhang FR, Huang W, Chen SM, Sun LD, Liu H, Li Y, Cui Y, Yan XX, Yang HT, Yang RD, et al. Genomewide association study of leprosy. N Engl J Med 2009; 361:2609-18; PMID:20018961; http://dx.doi.org/10.1056/NEJMoa0903753
- Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 2010; 138:1338-45, 45 e1-7; PMID:20060832; http://dx.doi.org/10.1053/j.gastro.2009.12.056
- Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet 2009; 41:591-5; PMID:19349983; http://dx.doi.org/10.1038/ng.348
- Bochud M. Genetics for clinicians: from candidate genes to whole genome scans (technological advances). Best Pract Res Clin Endocrinol Metab 2012; 26:119-32; PMID:22498243; http://dx.doi.org/10.1016/j.beem.2011.09.001
- Bochud PY, Bochud M, Telenti A, Calandra T. Innate immunogenetics: a tool for exploring new frontiers of host defence. Lancet Infect Dis 2007; 7:531-42; PMID:17646026; http://dx.doi.org/10.1016/S1473-3099(07)70185-8
- Chien JW, Boeckh MJ, Hansen JA, Clark JG. Lipopolysaccharide binding protein promoter variants influence the risk for Gram-negative bacteremia and mortality after allogeneic hematopoietic cell transplantation. Blood 2008; 111:2462-9; PMID:18056482; http://dx.doi.org/10.1182/blood-2007-09-101709
- Heumann D, Roger T. Initial responses to endotoxins and Gram-negative bacteria. Clin Chim Acta 2002; 323:59-72; PMID:12135807; http://dx.doi.org/10.1016/S0009-8981(02)00180-8
- Park BS, Lee JO. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med 2013; 45:e66; PMID:24310172; http://dx.doi.org/10.1038/emm.2013.97
- Schumann RR. Old and new findings on lipopolysaccharide-binding protein: a soluble pattern-recognition molecule. Biochem Soc Trans 2011; 39:989-93; PMID:21787335; http://dx.doi.org/10.1042/BST0390989
- Guinan EC, Palmer CD, Mancuso CJ, Brennan L, Stoler-Barak L, Kalish LA, Suter EE, Gallington LC, Huhtelin DP, Mansilla M, et al. Identification of single nucleotide polymorphisms in hematopoietic cell transplant patients affecting early recognition of, and response to, endotoxin. Innate Immun 2014; 20:697-711; PMID:24107515
- Flores C, Perez-Mendez L, Maca-Meyer N, Muriel A, Espinosa E, Blanco J, Sanguesa R, Muros M, Garcia JG, Villar J, et al. A common haplotype of the LBP gene predisposes to severe sepsis. Crit Care Med 2009; 37:2759-66; PMID:19707138; http://dx.doi.org/10.1097/CCM.0b013e3181a57b90
- Jaskula E, Lange A, Kyrcz-Krzemien S, Markiewicz M, Dzierzak-Mietla M, Jedrzejczak WW, Czajka P, Mordak-Domagala M, Lange J, Gronkowska A, et al. NOD2/CARD15 single nucleotide polymorphism 13 (3020insC) is associated with risk of sepsis and single nucleotide polymorphism 8 (2104C>T) with herpes viruses reactivation in patients after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 2014; 20:409-14; PMID:24345423; http://dx.doi.org/10.1016/j.bbmt.2013.12.558
- Grube M, Brenmoehl J, Rogler G, Hahn J, Herr W, Holler E. Donor nucleotide-binding oligomerization-containing protein 2 (NOD2) single nucleotide polymorphism 13 is associated with septic shock after allogeneic stem cell transplantation. Biol Blood Marrow Transplant 2015; 21:1399-404; PMID:25988661; http://dx.doi.org/10.1016/j.bbmt.2015.05.011
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 2001; 411:603-6; PMID:11385577; http://dx.doi.org/10.1038/35079114
- Moreto A, Farinas-Alvarez C, Puente M, Ocejo-Vinyals JG, Sanchez-Velasco P, Horcajada JP, Batlle A, Montes C, Santos F, Conde E, et al. Mannose-binding lectin gene variants and infections in patients receiving autologous stem cell transplantation. BMC Immunol 2014; 15:17; PMID:24886325; http://dx.doi.org/10.1186/1471-2172-15-17
- Molle I, Peterslund NA, Thiel S, Steffensen R. MBL2 polymorphism and risk of severe infections in multiple myeloma patients receiving high-dose melphalan and autologous stem cell transplantation. Bone Marrow Transplant 2006; 38:555-60; PMID:16953214; http://dx.doi.org/10.1038/sj.bmt.1705466
- Horiuchi T, Gondo H, Miyagawa H, Otsuka J, Inaba S, Nagafuji K, Takase K, Tsukamoto H, Koyama T, Mitoma H, et al. Association of MBL gene polymorphisms with major bacterial infection in patients treated with high-dose chemotherapy and autologous PBSCT. Genes Immun 2005; 6:162-6; PMID:15674393; http://dx.doi.org/10.1038/sj.gene.6364165
- Mullighan CG, Heatley S, Doherty K, Szabo F, Grigg A, Hughes TP, Schwarer AP, Szer J, Tait BD, Bik To L, et al. Mannose-binding lectin gene polymorphisms are associated with major infection following allogeneic hemopoietic stem cell transplantation. Blood 2002; 99:3524-9; PMID:11986203; http://dx.doi.org/10.1182/blood.V99.10.3524
- Golshayan D, Wojtowicz A, Bibert S, Pyndiah N, Manuel O, Binet I, Buhler LH, Huynh-Do U, Mueller T, Steiger J, et al. Polymorphisms in the lectin pathway of complement activation influence the incidence of acute rejection and graft outcome after kidney transplantation. Kidney Int 2016; 89:927-38; PMID:26924055; http://dx.doi.org/10.1016/j.kint.2015.11.025
- Bochud PY, Chien JW, Marr KA, Leisenring WM, Upton A, Janer M, Rodrigues SD, Li S, Hansen JA, Zhao LP, et al. Toll-like receptor 4 polymorphisms and aspergillosis in stem-cell transplantation. N Engl J Med 2008; 359:1766-77; PMID:18946062; http://dx.doi.org/10.1056/NEJMoa0802629
- Cunha C, Di Ianni M, Bozza S, Giovannini G, Zagarella S, Zelante T, D'Angelo C, Pierini A, Pitzurra L, Falzetti F, et al. Dectin-1 Y238X polymorphism associates with susceptibility to invasive aspergillosis in hematopoietic transplantation through impairment of both recipient- and donor-dependent mechanisms of antifungal immunity. Blood 2010; 116:5394-402; PMID:20807886; http://dx.doi.org/10.1182/blood-2010-04-279307
- Cunha C, Aversa F, Lacerda JF, Busca A, Kurzai O, Grube M, Loffler J, Maertens JA, Bell AS, Inforzato A, et al. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N Engl J Med 2014; 370:421-32; PMID:24476432; http://dx.doi.org/10.1056/NEJMoa1211161
- Koldehoff M, Beelen DW, Elmaagacli AH. Increased susceptibility for aspergillosis and post-transplant immune deficiency in patients with gene variants of TLR4 after stem cell transplantation. Transpl Infect Dis 2013; 15:533-9; PMID:23890253
- de Boer MG, Jolink H, Halkes CJ, van der Heiden PL, Kremer D, Falkenburg JH, van de Vosse E, van Dissel JT. Influence of polymorphisms in innate immunity genes on susceptibility to invasive aspergillosis after stem cell transplantation. PloS One 2011; 6:e18403; PMID:21483748
- Ferwerda B, McCall MB, Verheijen K, Kullberg BJ, van der Ven AJ, Van der Meer JW, Netea MG. Functional consequences of toll-like receptor 4 polymorphisms. Mol Med 2008; 14:346-52; PMID:18231573; http://dx.doi.org/10.2119/2007-00135.Ferwerda
- Chai LY, de Boer MG, van der Velden WJ, Plantinga TS, van Spriel AB, Jacobs C, Halkes CJ, Vonk AG, Blijlevens NM, van Dissel JT, et al. The Y238X stop codon polymorphism in the human beta-glucan receptor dectin-1 and susceptibility to invasive aspergillosis. J Infect Dis 2011; 203:736-43; PMID:21242599; http://dx.doi.org/10.1093/infdis/jiq102
- Gessner MA, Werner JL, Lilly LM, Nelson MP, Metz AE, Dunaway CW, Chan YR, Ouyang W, Brown GD, Weaver CT, et al. Dectin-1-dependent interleukin-22 contributes to early innate lung defense against Aspergillus fumigatus. Infect Immun 2012; 80:410-7; PMID:22038916; http://dx.doi.org/10.1128/IAI.05939-11
- Steele C, Rapaka RR, Metz A, Pop SM, Williams DL, Gordon S, Kolls JK, Brown GD. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog 2005; 1:e42; PMID:16344862; http://dx.doi.org/10.1371/journal.ppat.0010042
- Werner JL, Metz AE, Horn D, Schoeb TR, Hewitt MM, Schwiebert LM, Faro-Trindade I, Brown GD, Steele C. Requisite role for the dectin-1 beta-glucan receptor in pulmonary defense against Aspergillus fumigatus. J Immunol 2009; 182:4938-46; PMID:19342673; http://dx.doi.org/10.4049/jimmunol.0804250
- Wojtowicz A, Lecompte TD, Bibert S, Manuel O, Rueger S, Berger C, Boggian K, Cusini A, Garzoni C, Hirsch H, et al. PTX3 polymorphisms and invasive mold infections after solid organ transplant. Clin Infect Dis 2015; 61:619-22; PMID:25977268; http://dx.doi.org/10.1093/cid/civ386
- Cunha C, Monteiro AA, Oliveira-Coelho A, Kuhne J, Rodrigues F, Sasaki SD, Schio SM, Camargo JJ, Mantovani A, Carvalho A, et al. PTX3-based genetic testing for risk of aspergillosis after lung transplant. Clin Infect Dis 2015; 61:1893-4; PMID:26261201; http://dx.doi.org/10.1093/cid/civ679
- Bottazzi B, Bastone A, Doni A, Garlanda C, Valentino S, Deban L, Maina V, Cotena A, Moalli F, Vago L, et al. The long pentraxin PTX3 as a link among innate immunity, inflammation, and female fertility. J Leukoc Biol 2006; 79:909-12; PMID:16478917; http://dx.doi.org/10.1189/jlb.1005557
- Garlanda C, Bottazzi B, Salvatori G, De Santis R, Cotena A, Deban L, Maina V, Moalli F, Doni A, Veliz-Rodriguez T, et al. Pentraxins in innate immunity and inflammation. Novartis Found Symp 2006; 279:80-6; discussion 6-91, 216-9; PMID:17278387; http://dx.doi.org/10.1002/9780470035399.ch7
- Mantovani A, Garlanda C, Doni A, Bottazzi B. Pentraxins in innate immunity: from C-reactive protein to the long pentraxin PTX3. J Clin Immunol 2008; 28:1-13; PMID:17828584; http://dx.doi.org/10.1007/s10875-007-9126-7
- Garlanda C, Hirsch E, Bozza S, Salustri A, De Acetis M, Nota R, Maccagno A, Riva F, Bottazzi B, Peri G, et al. Non-redundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature 2002; 420:182-6; PMID:12432394; http://dx.doi.org/10.1038/nature01195
- Gaziano R, Bozza S, Bellocchio S, Perruccio K, Montagnoli C, Pitzurra L, Salvatori G, De Santis R, Carminati P, Mantovani A, et al. Anti-Aspergillus fumigatus efficacy of pentraxin 3 alone and in combination with antifungals. Antimicrob Agents Chemother 2004; 48:4414-21; PMID:15504871; http://dx.doi.org/10.1128/AAC.48.11.4414-4421.2004
- Jaillon S, Peri G, Delneste Y, Fremaux I, Doni A, Moalli F, Garlanda C, Romani L, Gascan H, Bellocchio S, et al. The humoral pattern recognition receptor PTX3 is stored in neutrophil granules and localizes in extracellular traps. J Exp Med 2007; 204:793-804; PMID:17389238; http://dx.doi.org/10.1084/jem.20061301
- Salvatori G, Campo S. Current understanding of PTX3 protective activity on Aspergillus fumigatus infection. Med Mycol 2012; 50:225-33; PMID:22309253; http://dx.doi.org/10.3109/13693786.2011.648215
- Jiang W, Johnson C, Jayaraman J, Simecek N, Noble J, Moffatt MF, Cookson WO, Trowsdale J, Traherne JA. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res 2012; 22:1845-54; PMID:22948769; http://dx.doi.org/10.1101/gr.137976.112
- Gardiner CM. NK cell function and receptor diversity in the context of HCV infection. Front Microbiol 2015; 6:1061; PMID:26483779
- Ivarsson MA, Michaelsson J, Fauriat C. Activating killer cell Ig-like receptors in health and disease. Front Immunol 2014; 5:184; PMID:24795726; http://dx.doi.org/10.3389/fimmu.2014.00184
- Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin Immunol 2008; 20:343-52; PMID:18635379; http://dx.doi.org/10.1016/j.smim.2008.06.003
- Cook M, Briggs D, Craddock C, Mahendra P, Milligan D, Fegan C, Darbyshire P, Lawson S, Boxall E, Moss P. Donor KIR genotype has a major influence on the rate of cytomegalovirus reactivation following T-cell replete stem cell transplantation. Blood 2006; 107:1230-2; PMID:16239436; http://dx.doi.org/10.1182/blood-2005-03-1039
- Chen C, Busson M, Rocha V, Appert ML, Lepage V, Dulphy N, Haas P, Socie G, Toubert A, Charron D, et al. Activating KIR genes are associated with CMV reactivation and survival after non-T-cell depleted HLA-identical sibling bone marrow transplantation for malignant disorders. Bone Marrow Transplant 2006; 38:437-44; PMID:16892071; http://dx.doi.org/10.1038/sj.bmt.1705468
- Sobecks RM, Askar M, Thomas D, Rybicki L, Kalaycio M, Dean R, Avery R, Mossad S, Copelan E, Bolwell BJ. Cytomegalovirus reactivation after matched sibling donor reduced-intensity conditioning allogeneic hematopoietic stem cell transplant correlates with donor killer immunoglobulin-like receptor genotype. Exp Clin Transplant 2011; 9:7-13; PMID:21605017
- Zaia JA, Sun JY, Gallez-Hawkins GM, Thao L, Oki A, Lacey SF, Dagis A, Palmer J, Diamond DJ, Forman SJ, et al. The effect of single and combined activating killer immunoglobulin-like receptor genotypes on cytomegalovirus infection and immunity after hematopoietic cell transplantation. Biol Blood Marrow Transplant 2009; 15:315-25; PMID:19203722; http://dx.doi.org/10.1016/j.bbmt.2008.11.030
- Gallez-Hawkins GM, Franck AE, Li X, Thao L, Oki A, Gendzekhadze K, Dagis A, Palmer J, Nakamura R, Forman SJ, et al. Expression of activating KIR2DS2 and KIR2DS4 genes after hematopoietic cell transplantation: relevance to cytomegalovirus infection. Biol Blood Marrow Transplant 2011; 17:1662-72; PMID:21596150; http://dx.doi.org/10.1016/j.bbmt.2011.04.008
- Xiao HW, Luo Y, Lai XY, Shi JM, Tan YM, He JS, Xie WZ, Zheng WY, Ye XJ, Yu XH, et al. Donor TLR9 gene tagSNPs influence susceptibility to aGVHD and CMV reactivation in the allo-HSCT setting without polymorphisms in the TLR4 and NOD2 genes. Bone Marrow Transplant 2014; 49:241-7; PMID:24121213; http://dx.doi.org/10.1038/bmt.2013.160
- Carvalho A, Cunha C, Carotti A, Aloisi T, Guarrera O, Di Ianni M, Falzetti F, Bistoni F, Aversa F, Pitzurra L, et al. Polymorphisms in Toll-like receptor genes and susceptibility to infections in allogeneic stem cell transplantation. Exp Hematol 2009; 37:1022-9; PMID:19539691; http://dx.doi.org/10.1016/j.exphem.2009.06.004
- Mezger M, Steffens M, Semmler C, Arlt EM, Zimmer M, Kristjanson GI, Wienker TF, Toliat MR, Kessler T, Einsele H, et al. Investigation of promoter variations in dendritic cell-specific ICAM3-grabbing non-integrin (DC-SIGN) (CD209) and their relevance for human cytomegalovirus reactivation and disease after allogeneic stem-cell transplantation. Clin Microbiol Infect 2008; 14:228-34; PMID:18076668; http://dx.doi.org/10.1111/j.1469-0691.2007.01902.x
- Saadi MI, Yaghobi R, Karimi MH, Geramizadeh B, Ramzi M, Zakerinia M. Association of the costimulatory molecule gene polymorphisms and active cytomegalovirus infection in hematopoietic stem cell transplant patients. Mol Biol Rep 2013; 40:5833-42; PMID:24057239; http://dx.doi.org/10.1007/s11033-013-2689-x
- Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol 2013; 13:227-42; PMID:23470321; http://dx.doi.org/10.1038/nri3405
- Sellathamby S, Lakshmi KM, Busson M, Viswabandya A, George B, Mathews V, Chandy M, Charron D, Krishnamoorthy R, Tamouza R, et al. Polymorphisms in the immunoregulatory genes are associated with hematopoietic recovery and increased susceptibility to bacterial infections in patients with thalassaemia major undergoing matched related hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 2012; 18:1219-26; PMID:22252124; http://dx.doi.org/10.1016/j.bbmt.2012.01.011
- Azarian M, Busson M, Rocha V, Ribaud P, Peffault de Latour R, Bleux H, Lepage V, Charron D, Toubert A, Socie G, et al. The PTPN22 R620W polymorphism is associated with severe bacterial infections after human leukocyte antigen geno-identical haematopoietic stem-cell transplantations. Transplantation 2008; 85:1859-62; PMID:18580482; http://dx.doi.org/10.1097/TP.0b013e31817729c4
- Lee KH, Park SS, Kim I, Kim JH, Ra EK, Yoon SS, Hong YC, Park S, Kim BK. P2X7 receptor polymorphism and clinical outcomes in HLA-matched sibling allogeneic hematopoietic stem cell transplantation. Haematologica 2007; 92:651-7; PMID:17488689; http://dx.doi.org/10.3324/haematol.10810
- Jaskula E, Dlubek D, Duda D, Bogunia-Kubik K, Mlynarczewska A, Lange A. Interferon gamma 13-CA-repeat homozygous genotype and a low proportion of CD4(+) lymphocytes are independent risk factors for cytomegalovirus reactivation with a high number of copies in hematopoietic stem cell transplantation recipients. Biol Blood Marrow Transplant 2009; 15:1296-305; PMID:19747638; http://dx.doi.org/10.1016/j.bbmt.2009.06.008
- Loeffler J, Steffens M, Arlt EM, Toliat MR, Mezger M, Suk A, Wienker TF, Hebart H, Nurnberg P, Boeckh M, et al. Polymorphisms in the genes encoding chemokine receptor 5, interleukin-10, and monocyte chemoattractant protein 1 contribute to cytomegalovirus reactivation and disease after allogeneic stem cell transplantation. J Clin Microbiol 2006; 44:1847-50; PMID:16672419; http://dx.doi.org/10.1128/JCM.44.5.1847-1850.2006
- Bravo D, Solano C, Gimenez E, Remigia MJ, Corrales I, Amat P, Navarro D. Effect of the IL28B Rs12979860 C/T polymorphism on the incidence and features of active cytomegalovirus infection in allogeneic stem cell transplant patients. J Med Virol 2014; 86:838-44; PMID:24374819; http://dx.doi.org/10.1002/jmv.23865
- Corrales I, Gimenez E, Solano C, Amat P, de la Camara R, Nieto J, Garcia-Noblejas A, Navarro D. Incidence and dynamics of active cytomegalovirus infection in allogeneic stem cell transplant patients according to single nucleotide polymorphisms in donor and recipient CCR5, MCP-1, IL-10, and TLR9 genes. J Med Virol 2015; 87:248-55; PMID:25132583; http://dx.doi.org/10.1002/jmv.24050
- Bogunia-Kubik K, Mlynarczewska A, Jaskula E, Lange A. The presence of IFNG 3/3 genotype in the recipient associates with increased risk for Epstein-Barr virus reactivation after allogeneic haematopoietic stem cell transplantation. Br J Haematol 2006; 132:326-32; PMID:16409297; http://dx.doi.org/10.1111/j.1365-2141.2005.05875.x
- Bogunia-Kubik K, Jaskula E, Lange A. The presence of functional CCR5 and EBV reactivation after allogeneic haematopoietic stem cell transplantation. Bone Marrow Transplant 2007; 40:145-50; PMID:17530006; http://dx.doi.org/10.1038/sj.bmt.1705703
- Bogunia-Kubik K, Mizia S, Polak M, Gronkowska A, Nowak J, Kyrcz-Krzemien S, Markiewicz M, Dzierzak-Mietla M, Koclega A, Sedzimirska M, et al. Beneficial effect of the CXCL12-3'A variant for patients undergoing hematopoietic stem cell transplantation from unrelated donors. Cytokine 2015; 76:182-6; PMID:25982843; http://dx.doi.org/10.1016/j.cyto.2015.05.001
- Plantinga TS, van der Velden WJ, Ferwerda B, van Spriel AB, Adema G, Feuth T, Donnelly JP, Brown GD, Kullberg BJ, Blijlevens NM, et al. Early stop polymorphism in human DECTIN-1 is associated with increased candida colonization in hematopoietic stem cell transplant recipients. Clin Infect Dis 2009; 49:724-32; PMID:19614557; http://dx.doi.org/10.1086/604714
- Carvalho A, De Luca A, Bozza S, Cunha C, D'Angelo C, Moretti S, Perruccio K, Iannitti RG, Fallarino F, Pierini A, et al. TLR3 essentially promotes protective class I-restricted memory CD8(+) T-cell responses to Aspergillus fumigatus in hematopoietic transplanted patients. Blood 2012; 119:967-77; PMID:22147891; http://dx.doi.org/10.1182/blood-2011-06-362582
- Grube M, Loeffler J, Mezger M, Kruger B, Echtenacher B, Hoffmann P, Edinger M, Einsele H, Andreesen R, Holler E. TLR5 stop codon polymorphism is associated with invasive aspergillosis after allogeneic stem cell transplantation. Med Mycol 2013; 51:818-25; PMID:23862689; http://dx.doi.org/10.3109/13693786.2013.809630
- Granell M, Urbano-Ispizua A, Suarez B, Rovira M, Fernandez-Aviles F, Martinez C, Ortega M, Uriburu C, Gaya A, Roncero JM, et al. Mannan-binding lectin pathway deficiencies and invasive fungal infections following allogeneic stem cell transplantation. Exp Hematol 2006; 34:1435-41; PMID:16982337; http://dx.doi.org/10.1016/j.exphem.2006.06.005
- Seo KW, Kim DH, Sohn SK, Lee NY, Chang HH, Kim SW, Jeon SB, Baek JH, Kim JG, Suh JS, et al. Protective role of interleukin-10 promoter gene polymorphism in the pathogenesis of invasive pulmonary aspergillosis after allogeneic stem cell transplantation. Bone Marrow Transplant 2005; 36:1089-95; PMID:16247433; http://dx.doi.org/10.1038/sj.bmt.1705181
- Mezger M, Steffens M, Beyer M, Manger C, Eberle J, Toliat MR, Wienker TF, Ljungman P, Hebart H, Dornbusch HJ, et al. Polymorphisms in the chemokine (C-X-C motif) ligand 10 are associated with invasive aspergillosis after allogeneic stem-cell transplantation and influence CXCL10 expression in monocyte-derived dendritic cells. Blood 2008; 111:534-6; PMID:17957030; http://dx.doi.org/10.1182/blood-2007-05-090928
- Carvalho A, Cunha C, Di Ianni M, Pitzurra L, Aloisi T, Falzetti F, Carotti A, Bistoni F, Aversa F, Romani L. Prognostic significance of genetic variants in the IL-23/Th17 pathway for the outcome of T cell-depleted allogeneic stem cell transplantation. Bone Marrow Transplant 2010; 45:1645-52; PMID:20173782; http://dx.doi.org/10.1038/bmt.2010.28
- Kesh S, Mensah NY, Peterlongo P, Jaffe D, Hsu K, M VDB O'Reilly R, Pamer E, Satagopan J, Papanicolaou GA. TLR1 and TLR6 polymorphisms are associated with susceptibility to invasive aspergillosis after allogeneic stem cell transplantation. AnnN Y Acad Sci 2005; 1062:95-103; PMID:16461792; http://dx.doi.org/10.1196/annals.1358.012
- Lupianez CB, Canet LM, Carvalho A, Alcazar-Fuoli L, Springer J, Lackner M, Segura-Catena J, Comino A, Olmedo C, Rios R, et al. Polymorphisms in Host Immunity-Modulating Genes and Risk of Invasive Aspergillosis: Results from the AspBIOmics Consortium. Infect Immun 2015; 84:643-57; PMID:26667837; http://dx.doi.org/10.1128/IAI.01359-15
- Cunha C, Giovannini G, Pierini A, Bell AS, Sorci G, Riuzzi F, Donato R, Rodrigues F, Velardi A, Aversa F, et al. Genetically-determined hyperfunction of the S100B/RAGE axis is a risk factor for aspergillosis in stem cell transplant recipients. PloS One 2011; 6:e27962; PMID:22114731; http://dx.doi.org/10.1371/journal.pone.0027962
- Zaas AK, Liao G, Chien JW, Weinberg C, Shore D, Giles SS, Marr KA, Usuka J, Burch LH, Perera L, et al. Plasminogen alleles influence susceptibility to invasive aspergillosis. PLoS Genet 2008; 4:e1000101; PMID:18566672; http://dx.doi.org/10.1371/journal.pgen.1000101
- Broen K, van der Waart AB, Greupink-Draaisma A, Metzig J, Feuth T, Schaap NP, Blijlevens NM, van der Velden WJ, Dolstra H. Polymorphisms in CCR6 are associated with chronic graft-versus-host disease and invasive fungal disease in matched-related hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 2011; 17:1443-9; PMID:21763254; http://dx.doi.org/10.1016/j.bbmt.2011.07.004
