ABSTRACT
Bacterial pathogenicity deeply depends on the ability to secrete virulence factors that bind specific targets on host cells and manipulate host responses. The Gram-positive bacterium Listeria monocytogenes is a human foodborne pathogen that remains a serious public health concern. To transport proteins across its cell envelope, this facultative intracellular pathogen engages a set of specialized secretion systems. Here we show that L. monocytogenes EGDe uses a specialized secretion system, named ESX-1, to secrete EsxA, a homolog of the virulence determinants ESAT-6 and EsxA of Mycobacterium tuberculosis and Staphylococcus aureus, respectively. Our data show that the L. monocytogenes ESX-1 secretion system and its substrates are dispensable for bacterial invasion and intracellular multiplication in eukaryotic cell lines. Surprisingly, we found that the EssC-dependent secretion of EsxA has a detrimental effect on L. monocytogenes in vivo infection.
Introduction
The capacity to secrete proteins is crucial for the pathogenesis of many bacteria. In Gram-positive bacteria, proteins are delivered by highly specialized secretion systems across cell envelope to reach specific targets.Citation1 A portion of these proteins, including virulence factors, are often secreted by Sec-independent systems. ESX-1, also called WXG100, is a Sec-independent secretion system first described in Mycobacterium tuberculosis.Citation2-4 This system allows the secretion of ≈100 amino-acid-long proteins that lack the classical signal peptide but contain a Trp-X-Gly motif (WXG100 proteins). M. tuberculosis ESAT-6 and CFP-10 are prototypes of WXG100 proteins, both encoded by the region of difference 1 (RD1), involved in virulence and described as highly immunogenic proteins.Citation5 Deletion of the esx-1 locus abrogates ESX-1-dependent secretion and strongly attenuates the virulence of M. tuberculosis.Citation6 Apart from mycobacterial species, ESX-1 systems are also found in Firmicutes, among which the S. aureus ESX-1 is one of the best-characterized.Citation7,8 It comprises genes encoding the canonical ESX-1 substrates (EsxA, EsxB), a membrane-anchored FtsK/SpoIIIE-like ATPase (EssC) essential for the secretion machinery, genes coding for membrane-embedded proteins (EssA, EssB, EssD) required for secretion of ESX-1 substrates, as well as staphylococci-specific ESX-1 substrates (EsxC, EsxD), and modulators of ESX-1 activity (EsaA, EsaB).Citation7-10 S. aureus EsxA and EsxB share features with the M. tuberculosis ESAT-6 and CFP-10, including the presence of a WXG motif and the co-dependent secretion. However, unlike ESAT-6 and CFP-10, EsxA and EsxB do not interact. EsxA dimerizes with itself or associates with EsxC, while EsxB interacts with EsxD.Citation8 Disruption of key components of the S. aureus ESX-1 secretion machinery (EssC) or deletion of esxA and esxB causes a significant reduction in the ability of S. aureus to establish kidney or liver abscesses.Citation7,9,11 Moreover, the S. aureus ESX-1 secretion system is required for nasal colonization and virulence in a murine lung pneumonia model.Citation12 EsxA was also shown to interfere with host cell apoptotic pathways, affecting bacterial survival and mediating S. aureus release from host cells.Citation13 Other ESX-1 substrates, such as EsaC, although dispensable for the establishment of acute infections, are required for the formation of persistent infection.Citation11 Functional ESX-1 secretion systems were also characterized in Bacillus anthracis, Bacillus subtilis, Actinobacterium and Streptomyces coelicolor.Citation14-17
Listeria monocytogenes (Lm) is a ubiquitous Gram-positive bacterium responsible for listeriosis, a severe opportunistic foodborne disease occurring mainly in immunocompromised individuals, newborn, elderly and pregnant women. Listeriosis is the most frequent cause of hospitalization and death due to the consumption of contaminated food in Europe, and involves high illness costs and quality life losses.Citation18 Clinical features of listeriosis includes septicemia, meningitis, meningoencephalitis and abortions. This facultative intracellular pathogen has evolved multiple strategies to survive inside phagocytic cells, invade non-phagocytic cells and spread from cell to cell.Citation19 Each step of its cell infection cycle depends on specific virulence determinants that play specific roles, most of them being surface or secreted proteins.Citation20,21 Genes encoding a potential ESX-1 secretion system were identified in LmCitation22 and the Lm EsxA homolog was previously shown dispensable for Lm mouse infection.Citation23 However, although ESX-1 was postulated to represent a broad Gram-positive secretion system,Citation1 the functionality of this apparatus and its role in infection were never previously investigated in Listeria.
Results
Lm encodes a putative ESX-1 secretion system
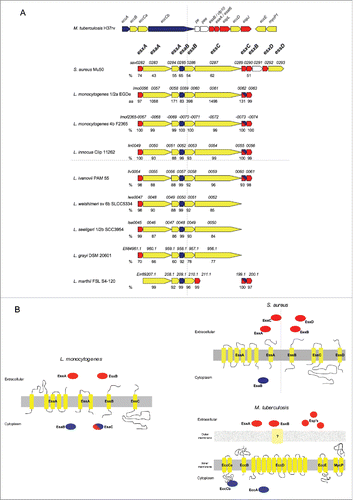
In agreement with the model proposed for the ESX-1 secretory apparatusCitation1,22 and following bioinformatic analyses (BLAST, TopPred2, ProDom) we determined that the Lm ESX-1 locus contains genes coding for: the 2 canonical WXG100 substrate paralogs EsxA (Lmo0056) and EsxB (Lmo0063), the integral membrane FtsK/SpoIIIE-type ATPase EssC (Lmo0061); EsaA (Lmo0057), a polytopic membrane protein with 5 predicted transmembrane helices; EssA (Lmo0058) and EssB (Lmo0060), 2 predicted membrane proteins with respectively one and 2 transmembrane domains; and EsaB (Lmo0059) and Lmo0062, 2 putative cytoplasmic proteins (). Membrane topology or soluble character of proteins encoded by the Lm ESX-1 locus was predicted and compared to M. tuberculosis and S. aureus ESX-1 secretion systems (). This revealed large similarities with the S. aureus ESX-1 systems. However, the 2 ESX-1 substrates (EsxC and EsxD) and the EssD transmembrane protein described in S. aureus are absent in Lm. Analysis of complete genome sequences available for different Listeria species revealed the high level of conservation of the ESX-1 locus within the Listeria genus (). Of note, esxB and lmo0062 are absent from 3 non-pathogenic species (L. welshimeri, L. seeligeri and L. grayi).
Figure 1. The ESX-1 locus. (A) Comparison of ESX-1 loci of M. tuberculosis, S. aureus, L. monocytogenes EGDe, and other Listeria species as indicated. Protein homology percentages relative to L. monocytogenes EGDe are indicated under each corresponding encoding gene. (B) Schematic representation showing membrane topology or soluble character of proteins encoded by the L. monocytogenes, S. aureus and M. tuberculosis ESX-1 locus. (A and B) Genes and proteins are colored following the same code: red corresponds to WXG100 encoding genes or proteins predicted to be secreted to the extracellular medium; blue indicates genes predicted as encoding soluble cytoplasmic proteins; yellow is related to genes or proteins predicted as transmembrane proteins.
Lm ESX-1 secretion system is functional although weakly expressed
To evaluate the expression of the Lm esx-1 locus, we analyzed by RT-PCR the transcription of genes encoding the 2 major ESX-1 effectors (esxA and esxB), and 2 integral membrane proteins essential for the secretion machinery (essB and essC).Citation7-10 RNAs extracted from bacteria in exponential growth phase in BHI at 37°C were processed for analysis and results showed that all the genes selected are transcribed in these conditions (). However, esxB, essC and essB appeared to be weakly expressed as compared to esxA and control genes (inlA, actA, iap) encoding known Lm virulence factors. We also observed that the expression of esxA appeared to decrease upon entry into stationary growth phase, which was confirmed by qRT-PCR (). We also attempted to assess by qRT-PCR the expression of esxB and essC during growth in exponential or stationary phases. However, transcript levels appeared insufficient to obtain quantifiable amplification, in particular during the stationary growth phase, thus underlining the weak level of expression of these genes. esxA was previously shown to be negatively regulated by SigB in S. aureus.Citation24 SigB being the major regulator of the stationary growth phase, we assessed if the decreased expression of esxA upon entry into this phase would be due to SigB regulation. We analyzed by qRT-PCR the expression of esxA in a sigB deletion mutant as compared to the WT strain during growth in exponential and stationary phases, and showed that in both growth phases esxA expression appeared independent of the presence of sigB (). bsh, which encodes a bile salt hydrolase, was used as a SigB-dependent control gene.Citation25
Figure 2. Lm ESX-1 system is weakly expressed but functional. (A) Expression of ESX-1 genes in standard growth conditions. The expression of esxA, esxB, essC and essB was analyzed by RT-PCR on total RNAs extracted from logarithmic cultures grown in BHI at 37°C. inlA, actA and iap were used as control genes. (B) Expression of esxA at exponential (Exp) and stationary (Stat) phase of growth measured by RT-PCR (left panel) and qRT-PCR (right panel). Expression value in stationary phase is expressed relative to the value obtained in exponential growth phase. (C) SigB-independent expression of esxA. qRT-PCRs performed on total RNAs extracted from WT and ΔsigB strains at the exponential (left panel) and stationary (right panel) phase of growth in BHI at 37°C. bsh was used as control gene whose expression is SigB-dependent. Gene expression levels in the ΔsigB mutant were normalized to those in the WT. (B and C) Values are mean ± SD (n = 3). (D) Secretion of EsxA is dependent on EssC. Detection of myc-tagged EsxA protein (EsxA-myc) in supernatants of Lm EGDe+esxA-myc (WT+esxA-myc) at different stages of growth (OD600nm = 0.7, 1.4 and 1.8) (Left panel), and in total bacterial lysates and supernatants from WT, WT+esxA-myc, ΔessC, ΔessC+esxA-myc and ΔessC+essC+esxA-myc strains in exponential growth phase (Right panel).
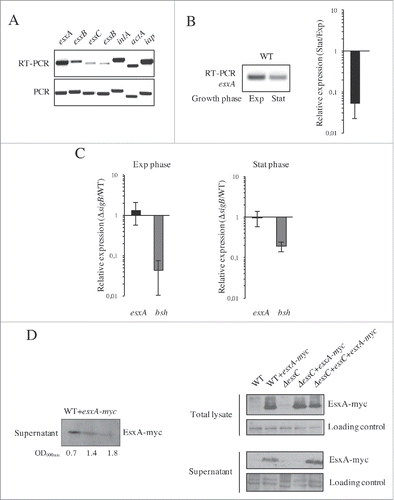
To analyze whether the putative Lm ESX-1 system was functional, we constructed a deletion mutant strain for essC (ΔessC), which encodes a structural protein essential for S. aureus ESX-1 functionality,Citation7 as well as the corresponding complemented strain (ΔessC+essC). Both strains were confirmed by PCR and sequencing, and their growth rate in BHI at 37°C was comparable to that of the WT (Fig. S1A-B). To test the functionality of the ESX-1 secretion system in Lm, we expressed a myc-tagged EsxA protein (EsxA-myc) in WT bacteria and analyzed its secretion during bacterial growth. Western blot of bacterial culture supernatants showed a band of 11 kDa, the expected size for EsxA-myc, with a higher intensity in the exponential growth phase ( left panel). These results indicate that Lm EsxA was secreted in these conditions. We then expressed EsxA-myc in the ΔessC and ΔessC+essC strains and performed the same western blot analysis on proteins from the WT, WT+esxA-myc, ΔessC, ΔessC+esxA-myc and ΔessC+essC+esxA-myc total bacterial lysates and culture supernatants in exponential growth phase. In the WT+esxA-myc strain, EsxA-myc was detected both in the total lysates and culture supernatants, indicating that the protein is produced and secreted ( right panel). In the ΔessC+esxA-myc strain, EsxA-myc was absent from the supernatant and retained in total lysates, demonstrating that EsxA is secreted in an EssC-dependent manner. This was confirmed by the complementation of the ΔessC+esxA-myc mutant that restored the secretion of EsxA-myc (ΔessC+essC+esxA-myc line). Altogether, these results show that Lm expresses a functional ESX-1 secretion system, albeit at low levels, and that EsxA secretion requires the putative membrane ATPase EssC.
Lm ESX-1 is dispensable for host cell invasion and intracellular multiplication
To investigate the role of the ESX-1 system in Lm cell invasion and intracellular multiplication, we constructed deletion mutants for esxA, that we showed to encode a substrate of Lm ESX-1, and esxB that encodes another putative ESX-1 substrate.Citation7 Mutants were confirmed by PCR and sequencing, and their growth rates observed in BHI at 37°C were comparable to that of the WT (Fig. S1A-B).
The WT, ΔesxA, ΔesxB and ΔessC strains were tested for their capacity to invade epithelial cell lines in which Lm entry is mainly mediated by internalin A (InlA) (Caco-2) or InlB (Vero). No significant difference in invasion was observed between mutant and WT bacteria in both cell lines (), suggesting that the ESX-1 system is not required for Lm invasion of epithelial cells.
Figure 3. The Lm ESX-1 secretion system is dispensable for epithelial cell invasion and intracellular multiplication in macrophages. (A) Entry of the WT, ΔesxA, ΔesxB and ΔessC into Caco-2 and Vero cell lines. Values are expressed relative to WT values arbitrarily fixed to 100%. (B) Intracellular replication behavior of the WT, ΔesxA, ΔesxB and ΔessC strains in J774 cells. Values are mean ± SD (n = 3).
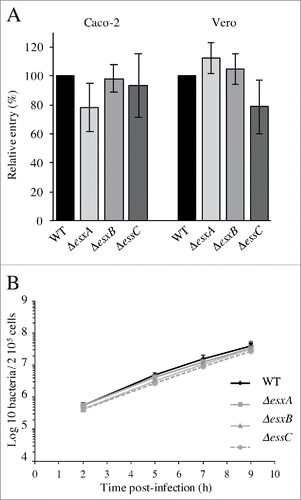
To analyze the role of ESX-1 in Lm intracellular multiplication, the behavior of the WT, ΔesxA, ΔesxB and ΔessC strains was studied after internalization in J774 murine macrophage-like cells. All strains grew with similar multiplication rates after uptake (), indicating that none of these genes is required for Lm intracellular replication in macrophage-like cells. Altogether these results indicate that the ESX-1 secretion system is dispensable for Lm cell invasion and intracellular multiplication.
ESX-1 activity impairs Lm infection
To analyze the involvement of the ESX-1 system in Lm infection in vivo, we monitored the number of bacteria in the liver and spleen of mice infected intravenously with ΔesxA, ΔesxB, ΔessC or WT bacteria. Unexpectedly, 72h post-infection, all the mutant strains showed a slight increase in bacterial counts in both organs as compared to WT, which was statistically significant for ΔesxB (), implying that ESX-1 activity might have a negative impact on Lm infection.
Figure 4. Lm ESX-1 secretion system is detrimental for in vivo infection. Bacterial counts for the WT, ΔesxA, ΔesxB and ΔessC strains, in spleens and livers of BALB/c mice (n = 5), 72h after (A) intravenous infection with 104 bacteria or (B) oral infection with 109 bacteria. Data are presented as scatter plots, which each animal represented by a dot and the mean is indicated by a horizontal line. *, p ≤ 0.05.
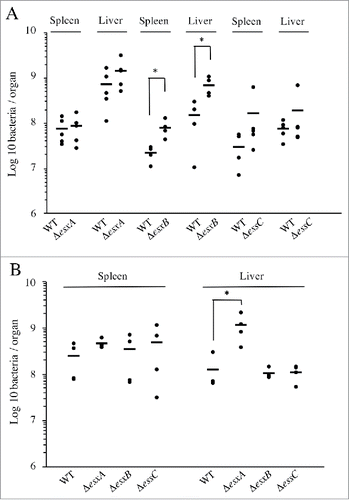
To investigate the potential role of ESX-1 in the gastrointestinal phase of the infectious process, we performed oral inoculation of mice with the WT, ΔesxA, ΔesxB and ΔessC strains. Three days post-inoculation, mutant strains appeared again to be slightly more virulent than the WT. This increased infection was statistically significant for ΔesxA in mouse livers ().
These data indicate that neither the ESX-1 apparatus nor its substrates are crucial for Lm infection in the mouse model. Conversely, they suggest that the expression/function of this secretion system causes an adverse effect in Lm pathogenicity.
Overexpression of esxA in the context of a functional ESX-1 system is detrimental to Lm infection
To further investigate if the ESX-1 function could have a negative effect on Lm infection, we performed intravenous infection of mice with Lm overexpressing esxA (+esxA), together with the WT and ΔesxA strains. The esxA overexpression in the +esxA strain was first confirmed by qRT-PCR (Fig. S2A). Growth rate of the +esxA strain in BHI or minimal medium at 37°C, as well as its cell adhesion and infection capacity were comparable to that of the WT strain (Fig. S2B-C), indicating that esxA overexpression has no significant impact on Lm growth and cellular infectious properties. Three days post infection the ΔesxA mutant appeared slightly more virulent as compared to the WT, as already observed (), whereas +esxA bacteria showed a significant number decrease in both mouse organs (). Inversely, esxA overexpression had no effect on the phenotype of a ΔessC mutant that, similarly to the ΔesxA mutant, also appeared to colonize more efficiently mouse organs than the WT strain (). These results demonstrate that the detrimental effect of ESX-1 on Lm pathogenicity is due to EsxA secretion and depends on a functional ESX-1 machinery.
Figure 5. EsxA secretion impairs Lm infection in vivo through an IFN-γ-independent mechanism. (A) Bacterial counts for WT, ΔesxA and +esxA strains, in spleens and livers of BALB/c mice (n = 5), 72h after intravenous infection with 104 bacteria. (B) Bacterial counts for WT and ΔessC+esxA strains, in spleens and livers of BALB/c mice (n = 5), 72h after intravenous infection with 104 bacteria. (C) Levels of IFN-γ and TNF-α transcripts measured by qRT-PCR in livers of mice 72h after intravenous infection with either WT or +esxA bacteria. Expression levels in +esxA-infected livers were normalized to those infected with the WT strain. Values are mean ± SD (n = 3). (D) Bacterial counts for WT and +esxA strains in spleens and livers of WT and IFN-γ knock-out mice (n = 5), 72h after intravenous infection with 104 bacteria. *, p ≤ 0.05; **, p ≤ 0.01.
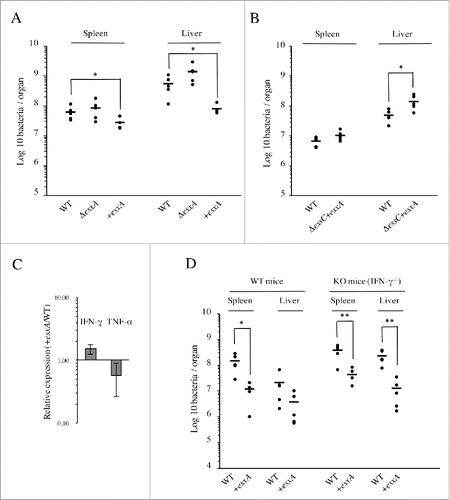
The production of IFN-γ and TNF-α by immune cells promotes bacterial clearance and is critical in controlling primary L. monocytogenes infections.Citation26 To investigate if the adverse effect of esxA overexpression on Lm infection is related with higher levels of host IFN-γ and/or TNF-α, we analyzed by qRT-PCR levels of IFN-γ and TNF-α transcripts in the liver of WT or +esxA-infected mice. No significant difference was observed regarding expression levels of IFN-γ and TNF-α (). In addition, to discard any role of IFN-γ in the increased resistance of mice to esxA overexpressing Lm, WT and IFN-γ knock-out mice (IFN-γ−/−) were intravenously infected with WT or +esxA bacteria. Three days post-infection, bacterial loads were overall higher in the organs of IFN-γ−/− than in WT mice (). These data indicated that IFN-γ deficient mice are more susceptible to Lm infection and confirmed the role of IFN-γ in the immune response against Lm.Citation27 In line with data observed in BALB/c mice (), the +esxA strain showed significant infection attenuation in both organs of WT mice as compared to Lm WT bacteria (). However, this infection defect was similar in IFN-γ deficient animals, suggesting that IFN-γ is not involved in the increased resistance of mice to Lm overexpressing esxA.
Discussion
The analysis of the bacterial ability to secrete proteins to or beyond their surface is crucial in the understanding of bacterial pathogenesis. In M. tuberculosis and S. aureus, ESX-1 and its substrates were shown to play an important role in virulence.Citation5,7 ESX-1 appears to be very conserved in Listeria, in particular among pathogenic species, and only partially present in the majority of non-pathogenic Listeria species.
We showed that the ESX-1 locus of L. monocytogenes EGDe is expressed in standard growth conditions, with esxA appearing more expressed than esxB, essB and essC. These results are in agreement with previous transcriptional analysesCitation28 and are consistent with the presence of a transcription terminator between esxA and esaA. This could suggest a different transcriptional regulation between esxA and the other ESX-1 genes. Regarding the expression of ESX-1 genes in different conditions, no change was observed when bacteria were grown at 37°C, 25°C or 7°C,Citation29,30 nor in presence of 6% NaCl,Citation29 nor when grown at pH 5,Citation31 nor when grown in culture media supplemented with glucose, cellobiose or glycerol.Citation32,33 In addition, as compared to bacteria grown in BHI at 37°C, the expression of the ESX-1 locus appears also unchanged in Listeria recovered from infected murine macrophages,Citation34,35 or from mouse intestinal lumenCitation36 and spleens.Citation37 Interestingly, the entire ESX-1 locus was shown as up-regulated after incubation of Lm in human blood,Citation36 and essC and esxC were also shown as upregulated during cell infection.Citation34 Altogether, these results indicate that the ESX-1 locus of L. monocytogenes EGDe is poorly or not expressed in most of the conditions, with some genes of the locus expressed in few conditions that could suggest a role of the ESX-1 apparatus in these specific environments.
Using a deletion mutant and complemented strain for essC, we demonstrated that ESX-1 is functional in Lm, at least for the secretion of EsxA. Even if EssC was shown to be an essential ESX-1 element also for the secretion of EsxB in M. tuberculosis and S. aureus,Citation5,7 the EssC-dependent secretion of EsxB remains to be confirmed in Lm. EsxA and EsxB are the only WXG100 proteins predicted to be encoded by the Lm genome. In addition to EsxA and EsxB, the EsxC protein is also a substrate for ESX-1 in S. aureus and confers pathogenic function to this bacterium.Citation11 However, an esxC ortholog is absent from Lm. Instead, an unrelated gene of unknown function (lmo0062) occupies the esxC position in the Lm genome (). Interestingly, Lmo0062 was recently predicted to be secreted,Citation38 therefore investigating its capacity to be secreted in an ESX-1-dependent manner could widen the spectrum of ESX-1 substrates to proteins that do not belong to the WXG100 family.
All the above observations induced a strong presumption for the involvement of ESX-1 in Lm pathogenicity. However, we demonstrated that this secretion system and its substrates are not required for Lm cell invasion, intracellular multiplication and in vivo infection. In agreement with our results, EsxA was also previously shown dispensable for Lm mouse infection.Citation23 Unexpectedly, we observed a slight increase in the infection level of mutants for ESX-1 components as compared to WT bacteria. This was also previously observed for a ΔesxA mutant,Citation23 suggesting a damaging role for ESX-1 in Listeria infectious capacity. In agreement with this hypothesis, overexpression of esxA resulted in a decrease infection of Lm in the mouse model, confirming the adverse effect of a functional ESX-1 secretion system. Despite our attempts to elucidate the reasons of this detrimental role, we failed to find differences regarding host immune responses upon infection by WT or esxA overexpressing Lm. ESAT-6 (the mycobacterial EsxA homolog) was shown to play a pro-apoptotic role in M. tuberculosis.Citation39 A comparable role of Lm EsxA could result in increased bacterial recognition and clearance by the host immune system that would explain the phenotype of esxA overexpressing bacteria. The absence of EssC in the bacterial membrane could also disturb cell envelope homeostasis, possibly resulting in the mislocalization of some surface proteins that could induce an increased virulence. However, no difference was observed regarding the capacity of mutants for ESX-1 components to invade or multiply inside host cells as compared to WT Lm.
L. monocytogenes is sensitive to a broad range of antibiotics. However, resistance to several antibiotics has been reported,Citation40 as well as multidrug-resistant strains.Citation41 The ESX-1 system could appear as a potential target for innovative anti-Listeria drugs that, by inducing ESX-1-dependent secretion, would be capable to impair the infectious capacity of bacteria but not their viability, significantly reducing the risk of resistance development.
Listeria has maintained this locus in its genome, suggesting that it may probably be helpful in certain conditions, such as resisting to stress encountered in specific environments. However, we were unable to find any difference between the WT and essC mutant regarding growth in stress conditions such as low pH (pH 5.5) and high salt concentration (4.5% NaCl) (Fig. S1C). Another reason for the conservation of this locus in the Listeria genome could be related to a strain issue. Indeed, we tested here the role of the ESX-1 system in only one specific Lm strain (L. monocytogenes EGDe), that is one of the most commonly used laboratory strains.Citation42,43 Even if EsxA was also shown to be dispensable for in vitro and in vivo growth of a different widely used Lm strain,Citation23,43 this locus could play important roles in the infection capacity of other Listeria strains from different serotypes.
In summary, we demonstrated here that the Lm genome encodes a functional ESX-1 secretion system required for the secretion of WXG100 proteins, such as EsxA. In addition, despite poorly expressed and dispensable for cell invasion, we showed that a working ESX-1 system is detrimental for Lm infection in vivo. Considering its wide distribution among Gram-positive bacteria and the lack of a convergent phenotypic trait for mutants in this pathway, ESX-1 certainly fulfils different functions that remain to be elucidated.
Materials and methods
Bacterial strains and media
Lm EGDe (ATCC-BAA-679) and E. coli strains were routinely cultured aerobically at 37 °C in brain heart infusion (BHI, Difco) and Lysogeny Broth (LB) media, respectively, with shaking.
The synthetic minimal medium (MM) was prepared as previously described.Citation44 When appropriate, the following antibiotics were included in culture media as selective agents: ampicilin (Amp), 100 μg/ml; chloramphenicol (Cm), 7 μg/ml (Lm) or 20 μg/ml (E. coli); erythromycin (Ery), 5 μg/ml. For genetic complementation purposes, colistin sulfate (Col) and nalidixic acid (Nax) were used at 10 and 50 μg/ml, respectively.
Construction and complementation of mutant strains
lmo0056, lmo0061 and lmo0063 deletions were performed in the EGD-e background through a process of double homologous recombination mediated by the suicide plasmid pMAD as describedCitation45 using the corresponding oligonucleotides (A-D; Table S1). Genetic complementation of the deletion mutant strains was performed using the phage-derived integrative plasmid pPL2 as describedCitation45 using the respective oligonucleotides (Table-S1). For overexpression, target genes were cloned into the pRB474 vector as described.Citation46 All plasmid constructs and strains were confirmed by PCR and DNA sequencing.
Western blot analysis of EsxA
Overnight bacterial cultures grown in static conditions (final OD660 ∼1.0) were centrifuged (10 000 g, 10 min, 4°C) and the supernatant filtered using Millipore 0.45 µm filters. A volume of 1.6 ml of 50% trichloroacetic acid was added to 6 ml of the filtered supernatant and incubated for 1 h at 4°C. The sample was centrifuged (30 000 g, 20 min, 4°C) and the pellet washed with cold acetone, repeating the centrifugation step in same conditions. The pellet was dried, suspended in 30 µl of PBS pH 7.4 and mixed with 15 µl of 4x Laemmli buffer. A volume of 15 µl was loaded in a 12% SDS-polyacrylamide gel. For total bacterial lysates, the bacterial pellet from 10 ml of culture was suspended in 0.8 ml of PBS pH 7.4 containing 100 µg/ml DNase and protease inhibitors. Bacteria were lysed in a FastPrep-24 homogenizer (MP Biomedicals) (30 s, maximum speed) and cell debris removed by centrifugation (3 000 g, 5 min, 4°C). A volume of 40 µl of 4x Laemmli buffer was added to 100 µl of supernatant of bacterial lysates, and 10 µl loaded into the gels. Western blotting was performed as describedCitation46 using anti-Myc tag mouse antibody (#clone 9B11, Cell Signaling #2276).
Gene expression analyses
Bacterial RNAs were isolated from 10 ml of cultures at the desired growth phase. For quantification of cytokine expression in mouse livers, organs were homogenized in RNAlater stabilization solution (Qiagen), quick-frozen in dry ice and stored at -80°C. Total RNAs were extracted by the phenol-chloroform method as previously described,Citation47 and treated with DNase I (Turbo DNA-free, Ambion) as recommended by the manufacturer. Purified RNAs (1 μg) were reverse-transcribed with random hexamers, using iScript cDNA Synthesis kit (Bio-Rad Laboratories). For qualitative analysis, PCR was performed in 20-μl reactions containing 2 μl of cDNA, 10 μl of MangoMix 2 × reaction mix (Bioline) and 0.5 μM of forward and reverse primers (Table S1), using the following protocol: 1 cycle at 95 °C (5 min), 25 cycles at 95 °C (30 s), 55 °C (30 s) and 72 °C (20 s), and 1 cycle at 72 °C (5 min). Amplification products were resolved in 1% (w/v) agarose gel and analyzed in a GelDoc XR+ System (Bio-Rad Laboratories). Quantitative real-time PCR (qRT-PCR) was performed in 20-μl reactions containing 2 μl of cDNA, 10 μl of SYBR Green Supermix (Bio-Rad Laboratories) and 0.25 μM of forward and reverse primers (Table S1), using the following cycling protocol: 1 cycle at 95 °C (3 min) and 40 cycles at 95 °C (30 s), 55 °C (30 s) and 72 °C (30 s). Each target gene was analyzed in triplicate and blank (water) and DNA contamination controls (unconverted DNase I-treated RNA) were included for each primer pair. Amplification data were analyzed by the comparative threshold (ΔΔCt) method, after normalization of the test and control sample expression values to a housekeeping reference gene (16S rRNA).
Adhesion and invasion assays
Adhesion and invasion assays were performed as described.Citation45 Briefly, Caco-2 (ATCC, HTB-37) and Vero (ATCC, CCL-81) cells were seeded (in triplicate) per 24-well plates (∼2 × 1055/well) in EMEM 20% foetal bovine serum and DMEM 10% foetal bovine serum, respectively (LONZA), and propagated for 48 h. Listeria were grown in BHI to OD600nm = 0.8, washed and inoculated at 50 bacteria-per-cell for 1h. For adhesion, cells were washed 3 times, lysed in 0.2% Triton X-100 and viable bacteria were enumerated after plating serial dilutions of the lysates in BHI agar media. For invasion assays, cells were infected for 1h and treated with 20 μg/ml gentamicin for 1h30 before lysis in 0.2% Triton X-100.
Intracellular multiplication
Mouse macrophage-like J774A.1 cells (ATCC TIB-67) were propagated in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and infection assays were performed as described.Citation37 Briefly, cells (∼2×1055/well) were infected for 45 min with exponential-phase bacteria at ˜10 bacteria/cell and treated afterwards with 20 μg/ml gentamicin for 75 min. At several time-points post-infection, cells were washed with PBS and lysed in cold 0.2% Triton X-100 for quantification of viable intracellular bacteria in BHI agar. One experiment was performed with triplicates for each strain and time-point.
Animal infections
Infections were performed in 6-to-8 week-old specific-pathogen-free females as described.Citation48 Briefly, wild-type BALB/c (Charles River Laboratories) or wild-type IFN-γ knockout C57BL/6J mice were infected intravenously with 104 CFUs in PBS, or starved 12h before gavage inoculation with 109 CFUs in PBS containing 150 mg/ml CaCO3. The infection was carried out for 72 h, at which point the animals were euthanized by general anesthesia. The spleen and liver were aseptically collected, homogenized in sterile PBS, and serial dilutions of the organ homogenates plated in BHI agar. Mice were maintained at the IBMC animal facilities, in high efficiency particulate air (HEPA) filter-bearing cages under 12-h light cycles, and were given sterile chow and autoclaved water ad libitum.
Ethics statement
All the animal procedures were in agreement with the guidelines of the European Commission for the handling of laboratory animals (directive 2010/63/EU), with the Portuguese legislation for the use of animals for scientific purposes (Decreto-Lei 113/2013), and were approved by the IBMC Animal Ethics Committee, as well as by the Direcção Geral de Veterinária, the Portuguese authority for animal protection, under license PTDC/SAU-MIC/111581/2009.
Statistical analyses
Statistical analyses were performed with Prism 6 (GraphPad Software). Unpaired two-tailed Student's t-test was used to compare the means of 2 groups; one-way ANOVA was used with Tukey's post-hoc test for pairwise comparison of means from more than 2 groups, or with Dunnett's post-hoc test for comparison of means relative to the mean of a control group. Mean differences were considered statistically non-significant (ns) when p value was above 0.05. For statistically significant differences: *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KVIR_S_1244589.zip
Download Zip (198.4 KB)Acknowledgments
We thank Rui Appelberg for PhD co-supervision of O.R., J.P. and F.C. and MT.A. We thank S. Lamas (Animal Facility), and P. Magalhaes (CCGEN) from IBMC facilities for technical assistance.
Funding
This work was supported for the DC lab by national funds through FCT - Fundação para a Ciência e a Tecnologia/MEC - Ministério da Educação e Ciência and co-funded by FEDER funds within the partnership agreement PT2020 related with the research unit number 4293, and within the research project Infect-ERA/0001/2013 PROANTILIS. O.R., J.P. and F.C. were supported by doctoral fellowships from FCT (SFRH/BD/28185/2006, SFRH/BD/86871/2012 and SFRH/BD/61825/2009), L.M. by the Santander International Mobility Program and SS by FCT Investigator program (COMPETE, POPH, and FCT). This work was supported for the F.G.-dP. lab by grants BIO2014-55238-R (to M.G.P.) and BIO2013-46281-P (to F.G.-dP.) from the Spanish Ministry of Economy and Competitiveness.
ORCID
Francisco García-del Portillo http://orcid.org/0000-0002-4120-0530
Sandra Sousa http://orcid.org/0000-0001-8578-0461
Didier Cabanes http://orcid.org/0000-0002-4001-1332
References
- Schneewind O, Missiakas DM. Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc Lond B Biol Sci 2012; 367:1123-39; PMID:22411983; http://dx.doi.org/10.1098/rstb.2011.0210
- Simeone R, Bottai D, Frigui W, Majlessi L, Brosch R. ESX/type VII secretion systems of mycobacteria: Insights into evolution, pathogenicity and protection. Tuberculosis 2015; 95 Suppl 1:S150-4; PMID:25732627; http://dx.doi.org/10.1016/j.tube.2015.02.019
- Houben EN, Korotkov KV, Bitter W. Take five - Type VII secretion systems of Mycobacteria. Biochim Biophys Acta 2014; 1843:1707-16; PMID:24263244; http://dx.doi.org/10.1016/j.bbamcr.2013.11.003
- Bottai D, Groschel MI, Brosch R. Type VII Secretion Systems in Gram-Positive Bacteria. Curr Top Microbiol Immunol 2016; pp. 1-31; PMID:26847354
- Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci U S A 2003; 100:13001-6; PMID:14557536; http://dx.doi.org/10.1073/pnas.2235593100
- Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, Behr MA, Sherman DR. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J Infect Dis 2003; 187:117-23; PMID:12508154; http://dx.doi.org/10.1086/345862
- Burts ML, Williams WA, DeBord K, Missiakas DM. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc Natl Acad Sci U S A 2005; 102:1169-74; PMID:15657139; http://dx.doi.org/10.1073/pnas.0405620102
- Anderson M, Aly KA, Chen YH, Missiakas D. Secretion of atypical protein substrates by the ESAT-6 secretion system of Staphylococcus aureus. Mol Microbiol 2013; 90:734-43; PMID:24033479; http://dx.doi.org/10.1111/mmi.12395
- Anderson M, Chen YH, Butler EK, Missiakas DM. EsaD, a secretion factor for the Ess pathway in Staphylococcus aureus. J Bacteriol 2011; 193:1583-9; PMID:21278286; http://dx.doi.org/10.1128/JB.01096-10
- Chen YH, Anderson M, Hendrickx AP, Missiakas D. Characterization of EssB, a protein required for secretion of ESAT-6 like proteins in Staphylococcus aureus. BMC Microbiol 2012; 12:219; PMID:23006124; http://dx.doi.org/10.1186/1471-2180-12-219
- Burts ML, DeDent AC, Missiakas DM. EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Mol Microbiol 2008; 69:736-46; PMID:18554323; http://dx.doi.org/10.1111/j.1365-2958.2008.06324.x
- Kneuper H, Cao ZP, Twomey KB, Zoltner M, Jager F, Cargill JS, Chalmers J, van der Kooi-Pol MM, van Dijl JM, Ryan RP, et al. Heterogeneity in ess transcriptional organization and variable contribution of the Ess/Type VII protein secretion system to virulence across closely related Staphylocccus aureus strains. Mol Microbiol 2014; 93:928-43; PMID:25040609; http://dx.doi.org/10.1111/mmi.12707
- Korea CG, Balsamo G, Pezzicoli A, Merakou C, Tavarini S, Bagnoli F, Serruto D, Unnikrishnan M. Staphylococcal Esx proteins modulate apoptosis and release of intracellular Staphylococcus aureus during infection in epithelial cells. Infect Immun 2014; 82:4144-53; PMID:25047846; http://dx.doi.org/10.1128/IAI.01576-14
- Garufi G, Butler E, Missiakas D. ESAT-6-like protein secretion in Bacillus anthracis. J Bacteriol 2008; 190:7004-11; PMID:18723613; http://dx.doi.org/10.1128/JB.00458-08
- Akpe San Roman S, Facey PD, Fernandez-Martinez L, Rodriguez C, Vallin C, Del Sol R, Dyson P. A heterodimer of EsxA and EsxB is involved in sporulation and is secreted by a type VII secretion system in Streptomyces coelicolor. Microbiology 2010; 156:1719-29; PMID:20223806; http://dx.doi.org/10.1099/mic.0.037069-0
- Baptista C, Barreto HC, Sao-Jose C. High levels of DegU-P activate an Esat-6-like secretion system in Bacillus subtilis. PLoS One 2013; 8:e67840; PMID:23861817; http://dx.doi.org/10.1371/journal.pone.0067840
- Huppert LA, Ramsdell TL, Chase MR, Sarracino DA, Fortune SM, Burton BM. The ESX system in Bacillus subtilis mediates protein secretion. PLoS One 2014; 9:e96267; PMID:24798022; http://dx.doi.org/10.1371/journal.pone.0096267
- European Food Safety Authority ECfDPaC. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. Euro Food Safety Authority, European Centre Dis Prevention Control EFSA J 2015; 13:3991.
- Cossart P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci U S A 2011; 108:19484-91; PMID:22114192; http://dx.doi.org/10.1073/pnas.1112371108
- Camejo A, Carvalho F, Reis O, Leitao E, Sousa S, Cabanes D. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2011; 2:379-94; PMID:21921683; http://dx.doi.org/10.4161/viru.2.5.17703
- Carvalho F, Sousa S, Cabanes D. How Listeria monocytogenes organizes its surface for virulence. Front Cell Infect Microbiol 2014; 4:48; PMID:24809022; http://dx.doi.org/10.3389/fcimb.2014.00048
- Desvaux M, Hebraud M. The protein secretion systems in Listeria: inside out bacterial virulence. FEMS Microbiol Rev 2006; 30:774-805; PMID:16911044; http://dx.doi.org/10.1111/j.1574-6976.2006.00035.x
- Way SS, Wilson CB. The Mycobacterium tuberculosis ESAT-6 homologue in Listeria monocytogenes is dispensable for growth in vitro and in vivo. Infect Immun 2005; 73:6151-3; PMID:16113338; http://dx.doi.org/10.1128/IAI.73.9.6151-6153.2005
- Schulthess B, Bloes DA, Berger-Bachi B. Opposing roles of sigmaB and sigmaB-controlled SpoVG in the global regulation of esxA in Staphylococcus aureus. BMC Microbiol 2012; 12:17; PMID:22272815; http://dx.doi.org/10.1186/1471-2180-12-17
- Zhang Q, Feng Y, Deng L, Feng F, Wang L, Zhou Q, Luo Q. SigB plays a major role in Listeria monocytogenes tolerance to bile stress. Int J Food Microbiol 2011; 145:238-43; PMID:21262551; http://dx.doi.org/10.1016/j.ijfoodmicro.2010.12.028
- Stavru F, Archambaud C, Cossart P. Cell biology and immunology of Listeria monocytogenes infections: novel insights. Immunol Rev 2011; 240:160-84; PMID:21349093; http://dx.doi.org/10.1111/j.1600-065X.2010.00993.x
- Harty JT, Bevan MJ. Specific immunity to Listeria monocytogenes in the absence of IFN gamma. Immunity 1995; 3:109-17; PMID:7621071; http://dx.doi.org/10.1016/1074-7613(95)90163-9
- Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Becavin C, Archambaud C, Cossart P, Sorek R. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol 2012; 8:583; PMID:22617957; http://dx.doi.org/10.1038/msb.2012.11
- Bergholz TM, Bowen B, Wiedmann M, Boor KJ. Listeria monocytogenes shows temperature-dependent and -independent responses to salt stress, including responses that induce cross-protection against other stresses. Appl Environ Microbiol 2012; 78:2602-12; PMID:22307309; http://dx.doi.org/10.1128/AEM.07658-11
- Garmyn D, Augagneur Y, Gal L, Vivant AL, Piveteau P. Listeria monocytogenes differential transcriptome analysis reveals temperature-dependent Agr regulation and suggests overlaps with other regulons. PLoS One 2012; 7:e43154; PMID:23024744; http://dx.doi.org/10.1371/journal.pone.0043154
- Neuhaus K, Satorhelyi P, Schauer K, Scherer S, Fuchs TM. Acid shock of Listeria monocytogenes at low environmental temperatures induces prfA, epithelial cell invasion, and lethality towards Caenorhabditis elegans. BMC Genomics 2013; 14:285; PMID:23622257; http://dx.doi.org/10.1186/1471-2164-14-285
- Marr AK, Joseph B, Mertins S, Ecke R, Muller-Altrock S, Goebel W. Overexpression of PrfA leads to growth inhibition of Listeria monocytogenes in glucose-containing culture media by interfering with glucose uptake. J Bacteriol 2006; 188:3887-901; PMID:16707681; http://dx.doi.org/10.1128/JB.01978-05
- Joseph B, Mertins S, Stoll R, Schar J, Umesha KR, Luo Q, Müller-Altrock S, Goebel W. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J Bacteriol 2008; 190:5412-30; PMID:18502850; http://dx.doi.org/10.1128/JB.00259-08
- Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T. Intracellular gene expression profile of Listeria monocytogenes. Infect Immun 2006; 74:1323-38; PMID:16428782; http://dx.doi.org/10.1128/IAI.74.2.1323-1338.2006
- Schultze T, Hilker R, Mannala GK, Gentil K, Weigel M, Farmani N, Windhorst AC, Goesmann A, Chakraborty T, Hain T. A detailed view of the intracellular transcriptome of Listeria monocytogenes in murine macrophages using RNA-seq. Front Microbiol 2015; 6:1199; PMID:26579105; http://dx.doi.org/10.3389/fmicb.2015.01199
- Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature 2009; 459:950-6; PMID:19448609; http://dx.doi.org/10.1038/nature08080
- Camejo A, Buchrieser C, Couve E, Carvalho F, Reis O, Ferreira P, Sousa S, Cossart P, Cabanes D. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog 2009; 5:e1000449; PMID:19478867; http://dx.doi.org/10.1371/journal.ppat.1000449
- Renier S, Micheau P, Talon R, Hebraud M, Desvaux M. Subcellular Localization of Extracytoplasmic Proteins in Monoderm Bacteria: Rational Secretomics-Based Strategy for Genomic and Proteomic Analyses. PLoS One 2012; 7:e42982; PMID:22912771; http://dx.doi.org/10.1371/journal.pone.0042982
- Aguilo JI, Alonso H, Uranga S, Marinova D, Arbues A, de Martino A, Anel A, Monzon M, Badiola J, Pardo J, et al. ESX-1-induced apoptosis is involved in cell-to-cell spread of Mycobacterium tuberculosis. Cell Microbiol 2013; 15:1994-2005; PMID:23848406; http://dx.doi.org/10.1111/cmi.12169
- Walsh D, Sheridan JJ, Duffy G, Blair IS, McDowell DA, Harrington D. Thermal resistance of wild-type and antibiotic-resistant Listeria monocytogenes in meat and potato substrates. J Appl Microbiol 2001; 90:555-60; PMID:11309067; http://dx.doi.org/10.1046/j.1365-2672.2001.01284.x
- Soni DK, Singh RK, Singh DV, Dubey SK. Characterization of Listeria monocytogenes isolated from Ganges water, human clinical and milk samples at Varanasi, India. Infect Genet Evol 2013; 14:83-91; PMID:23201044; http://dx.doi.org/10.1016/j.meegid.2012.09.019
- Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, et al. Comparative genomics of Listeria species. Science 2001; 294:849-52; PMID:11679669
- Becavin C, Bouchier C, Lechat P, Archambaud C, Creno S, Gouin E, Wu Z, Kühbacher A, Brisse S, Pucciarelli MG, et al. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. MBio 2014; 5:e00969-14; PMID:24667708; http://dx.doi.org/10.1128/mBio.00969-14
- Tsai HN, Hodgson DA. Development of a synthetic minimal medium for Listeria monocytogenes. Appl Environ Microbiol 2003; 69:6943-5; PMID:14602660; http://dx.doi.org/10.1128/AEM.69.11.6943-6945.2003
- Reis O, Sousa S, Camejo A, Villiers V, Gouin E, Cossart P, Cabanes D. LapB, a novel Listeria monocytogenes LPXTG surface adhesin, required for entry into eukaryotic cells and virulence. J Infect Dis 2010; 202:551-62; PMID:20617901; http://dx.doi.org/10.1086/654880
- Lecuit M, Ohayon H, Braun L, Mengaud J, Cossart P. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect Immun 1997; 65:5309-19.
- Garcia-del Portillo F, Calvo E, D'Orazio V, Pucciarelli MG. Association of ActA to peptidoglycan revealed by cell wall proteomics of intracellular Listeria monocytogenes. J Biol Chem 2011; 286:34675-89; PMID:21846725; http://dx.doi.org/10.1074/jbc.M111.230441
- Cabanes D, Lecuit M, Cossart P. Animal models of Listeria infection. Curr Protoc Microbiol 2008; Chapter 9: 9B.1.1-9B.1.17; Unit9B 1; PMID:18729060
