ABSTRACT
Studies of the microbiome in the setting of haematopoietic stem cell transplantation (SCT) have shown evidence that intestinal microbes appear to play a particularly important role in determining the outcome of treatment, impacting complications such as infection or graft-versus-host disease. Past studies may vary in terms of the level at which the microbiome is examined, leading to different but overlapping systems of taxonomy or nomenclature, which may be difficult for non-specialists to understand. This article will review the current body of work examining the clinical impact of the microbiome on SCT, and will provide a basic framework for the bacterial phylogenetic structure upon which the results of these studies rest. With this framework it can be shown that recurring patterns do emerge in prior studies identifying the microbes that confer benefit in this population.
Introduction
Numerous studies have shown evidence that the intestinal microbiome has close ties to their human hosts under normal circumstances. This long-lived relationship results in a number of benefits for the host, including absorption of nutrients, prevention of overgrowth by potential pathogens, maintenance of epithelial barrier function, and stimulation and regulation of the immune system.Citation1,2
The premise that intestinal bacteria are important mediators in determining the outcome of allogeneic haematopoietic stem cell transplantation (SCT) is one that has been hypothesized by researchers as far back as the 1970s.Citation3,4 Some early work suggested potential benefit from suppressing the intestinal microbiota in an effort to reduce infections and graft-versus-host disease (GVHD);Citation5,6 however these strategies were not consistently beneficial, were difficult to maintain, and over time eventually fell out of favor.Citation7,8 It was during this time that van der WaaijCitation9 described the concept of colonization resistance, referring to resistance of a microbial population to colonization by a pathogen. These initial studies showed that anaerobic flora were believed to prevent long term colonization by potentially pathogenic aerobic bacteria.
More recent studies of the intestinal microbiota now indicate that specific anaerobic gut microbes may be particularly important in allogeneic SCT, exhibiting evidence of colonization resistance, but also demonstrating evidence of impact on important clinical outcomes such as overall survival and transplant related mortality.
A possible explanation for the apparent importance of the microbiota in this population is that the immunoregulatory and colonization resistance benefits of a healthy microbiome may be particularly beneficial in a patient with a newly transplant stem cell graft and susceptibility to poor graft function. Exposure to healthy gut microbes may serve to induce immune components such as colonic regulatory T-cells, promote immune tolerance, resist expansion by potential pathogens, and improve epithelial barrier integrity and function.Citation10 These benefits may ultimately lead to decreased transplant-related complications such as infection and GVHD.
This article will summarize the body of work examining the changes and impact of the intestinal microbiota in recipients of allogeneic SCT. Important recurring microbial ecology concepts that appear in past work will be reviewed and discussed. Particular focus will given to over-arching patterns in microbiome studies that have identified beneficial microbes. A basic overview of relevant taxonomic structure will be provided, upon which the results of prior microbiome studies can be understood and compared.
Loss of microbial diversity occurs during early SCT and impact outcomes
Diversity has been used in microbiome studies as a simple indicator of the overall “health” of the microbiome. Approaches and concepts for assessing microbial diversity are largely derived from conventional ecology. These ecologic measures may assess α diversity, which describes the composition of a single population or specimen (e.g. Shannon index, inverse Simpson index), or β diversity, which measures the overall compositional differences between 2 populations or samples (e.g., Bray-Curtis dissimilarity index, Unifrac distance).Citation11
During the early portion of allogeneic SCT, microbial diversity in the intestinal tract has been shown to decrease substantially during the first few weeks of SCT, where systemic chemotherapy, total body irradiation, and/or broad spectrum antibiotics are given. One study of a cohort of 94 patients undergoing allogeneic SCT showed an overall drop in α diversity, measured by Shanon index, though individual experiences that comprised this trend varied considerably.Citation12 In another study of allogeneic SCT at the same institution, average Bray-Curtis dissimilarity was used to measure β diversity differences in successive pairs of stool samples, collected serially over time, as an estimate of microbial dysbiosis or “chaos.”Citation13 Using this metric, this study showed that patients who experienced more microbial fluctuations during early allogeneic SCT were more likely to develop subsequent GVHD.
The clinical importance of the intestinal microbiome was shown in a study examining the diversity of gut microbiota in allogeneic SCT patients through stool samples collected at the time of stem cell engraftment. Alpha diversity at engraftment was measured by inverse Simpson index,Citation11 and patients were divided into groups of low, intermediate, and high α diversity, based on defined index cutoffs. Patients with low diversity at engraftment were 5 times more likely to die of transplant-related causes compared with those with intermediate or high diversity. Microbial diversity was shown to be a strong predictor of transplant related mortality, independent of other factors, including pretransplant comorbidities and antibiotic administration.Citation14
Another study of allogeneic SCT showed that urinary levels of indoxyl sulfate, a compound predominantly produced by beneficial gut microbes, could be used as a surrogate marker for bacterial diversity.Citation15 These authors showed that indoxyl sulfate levels decreased with antibiotic administration, and were also low in patients with subsequent GVHD. A subsequent study by this group also found that low levels of urinary indoxyl sulfate were correlated with higher transplant related mortality.Citation16 In turn, levels of indoxyl sulfate were notably shaped by antibiotic administration and the approach to gut decontamination.
Currently it is still uncertain exactly how beneficial gut microbes can serve to ultimately enhance protection against transplant complications in allogeneic SCT recipients; this continues to be an area of active study. Mechanistic studies have sought to understand the complex underpinnings of host-microbe mutualism, and thus far offer a number of possible explanations for observed importance of the microbiota in this setting.
Many elements within the intestinal microbiota participate deeply in the development and training of the host immune system. Overall, the immune system is reliant on tonic signaling from an intact microbiota in order to function optimally.Citation17 Immune elements influenced by the microbiome include neutrophils, dendritic cells of various lymphoid compartments, localized and systemic T-cell and B-cell subsets, and NK cells.Citation2,17–20 Following haematopoietic stem cell transplantation, all of these components are initially deficient. Immunologic recovery occurs eventually, over time, and can be highly variable from patient to patient. In this scenario, an intact microbiota may serve to enhance and accelerate immune training and development of transplanted haematopoietic cells, allowing recipients to achieve immune recovery, avoiding complications which occur as a result of an immature and/or dysfunctional immunity, namely opportunistic infections and GVHD.
A complex system between microbiota and host tissues exists to maintain intestinal homeostasis. This homeostasis promotes colonization resistance, which resists overcolonization by opportunistic pathogens. The microbiota achieves colonization resistance through a variety of mechanisms. It may do so directly, through bacteria-to-bacteria mechanisms, such as outcompeting pathogens for nutrients, or production of inhibitory substances (e.g. bacteriocins).Citation21
The microbiota also exert homeostatic balance through routine stimulation of the host pattern recognition receptors such as Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD) receptors, which activates compensatory immune responses and release of antimicrobial peptides, such as defensins and RegIIIγ, which act by killing pathogenic bacteria.Citation2,17,21 The host also exerts control of bacterial translocation by sampling bacteria from the overlying epithelium, and interacting with Peyer's patches, which produce IgA directed against invading pathogens within the intestinal lumen.2 In stem cell transplantation, major damage to the epithelial mucosa occurs due to myeloablative chemotherapy and radiation. As a result these well-orchestrated homeostatic mechanisms are disrupted, leaving the host susceptible to invasion through bacterial translocation, with infection spurred on by overgrowth of pathogenic species, left unchecked due to compromised colonization resistance.
Members of the microbiome produce short chain fatty acids, such as butyrate, which are a primary source of fuel for intestinal epithelial cells, and offer a variety of benefits important for preservation of gut homeostasis, such as inhibition of intestinal inflammation, promotion of epithelial integrity, mucin production, and promotion of direct colonization resistance against pathogens such as C. difficile.Citation22,23 Notably, SCFA have been shown to induce Tregs,Citation24–26 which are important for immune tolerance and therefore may be important for mitigation of GVHD. Indeed, one study of murine GVHD demonstrated that administration with butyrate serves to protect the GI epithelium and mitigate GVHD disease.Citation27
Intestinal domination leads to systemic bacterial infection and is shaped by antibiotics
Concurrent with the loss in bacterial diversity described above, the microbiome may experience overgrowth or expansion of a single potential pathogenic organism. This process was referred to as intestinal “domination” by Ubeda and colleagues,Citation28 noting a dramatic rise of vancomycin-resistant Enterococcus (VRE) in 2 out of 5 individuals undergoing allogeneic SCT. This study further showed that these 2 individuals went on to develop bloodstream infection with VRE afterwards.
Those initial observations were further confirmed in the 94-patient cohort;Citation12 almost all cases of VRE bloodstream infection in the cohort were preceded by intestinal domination by Enterococcus in serially collected stool samples. This was observed during early allogeneic SCT, prior to stem cell engraftment, during neutropenia, where patients are at risk for mucosal barrier injury and systemic infection by gut translocation. The study also showed analogously that gram negative bloodstream infections were preceded by intestinal domination by Proteobacteria, the bacterial phylum that contains almost all gram negative bloodstream pathogens encountered during neutropenia.
The 94-patient cohort study also demonstrated that specific antibiotics can serve to either cause or prevent intestinal domination. Administration with metronidazole, a potent inhibitor of anaerobic bacteria, was identified as a significant driver of subsequent enterococcal domination. This work supported from culture-based studies of VRE colonization, which also suggested that anti-anaerobic antibiotics were the strongest drivers of VRE overgrowth.Citation29–31 In contrast to metronidazole and Enterococcus, administration with fluoroquinolone antibiotics was associated with lower risk of domination by Proteobacteria. Together these results suggest that judicious use of targeted antibiotics can successfully prevent infections, but still preserve important microbes that confer beneficial effects and protect against further pathogen expansion through colonization resistance. The study did not find an association between the intensity of conditioning chemotherapy and the development of subsequent intestinal domination. However, it is still reasonable to assume that conditioning chemotherapy plays a substantial role in disruption of the microbiota, either directly or indirectly through damage to the intestinal epithelium (e.g., mucositis), which in turn leads to impaired host-microbe homeostasis.Intestinal domination was further studied by Holler and colleagues,Citation15 who found that domination by Enterococcus occurred after transplantation, and was shaped by antibiotics. Furthermore, enterococcal domination was associated with subsequent gastrointestinal GVHD. Whether enterococci directly contribute to GVHD or is merely an indicator of loss of diversity is unclear. Since they also observed decreased microbial diversity and urinary indoxyl sulfate during times of enterococcal domination, the authors concluded that Enterococcus was more likely to be an indicator rather than direct cause. However, there is some murine data which suggests that Enterococcus could potentially contribute to worsening GVHD through enterococcal epitheliolysins and other toxins that could cause damage to gut epithelium.Citation32,33
Finally, Harris and colleaguesCitation34 recently re-examined in the original 94-patient cohort in the clinical context of pulmonary complications. These authors found that intestinal domination by Gammaproteobacteria, a major class within Proteobacteria containing Enterobacteriaceae, was a significant predictor of pulmonary complications following stem cell infusion, independent of pre-transplant comorbidity and antibiotic administration. Intestinal domination by this class also correlated with overall mortality.
Bacteria
Many studies of the bacterial microbiome, including those in SCT, have relied on a wide variety of approaches to microbial classification. These include taxonomic ranks such as phylum, class, order, family, genus, and species, but also non-taxonomic designations such as clostridium clusters (I through XIX), a phylogenetic system of classification for various clostridial species.Citation35 These classifications vary greatly in terms of scope, depth, and total number of groups. Currently there is no consensus as to which level is optimal for study or most relatable clinically.
Studies of the microbiome in SCT are summarized here. shows a phylogenetic tree of typical bacteria that can be found in the human intestinal tract, and highlights several studies of SCT. Despite the numerous levels of classification used in various studies, it is apparent that certain portions of the bacterial phylogeny have repeatedly shown themselves to be important members in outcomes for SCT. These members are described further and discussed below.
Figure 1. Significant microbiome findings in stem cell transplantation. A phylogenetic tree of intestinal bacterial species is shown. Results of microbiome studies in SCT recipients is summarized; bacterial clades (dotted lines with shading) are shown at various levels of taxonomic classification. Highlighted clades summarize the findings of each study (blue shading denotes association with beneficial effects, red shading denotes association with negative effects).
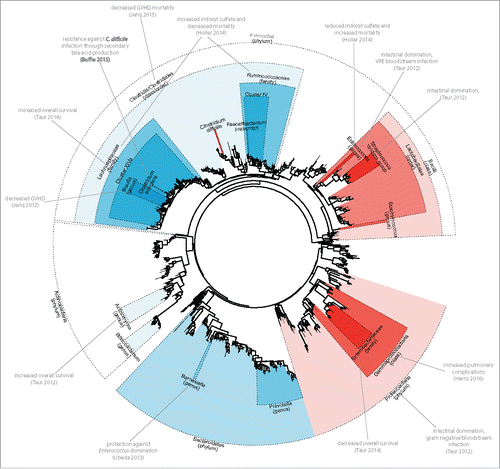
Firmicutes/clostridia
The Firmicutes phylum is a diverse group of bacteria that ranges from highly beneficial microbes to potential pathogens. A predominant group of within the gut is the Clostridia class, a group of obligate anaerobic bacteria. An equivalent designation is the order Clostridiales, which microbiome studies have also used. Though Clostridia include potential pathogens such as Clostridium difficile, 2 bacterial families that appear to be purely comprised of beneficial microbes are Lachnospiraceae and Ruminococcaceae. These bacteria roughly correspond to Clostridium clusters XIVa and IV, respectively;Citation36 the Clostridium clusters systems was proposed as a system to group Clostridia into meaningful phylogenetic groups,Citation35 and has been used by many microbiome investigators.
At various levels of phylogeneic designation, these bacterial groups appear particularly associated with positive outcomes in allogeneic SCT. Indeed, they have been shown to exhibit a number of beneficial effects to the host, including stimulation of T-cell immunity and promotion of colonization resistance.Citation1 Clostridial species such as those in cluster IV and XIVa are notable for generating high amounts of SCFA such as butyrate and inducing T-regs, which may be important for mitigating GVHD.Citation10,20
At the level of order, Clostridiales bacteria was associated with protection against GVHD, in one fecal microbiome study of human allogeneic SCT recipients.Citation13 In another study of the same biospecimen cohort, colonization with the family Lachnospiraceae was associated with improved overall survival following allogeneic SCT.Citation14 Another fecal microbiome study in Europe found levels of urinary indoxyl sulfate to be associated with improved overall survival; which is principally produced by Lachnospiraceae and Ruminococcaceae.Citation16
Within Lachospiraceae, the genus Blautia was observed to be associated with reduced death due to GVHD.Citation37 Also within Lachnospiraceae, Clostridium scindens was associated with protection against Clostridium difficile infection in a study of allogeneic SCT; that study further demonstrated in mice that the mechanism of protection was through generation of secondary bile acids, which inhibit vegetative growth of and toxin production of Clostridium difficile.Citation38
Bacteroidetes
The Bacteroidetes phylum are gram-negative obligate anaerobic bacteria that are highly suited for the gut. These bacteria enjoy close ties to host immunity; a number of host factors appear to specifically select members of Bacteroidetes for intestinal colonization.Citation39 This may serve to explain why it is felt to be the most stable component of the gut microbiota in healthy adults.Citation40
In allogeneic SCT, one study suggested possible benefit from colonization with Bacteroidetes bacteria: a follow-up study to the 94-patient cohort showed that colonization with Barnesiella, a genus within Bacteroidetes, conferred protection against subsequent Enterococcus (VRE) intestinal domination.Citation41
Given its relative microbial stability and host-specificity, as well as its established benefits in other patient populations, it may surprising that more benefit has not been observed for the Bacteroidetes phylum in microbiome studies of SCT patients. It could be that Bacteroidetes confer a great deal of benefits, but do not offer specific benefits to host immunity as do their gram-positive counterparts Lachnospiraceae and Ruminococcaceae, and therefore do not substantially affect outcomes related to transplant complications, such as GVHD, GVHD-mortality, and transplant-related mortality (TRM). However, another possible explanation is that Bacteroidetes are generally not abundant in SCT patients,Citation12 potentially making it difficult for microbiome studies to be sufficiently powered to detected their benefits. Since most SCT have generally been exposed to prior treatment with antibiotics and chemotherapy, Bacteroidetes may have been lost in these patients prior to allogeneic SCT and not easily regained. Indeed, studies of infant gut colonization indicate that Bacteroidetes does appear in high abundance in the human intestinal tract until several months after birth.Citation42–45
Actinobacteria
Actinobacteria are gram positive bacteria that represent a common constituent of the human microbiota. Though they appear highest abundance in early gut colonization during infancy,Citation44 they are generally less abundant than Firmicutes and Bacteroidetes in a stable microbiome within adults.Citation46 Though they appear to be associated with several benefits, it is not clear that these benefits exceed those from Clostridia or Bacteroidetes. Certain members such as Bifidobacterium spp. are presumed to be beneficial and have been formulated in many probiotic supplements. In allogeneic SCT, colonization with bacteria within Actinobacteria, specifically Actinomycetaceae, were found to be associated with decreased overall mortality.Citation14
The controversy of bacterial decontamination in allogeneic SCT
Despite the observed benefits of the microbiome in the studies described, these results contrast with prior work which has demonstrated improved outcomes in association with intestinal decontamination. For example, Beelen and colleaguesCitation47 conducted a single-center randomized trial of bacterial decontamination, which compared ciprofloxacin monotherapy with combination therapy with ciprofloxacin plus metronidazole. This study demonstrated a 50 % reduction in GVHD in the combination therapy group compared with monotherapy group. Based on these results, some transplant centers continue to practice routine bacterial decontamination, with regimens that target anaerobic bacteria. The merits of anaerobic bacterial decontamination are therefore still a matter of controversy. In one recent study, Weber and colleaguesCitation48 compared the bacterial decontamination with rifaximin, a nonabsorbable antibiotic used for travelers' diarrhea, with conventional decontamination with ciprofloxacin plus metronidazole; this study found that patients who received rifaximin had lower transplant-related mortality and high overall survival, compared with the combination therapy group. As discussed by the authors, rifaximin achieves some degree of decolonization but at the same time has been shown to preserve certain beneficial anaerobic gut microbes, and does not affect the overall composition of the microbiota.Citation49 This was evidenced by higher levels of urinary indoxyl sulfate in the rifaxamin group.
Ongoing challenges and future directions
Currently, there are ongoing challenges for understanding the full implications of the intestinal microbiota in the setting of stem cell transplantation (). These questions may need to be answered in order to translate current findings into clinical practice.
Table 1. Ongoing Challenges and Future Questions.
Since almost all human studies of microbiome in SCT have been done using collected stool samples, it is unclear whether differences between microbiota in various sites across the gut are important, and are under-appreciated. It is well-known that microbiome populations are vastly different in various locations within the intestinal tract, such as mouth, esophagus, small intestine, and large intestine.Citation28,46 One example of how gut geography is important in allogeneic SCT is in systemic bacterial infections during neutropenia. These infections are thought to be dependent on the location of mucosal injury. For instance, systemic infection with viridans streptococci during neutropenia are known to be specifically associated with chemotherapeutic agents (e.g. high-dose cytarabine) that are likely to cause oral mucositis.Citation50,51 This is not necessarily surprising, since viridans streptococci are known to preferentially inhabit the upper gut, such as the oral cavity, rather than lower gut.Citation46 Fecal biospecimen studies currently fail to distinguish differences in gut locale, but may turn out to be considerably important in SCT.
The use of other ‘omic’ approaches may be important for completing our understanding of the microbiome's impact in the allogeneic SCT recipient. For instance, detection of small molecules present in the intestinal lumen through metabolomics may help to catalog and identify small molecule profiles associated with specific microbiome patterns. Identification of bacterial by-products such as SCFA and primary and secondary bile salts, as well as presence of administered drugs such as antibiotics could provide an additional dimension to consider in the host-microbe landscape of SCT.
The bacterial microbiome has been studied for the most part, but much less is known about the mycobiome and virome. Similar to bacteria, fungi and viruses appear to enjoy a high diversity in the gut, and show evidence that they also interact with host immunity.Citation52–56 However, currently it is unclear what roles they might play in host-microbe homeostasis, and how important they are for human health. Some preliminary studies have examined the mycobiome and virome in the setting of immunocompromised patients, and have reported links with antiviral administration and the degree of immune suppression.Citation53,57,58 However, little work has been done in SCT recipients, who are uniquely at risk for infection by various fungi and viruses, due to their immune compromise. Thus, study of these populations could particularly worthwhile for understanding the inherent risks these patients experience.
Conclusions
The studies described have shown evidence of the intestinal microbiota's importance in allogeneic SCT, affecting important outcomes in infections, pulmonary complications, GVHD, and transplant-related mortality. However, future studies will need to further clarify the precise nature and mechanism of benefit of the microbiome in allogeneic SCT.
Additional insights from these studies may enable physicians to intelligently use antibiotics in a targeted manner which preserves important bacterial groups such as Lachnospiracaeae, Ruminococcaceae, or Bacteroidetes. If loss of these microbes is unavoidable, re-population through fecal microbiota transplantation or bacteriotherapy with microbial consortia might exist as a potential interventions aimed at restoring specific beneficial microbes. Currently, a randomized trial of autologous fecal microbiota transplantation for prevention of transplant complications is underway.Citation59 These interventions may serve to optimize outcomes in allogeneic SCT and reduce potential complications that currently limit its use.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Institutes of Health (grants K23 AI095398-01, RO1 AI42135, U01 AI124275-01, and P30 CA008748).
References
- Nishio J, Honda K. Immunoregulation by the gut microbiota. Cell Mol Life Sci 2012; 69:3635-50; PMID:22527722; http://dx.doi.org/10.1007/s00018-012-0993-6
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science 2012; 336:1268-73; PMID:22674334; http://dx.doi.org/10.1126/science.1223490
- Van Bekkum D, Roodenburg J, Heidt P, Van der Waaij D. Mitigation of secondary disease of allogeneic mouse radiation chimeras by modification of the intestinal microflora. J Natl Cancer Inst 1974; 52:401-4; PMID:4150164
- Jones JM, Wilson R, Bealmear PM. Mortality and gross pathology of secondary disease in germfree mouse radiation chimeras. Radiat Res 1971; 45:577-88; PMID:4396814; http://dx.doi.org/10.2307/3573066
- Vossen J, Heidt P, Van Den Berg H, Gerritsen E, Hermans J, Dooren L. Prevention of infection and graft-vs.-host disease by suppression of intestinal microflora in children treated with allogeneic bone marrow transplantation. Euro J Clin Microbiol Infect Dis 1990; 9:14-23; PMID:2105890; http://dx.doi.org/10.1007/BF01969527
- Storb R, Prentice RL, Buckner CD, Clift R, Appelbaum F, Deeg J, Doney K, Hansen JA, Mason M, Sanders JE, et al. Graft-versus-host disease and survival in patients with aplastic anemia treated by marrow grafts from HLA-identical siblings: beneficial effect of a protective environment. N Engl J Med 1983; 308:302-7; PMID:6337323; http://dx.doi.org/10.1056/NEJM198302103080602
- Armstrong D. Protected environments are discomforting and expensive and do not offer meaningful protection. Am J Med 1984; 76:685-9; PMID:6424468; http://dx.doi.org/10.1016/0002-9343(84)90295-X
- Bodey GP. Fever and neutropenia: the early years. J Antimicrob Chemother 2009; 63:i3-i13; PMID:19372179; http://dx.doi.org/10.1093/jac/dkp074
- Van der Waaij D, Berghuis-de Vries J, Lekkerkerk-Van der Wees J. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hygiene 1971; 69:405-11; PMID:4999450; http://dx.doi.org/10.1017/S0022172400021653
- Shono Y, Docampo MD, Peled JU, Perobelli SM, Jenq RR. Intestinal microbiota-related effects on graft-versus-host disease. Int J Hematol 2015; 101:428-37; PMID:25812838; http://dx.doi.org/10.1007/s12185-015-1781-5
- Magurran AE. Measuring biological diversity. Malden, MA: Blackwell Publishing; 2004
- Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, et al. Intestinal Domination and the Risk of Bacteremia in Patients Undergoing Allogeneic Hematopoietic Stem Cell Transplantation. Clin Infect Dis 2012; 55:905-14; PMID:22718773; http://dx.doi.org/10.1093/cid/cis580
- Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, Liu C, West ML, Singer NV, Equinda MJ, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med 2012; 209:903-11; PMID:22547653; http://dx.doi.org/10.1084/jem.20112408
- Taur Y, Jenq RR, Perales M-A, Littmann ER, Morjaria S, Ling L, No D, Gobourne A, Viale A, Dahi PB, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014; 124:1174-82; PMID:24939656; http://dx.doi.org/10.1182/blood-2014-02-554725
- Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, Zhu W, Sporrer D, Hehlgans T, Kreutz M, et al. Metagenomic Analysis of the Stool Microbiome in Patients Receiving Allogeneic Stem Cell Transplantation: Loss of Diversity Is Associated with Use of Systemic Antibiotics and More Pronounced in Gastrointestinal Graft-versus-Host Disease. Biol Blood Marrow Transplant 2014; 20:640; PMID:24492144; http://dx.doi.org/10.1016/j.bbmt.2014.01.030
- Weber D, Oefner PJ, Hiergeist A, Koestler J, Gessner A, Weber M, Hahn J, Wolff D, Stämmler F, Spang R, et al. Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood 2015; 126:1723-8; PMID:26209659; http://dx.doi.org/10.1182/blood-2015-04-638858
- Becattini S, Taur Y, Pamer EG. Antibiotic-Induced Changes in the Intestinal Microbiota and Disease. Trends Mol Med 2016; 22:458-78; PMID:27178527; http://dx.doi.org/10.1016/j.molmed.2016.04.003
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011; 331:337-41
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139:485-98; PMID:19836068; http://dx.doi.org/10.1016/j.cell.2009.09.033
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013; 500:232-6; PMID:23842501; http://dx.doi.org/10.1038/nature12331
- Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 2013; 13:790-801; PMID:24096337; http://dx.doi.org/10.1038/nri3535
- Van Vliet MJ, Harmsen HJ, de Bont ES, Tissing WJ. The role of intestinal microbiota in the development and severity of chemotherapy-induced mucositis. PLoS Pathog 2010; 6:e1000879; PMID:20523891; http://dx.doi.org/10.1371/journal.ppat.1000879
- Rolfe RD. Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infect Immun 1984; 45:185-91; PMID:6735467
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504:446-50.
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341:569-73; PMID:23828891; http://dx.doi.org/10.1126/science.1241165
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013; 504:451-5; PMID:24226773
- Mathewson ND, Jenq R, Mathew AV, Koenigsknecht M, Hanash A, Toubai T, Oravecz-Wilson K, Wu S-R, Sun Y, Rossi C, et al. Corrigendum: Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol 2016; 17:1235; PMID:27648549; http://dx.doi.org/10.1038/ni1016-1235b
- Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 2010; 120:4332; PMID:21099116; http://dx.doi.org/10.1172/JCI43918
- Donskey CJ, Hanrahan JA, Hutton RA, Rice LB. Effect of parenteral antibiotic administration on persistence of vancomycin-resistant Enterococcus faecium in the mouse gastrointestinal tract. J Infect Dis 1999; 180:384-90; PMID:10395853; http://dx.doi.org/10.1086/314874
- Edmond MB, Ober JF, Weinbaum DL, Pfaller MA, Hwang T, Sanford MD, Wenzel RP. Vancomycin-resistant Enterococcus faecium bacteremia: risk factors for infection. Clin Infect Dis 1995; 20:1126-33; PMID:7619987; http://dx.doi.org/10.1093/clinids/20.5.1126
- Pultz NJ, Stiefel U, Subramanyan S, Helfand MS, Donskey CJ. Mechanisms by which anaerobic microbiota inhibit the establishment in mice of intestinal colonization by vancomycin-resistant Enterococcus. J Infect Dis 2005; 191:949-56; PMID:15717271; http://dx.doi.org/10.1086/428090
- Sava IG, Heikens E, Huebner J. Pathogenesis and immunity in enterococcal infections. Clin Microbiol Infect 2010; 16:533-40; PMID:20569264; http://dx.doi.org/10.1111/j.1469-0691.2010.03213.x
- Cox CR, Coburn PS, Gilmore MS. Enterococcal cytolysin: a novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr Protein Pept Sci 2005; 6:77-84; PMID:15638770; http://dx.doi.org/10.2174/1389203053027557
- Harris B, Morjaria SM, Littmann ER, Geyer AI, Stover DE, Barker JN, Giralt SA, Taur Y, Pamer EG. Gut Microbiota Predict Pulmonary Infiltrates after Allogeneic Hematopoietic Cell Transplantation. Am J Respir Crit Care Med 2016; 194:450-63; PMID:26886180; http://dx.doi.org/10.1164/rccm.201507-1491OC
- Collins M, Lawson P, Willems A, Cordoba J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 1994; 44:812-26; PMID:7981107; http://dx.doi.org/10.1099/00207713-44-4-812
- Rajilić-Stojanović M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 2014; 38:996-1047; PMID:24861948; http://dx.doi.org/10.1111/1574-6976.12075
- Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, Littmann ER, Ling L, Gobourne AC, Miller LC, et al. Intestinal Blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant 2015; 21:1373-83
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015; 517:205-8; PMID:25337874; http://dx.doi.org/10.1038/nature13828
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 2013; 501:426-9; PMID:23955152; http://dx.doi.org/10.1038/nature12447
- Rajili'c-Stojanovi'c M, Heilig HG, Tims S, Zoetendal EG, Vos WM. Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol 2013; 15:1146-59; http://dx.doi.org/10.1111/1462-2920.12023
- Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, Equinda M, Lipuma L, Ling L, Gobourne A, No D, et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun 2013; 81:965-73; PMID:23319552; http://dx.doi.org/10.1128/IAI.01197-12
- Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev 2007; 20:593-621; PMID:17934076; http://dx.doi.org/10.1128/CMR.00008-07
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 2010; 107:11971-5; PMID:20566857; http://dx.doi.org/10.1073/pnas.1002601107
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci 2011; 108:4578-85; http://dx.doi.org/10.1073/pnas.1000081107
- Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015; 17:690-703; http://dx.doi.org/10.1016/j.chom.2015.04.004
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, Fitzgerald MG, Fulton RS, et al. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207-14; PMID:22699609; http://dx.doi.org/10.1038/nature11234
- Beelen DW, Elmaagacli A, Müller KD, Hirche H, Schaefer UW. Influence of intestinal bacterial decontamination using metronidazole and ciprofloxacin or ciprofloxacin alone on the development of acute graft-versus-host disease after marrow transplantation in patients with hematologic malignancies: final results and long-term follow-up of an open-label prospective randomized trial. Blood 1999; 93:3267-75; PMID:10233878
- Weber D, Oefner PJ, Dettmer K, Hiergeist A, Koestler J, Gessner A, Weber M, Stämmler F, Hahn J, Wolff D, et al. Rifaximin preserves intestinal microbiota balance in patients undergoing allogeneic stem cell transplantation. Bone Marrow Transplant 2016; 51:1087-92; PMID:26999466
- Maccaferri S, Vitali B, Klinder A, Kolida S, Ndagijimana M, Laghi L, Calanni F, Brigidi P, Gibson GR, Costabile A. Rifaximin modulates the colonic microbiota of patients with Crohn's disease: an in vitro approach using a continuous culture colonic model system. J Antimicrob Chemother 2010; 65:2556-65; PMID:20852272
- Richard P, Felice M, Daeschler T, Richet H, Amador Del Valle G, Moreau P, Milpied N, Harousseau J. Viridans streptococcal bacteraemia in patients with neutropenia. Lancet 1995; 345:1607-9; http://dx.doi.org/10.1016/S0140-6736(95)90117-5
- Tunkel AR, Sepkowitz KA. Infections caused by viridans streptococci in patients with neutropenia. Clin Infect Dis 2002; 34:1524-9; PMID:12015700; http://dx.doi.org/10.1086/340402
- Cui L, Morris A, Ghedin E. The human mycobiome in health and disease. Genome Med 2013; 5:1; PMID:23311897; http://dx.doi.org/10.1186/gm467
- Wylie KM, Weinstock GM, Storch GA. Emerging view of the human virome. Transl Res 2012; 160:283-90; PMID:22683423; http://dx.doi.org/10.1016/j.trsl.2012.03.006
- Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proc Natl Acad Sci 2013; 110:12450-5; http://dx.doi.org/10.1073/pnas.1300833110
- Moon C, Stappenbeck TS. Viral interactions with the host and microbiota in the intestine. Curr Opin Immunol 2012; 24:405-10; PMID:22626624
- Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 2014; 14:405-16; PMID:24854590; http://dx.doi.org/10.1038/nri3684
- De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, Okamoto J, Snyder TM, Cornfield DN, Nicolls MR, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 2013; 155:1178-87; PMID:24267896; http://dx.doi.org/10.1016/j.cell.2013.10.034
- Shelburne SA, Ajami NJ, Chibucos MC, Beird HC, Tarrand J, Galloway-Peña J, Albert N, Chemaly RF, Ghantoji SS, Marsh L, et al. Implementation of a Pan-Genomic Approach to Investigate Holobiont-Infecting Microbe Interaction: A Case Report of a Leukemic Patient with Invasive Mucormycosis. PLoS One 2015; 10:e0139851; PMID:26556047; http://dx.doi.org/10.1371/journal.pone.0139851
- Memorial Sloan Kettering Cancer Center. Autologous Fecal Microbiota Transplantation (Auto-FMT) for Prophylaxis of Clostridium Difficile Infection in Recipients of Allogeneic Hematopoietic Stem Cell Transplantation [Internet]. 2000; Available from: https://clinicaltrials.gov/ct2/show/NCT02269150
