Clostridium difficile is an Gram positive enteropathogen that can cause opportunistic infections resulting in colitis.Citation1,2 It is predominantly associated with antibiotic treatment, but it is increasingly recognized as the causative agent of symptoms in patients that lack this risk factor.Citation1,3 Its classification as an urgent antibiotic resistance threat by the US Centers for Disease Control and PreventionCitation4 is based on the fact that the pathogen affects an already vulnerable population that is treated by antibiotics for other infections or prophylactically, rather than resistance of C. difficile against clinically used antibiotics, which is limited.Citation1 C. difficile can be identified in livestock and companion animals and it has been shown that strains from animal and human reservoirs are identical, suggesting a clear zoonotic potential.Citation5-7
Initially identified as Bacillus difficilis as part of the microbiome of healthy infants,Citation8 it gained notoriety as Clostridium difficile and the disease it causes is generally referred to as Clostridium difficile infection (CDI). Genomic analyses however indicated that C. difficile should be placed in the family Peptostreptococcaceae rather than Clostridiaceae, and to reflect this the name Peptoclostridium difficile was proposed.Citation9 Though this was unilaterally adopted by the National Center for Biotechnology Information (NCBI), the proposal lacked a formal definition of the type species and the name was not widely adopted by the community. A formal reclassification was published in 2016Citation10 and the new nomenclature Clostridioides difficile allows the continued use of CDI, as well as the colloquialism Cdiff.
The symptoms of CDI are the ultimate result of toxins produced by the C. difficile bacteria.Citation11 The genes encoding these toxins are located on a mobile pathogenicity locus.Citation12-14 Indeed, strains lacking the pathogenicity island are non-toxigenic. Most pathogenic C. difficile strains encode 2 high molecular weight toxins, TcdA and TcdB, and the relative contribution of these toxins to pathogenesis has been subject of controversy.Citation15-18 Similarly, conflicting findings have been reported with respect to the function of other proteins (TcdC, TcdE) encoded on the pathogenicity locus.Citation1,19 Further, certain C. difficile strains encode a binary toxin that contributes to pathogenesis.Citation1,11,19-21 Overall, virulence, fitness and transmissibility of the pathogen appear to be multifactorial.Citation19,22,23
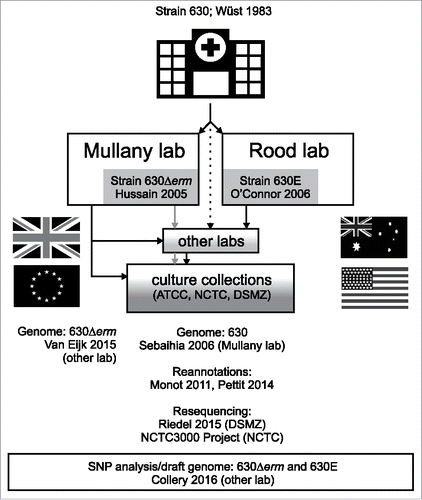
In this issue of Virulence, Collery and coworkers attempt to identify a possible cause of the discrepancies observed between various laboratories studying C. difficile.Citation24 To appreciate the intricacies of this work, it is necessary to understand the background of the strains that were investigated. The first strain of C. difficile to be sequenced was isolated from a patient suffering pseudomembranous colitis and was responsible for an outbreak of CDI in the hospital in Zürich.Citation25,26 This strain, called 630, demonstrated transferable resistance to the antimicrobial erythromycin and is also resistant to several other drugs.Citation25,26 The strain was redistributed to several other labs, and the annotation of the genome sequence has been updated several times.Citation27,28 In order to facilitate genetic studies on C. difficile, 2 groups independently derived an erythromycin sensitive strain by serial culturing on non-selective media: the Mullany laboratory (University College London, London, UK) generated strain 630Δerm,Citation29 and the Rood laboratory (Monash University, Victoria, Australia) generated 630E (also known as JIR8094).Citation30 These strains – harboring an identical 2.4 kb deletion in the mobile element Tn5398,Citation31-33 allowed the use of the ermB gene (conferring erythromycin resistance) as a selectable marker in C. difficile. Both 630E and 630Δerm were provided to other laboratories. Notably, both 630 and 630Δerm have been deposited directly or indirectly in various culture collections [ATCC (https://www.atcc.org), NCTC (https://www.phe-culturecollections.org.uk) and DSMZ (https://www.dsmz.de)] which – in turn – provide bacterial strains to other scientists (). Whereas the DNA from the 2006 genome sequence of strain 630 came from the Mullany laboratory,Citation25 DNA from 2 independent resequencing projects was derived from the isolates banked by the NCTC (NCTC3000 Project, https://www.phe-culturecollections.org.uk/collections/nctc-3000-project.aspx) and the DSMZ.Citation34 The latter genome sequence shows some peculiar features, including the apparent loss (i.e. not detected in their analyses) of plasmid pCD630 and transposon Tn5397 and acquisition of an additional rRNA cluster, that seem to suggest extensive sub-culturing. Of note, the DSMZ strain was obtained from the NCTC, that in turn received its isolate from the Mullany laboratory. A single study has reported a complete genome sequence for strain 630ΔermCitation33 prior to the study of Collery and coworkers.Citation24 Strikingly, the authors identified many more differences from strain 630 than the deletion of an ermB gene in Tn5398, including a transposition of the conjugative transposon CTn5 and an additional rRNA cluster, similar to the resequenced 630 strain.Citation34 Thus, despite a common ancestry, the strains differ vastly.
Figure 1. Schematic representation of the genealogy of strain 630, 630Δerm and 630E (JIR8094) and their related genome sequences. Strain 630 was deposited by B. Wren/H. Maschler (ATCC BAA-1382), P. Mullany (NCTC 13307). The DSMZ lists the provenance for 630 (DSMZ 27543) as obtained from P.Bracegirdle (NCTC) and for 630Δerm (DSMZ 28645) as H. Hussain (Mullany laboratory) > N. Minton > R. Gerhard. Figure also highlights that, historically, 630Δerm was the dominant strain used in European C. difficile laboratories, whereas 630E (JIR8094) was mainly used in Australia and the USA. The sources of the sequenced DNA are indicated in brackets.
The study of Collery and coworkers is noteworthy for several reasons. First, it represents a multi-laboratory effort to determine if differences reported in literature are due to the different 630 derivatives used (630Δerm versus 630E). Second, in a comprehensive approach, the authors try to define the contributions of a selected set of single nucleotide polymorphisms (SNPs) to the phenotypic and transcriptomic differences. Instances where a single SNP defines major phenotypic changes are scarce, but not unprecedented; e.g. for C. difficile, SNPs in the gyrA gene that result in fluoroquinolone resistance underlie the expansion of the epidemic 027/BI/NAP1 strainCitation35 and for Campylobacter jejuni hypervirulence has been linked to SNPs in the outer membrane protein PorA.Citation36 Also for non-pathogenic model bacteria SNPs can be linked to specific phenotypic changes, related to domesticationCitation37 or adaptation,Citation38 for instance.
However, considering the number of SNPs, it should not come as a surprise that the authors failed to link SNPs to specific phenotypic differences. Both derivatives differ significantly from strain 630, and though one can argue that 630Δerm more closely resembles the ancestral strain,Citation24 it is an illusion to consider the findings obtained with this strain to be directly representative for strain 630. These findings are also consistent with other studies that observe substantial phenotypic variation with a specific type of C. difficile with respect to, for instance, sporulation.Citation39-41
Should we then move away from laboratory strains and research only clinical isolates? While this may address the fact that some regulatory interactions are only observed in certain clinical strains,Citation42,43 other major issues, such as passaging in laboratories, remain problematic. Moreover, it will further increase the inter-laboratory variation, as each laboratory would have its own “wild type.” There is therefore value in the use of a standard strain, and the authors argue that this could be 630Δerm.Citation24
In what way could some of the challenges in linking SNPs and phenotypes be addressed? One strategy is to expand the number of strains analyzed to allow for genome wide association studies (GWAS); such studies have demonstrated for instance the relation between SNPs and β-lactam resistance in Streptococcus pneumoniaCitation44 and predicted virulence in MRSA from genome data.Citation45 With a limited set of 14 genome sequences it has already been possible to identify SNPs associated with the epidemic group BI/NAP1/027 that can cause severe disease.Citation46 With a broader analysis of phenotypic and clinical characteristics and an increasing number of C. difficile genome sequences available (on Oct 11, 2016 the number of genome assemblies in Genbank was 647; https://www.ncbi.nlm.nih.gov/genome/?term=clostridioides+difficile), GWAS analysis could contribute significantly to our understanding of this important pathogen.
In summary, the work on C. difficile genomes and strains by Collery and coworkers,Citation24 as well as several others,Citation39,42,43 should be a caveat to many researchers; their findings may apply only to their specific isolate or strain and should encourage them to be careful with generalizations. Also, researchers should exercise caution in repeated propagation of strains under laboratory conditions and document the provenance even when strains are obtained from reputable sources.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
I thank Jeroen Corver and Adam P. Roberts for helpful discussions.
Funding
WKS is supported, in part, by a VIDI fellowship from the Netherlands Organization for Scientific Research and a Gisela Thier Fellowship from the Leiden University Medical Center.
References
- Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. Clostridium difficile infection. Nat Rev Dis Primers (2016); 2:16020; PMID:27158839; http://dx.doi.org/10.1038/nrdp.2016.20
- Abt MC, McKenney PT, Pamer EG. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol (2016); 14:609-20; PMID:27573580; http://dx.doi.org/10.1038/nrmicro.2016.108
- Martin JS, Monaghan TM, Wilcox MH. Clostridium difficile infection: epidemiology, diagnosis and understanding transmission. Nat Rev Gastroenterol Hepatol (2016); 13:206-16; PMID:26956066; http://dx.doi.org/10.1038/nrgastro.2016.25
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013. (2013); http://www.cdc.gov/drugresistance/threat-report-2013/
- Knetsch CW, Connor TR, Mutreja A, van Dorp SM, Sanders IM, Browne HP, Harris D, Lipman L, Keessen EC, Corver J, et al. Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011. Euro Surveill (2014); 19:20954; PMID:25411691; http://dx.doi.org/10.2807/1560-7917.ES2014.19.45.20954
- Rodriguez C, Taminiau B, Van BJ, Delmee M, Daube G. Clostridium difficile in Food and Animals: A Comprehensive Review. Adv Exp Med Biol (2016); 932:65-92; PMID:27350639
- Hensgens MP, Keessen EC, Squire MM, Riley TV, Koene MG, de BE, Lipman LJ, Kuijper EJ. Clostridium difficile infection in the community: a zoonotic disease? Clin Microbiol Infect (2012); 18:635-45; PMID:22536816; http://dx.doi.org/10.1111/j.1469-0691.2012.03853.x
- Hall IC, O'Toole E. Intestinal flora in new-born infants: with a description of a new pathogenic anaerobe, Bacillus difficilis. Am J Child Dis (1935); 49:390-402; http://dx.doi.org/10.1001/archpedi.1935.01970020105010
- Yutin N, Galperin MY. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol (2013); 15:2631-41; PMID:23834245
- Lawson PA, Citron DM, Tyrrell KL, Finegold SM. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prevot 1938. Anaerobe (2016); 40:95-99; PMID:27370902; http://dx.doi.org/10.1016/j.anaerobe.2016.06.008
- Shen A. Clostridium difficile toxins: mediators of inflammation. J Innate Immun (2012); 4:149-58; PMID:22237401; http://dx.doi.org/10.1159/000332946
- Monot M, Eckert C, Lemire A, Hamiot A, Dubois T, Tessier C, Dumoulard B, Hamel B, Petit A, Lalande V, et al. Clostridium difficile: New Insights into the Evolution of the Pathogenicity Locus. Sci Rep (2015); 5:15023; PMID:26446480; http://dx.doi.org/10.1038/srep15023
- Braun V, Hundsberger T, Leukel P, Sauerborn M, von Eichel-Streiber C. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene (1996); 181:29-38; PMID:8973304; http://dx.doi.org/10.1016/S0378-1119(96)00398-8
- Brouwer MS, Roberts AP, Hussain H, Williams RJ, Allan E, Mullany P. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat Commun (2013); 4:2601; PMID:24131955; http://dx.doi.org/10.1038/ncomms3601
- Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. The role of toxin A and toxin B in Clostridium difficile infection. Nature (2010); 467:711-3; PMID:20844489; http://dx.doi.org/10.1038/nature09397
- Kuehne SA, Collery MM, Kelly ML, Cartman ST, Cockayne A, Minton NP. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis (2014); 209:83-6; PMID:23935202; http://dx.doi.org/10.1093/infdis/jit426
- Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, Howarth P, Clare S, Cunningham B, Sambol SP, et al. Defining the Roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during clostridium difficile infections. MBio (2015); 6:e00551; PMID:26037121; http://dx.doi.org/10.1128/mBio.00551-15
- Lyras D, O'Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, et al. Toxin B is essential for virulence of Clostridium difficile. Nature (2009); 458:1176-9; PMID:19252482; http://dx.doi.org/10.1038/nature07822
- Smits WK. Hype or hypervirulence: A reflection on problematic C. difficile strains. Virulence (2013); 4:26297 [pii]; http://dx.doi.org/10.4161/viru.26297
- Stubbs S, Rupnik M, Gibert M, Brazier J, Duerden B, Popoff M. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol Lett (2000); 186:307-12; PMID:10802189; http://dx.doi.org/10.1111/j.1574-6968.2000.tb09122.x
- Cowardin CA, Buonomo EL, Saleh MM, Wilson MG, Burgess SL, Kuehne SA, Schwan C, Eichhoff AM, Koch-Nolte F, Lyras D, Aktories K, Minton NP, Petri WA, Jr. The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat Microbiol (2016); 1:16108; PMID:27573114; http://dx.doi.org/10.1038/nmicrobiol.2016.108
- Vedantam G, Clark A, Chu M, McQuade R, Mallozzi M, Viswanathan VK. Clostridium difficile infection: toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes (2012); 3:121-34; PMID:22555464; http://dx.doi.org/10.4161/gmic.19399
- Pechine S, Collignon A. Immune responses induced by Clostridium difficile. Anaerobe (2016); 41:68-78; PMID:27108093; http://dx.doi.org/10.1016/j.anaerobe.2016.04.014
- Collery MM, Kuehne SA, McBride SM, Kelly ML, Monot M, Cockayne A, Dupuy B, Minton NP. What's a SNP between friends: The influence of single nucleotide polymorphisms on virulence and phenotypes of Clostridium difficile strain 630 and derivatives. Virulence (2016); [in press].
- Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet (2006); 38:779-86; PMID:16804543; http://dx.doi.org/10.1038/ng1830
- Wust J, Hardegger U. Transferable resistance to clindamycin, erythromycin, and tetracycline in Clostridium difficile. Antimicrob Agents Chemother (1983); 23:784-6; PMID:6870225; http://dx.doi.org/10.1128/AAC.23.5.784
- Monot M, Boursaux-Eude C, Thibonnier M, Vallenet D, Moszer I, Medigue C, Martin-Verstraete I, Dupuy B. Reannotation of the genome sequence of Clostridium difficile strain 630. J Med Microbiol (2011); 60:1193-9; PMID:21349987; http://dx.doi.org/10.1099/jmm.0.030452-0
- Pettit LJ, Browne HP, Yu L, Smits WK, Fagan RP, Barquist L, Martin MJ, Goulding D, Duncan SH, Flint HJ, et al. Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC Genomics (2014); 15:160; PMID:24568651; http://dx.doi.org/10.1186/1471-2164-15-160
- Hussain HA, Roberts AP, Mullany P. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Deltaerm) and demonstration that the conjugative transposon Tn916DeltaE enters the genome of this strain at multiple sites. J Med Microbiol (2005); 54:137-41; PMID:15673506; http://dx.doi.org/10.1099/jmm.0.45790-0
- O'Connor JR, Lyras D, Farrow KA, Adams V, Powell DR, Hinds J, Cheung JK, Rood JI. Construction and analysis of chromosomal Clostridium difficile mutants. Mol Microbiol (2006); 61:1335-51; PMID:16925561; http://dx.doi.org/10.1111/j.1365-2958.2006.05315.x
- Farrow KA, Lyras D, Rood JI. The macrolide-lincosamide-streptogramin B resistance determinant from Clostridium difficile 630 contains two erm(B) genes. Antimicrob Agents Chemother (2000); 44:411-3; PMID:10639372; http://dx.doi.org/10.1128/AAC.44.2.411-413.2000
- Farrow KA, Lyras D, Rood JI. Genomic analysis of the erythromycin resistance element Tn5398 from Clostridium difficile. Microbiology (2001); 147:2717-28; PMID:11577151; http://dx.doi.org/10.1099/00221287-147-10-2717
- van Eijk E, Anvar SY, Browne HP, Leung WY, Frank J, Schmitz AM, Roberts AP, Smits WK. Complete genome sequence of the Clostridium difficile laboratory strain 630Deltaerm reveals differences from strain 630, including translocation of the mobile element CTn5. BMC Genomics (2015); 16:31; PMID:25636331; http://dx.doi.org/10.1186/s12864-015-1252-7
- Riedel T, Bunk B, Thurmer A, Sproer C, Brzuszkiewicz E, Abt B, Gronow S, Liesegang H, Daniel R, Overmann J. Genome resequencing of the virulent and multidrug-resistant reference strain clostridium difficile 630. Genome Announc (2015); 3 3/2/:e00276-15 [pii];10.1128/genomeA.00276-15 [doi].
- He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet (2013); 45:109-13; PMID:23222960; http://dx.doi.org/10.1038/ng.2478
- Wu Z, Periaswamy B, Sahin O, Yaeger M, Plummer P, Zhai W, Shen Z, Dai L, Chen SL, Zhang Q. Point mutations in the major outer membrane protein drive hypervirulence of a rapidly expanding clone of Campylobacter jejuni. Proc Natl Acad Sci U S A (2016); 113:10690-5; PMID:27601641; http://dx.doi.org/10.1073/pnas.1605869113
- McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol (2011); 193:2027-34; PMID:21278284; http://dx.doi.org/10.1128/JB.01542-10
- Tenaillon O, Barrick JE, Ribeck N, Deatherage DE, Blanchard JL, Dasgupta A, Wu GC, Wielgoss S, Cruveiller S, Medigue C, et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature (2016); 536:165-70; PMID:27479321; http://dx.doi.org/10.1038/nature18959
- Burns DA, Heap JT, Minton NP. The diverse sporulation characteristics of Clostridium difficile clinical isolates are not associated with type. Anaerobe (2010); 16:618-22; PMID:20950700; http://dx.doi.org/10.1016/j.anaerobe.2010.10.001
- Burns DA, Heeg D, Cartman ST, Minton NP. Reconsidering the sporulation characteristics of hypervirulent Clostridium difficile BI/NAP1/027. PLoS One (2011); 6:e24894; PMID:21949780; http://dx.doi.org/10.1371/journal.pone.0024894
- Heeg D, Burns DA, Cartman ST, Minton NP. Spores of Clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS One (2012); 7:e32381; PMID:22384234; http://dx.doi.org/10.1371/journal.pone.0032381
- Mackin KE, Carter GP, Howarth P, Rood JI, Lyras D. Spo0A differentially regulates toxin production in evolutionarily diverse strains of Clostridium difficile. PLoS One (2013); 8:e79666; PMID:24236153; http://dx.doi.org/10.1371/journal.pone.0079666
- Lyon SA, Hutton ML, Rood JI, Cheung JK, Lyras D. CdtR Regulates TcdA and TcdB Production in Clostridium difficile. PLoS Pathog (2016); 12:e1005758; PMID:27414650; http://dx.doi.org/10.1371/journal.ppat.1005758
- Chewapreecha C, Marttinen P, Croucher NJ, Salter SJ, Harris SR, Mather AE, Hanage WP, Goldblatt D, Nosten FH, Turner C, et al. Comprehensive identification of single nucleotide polymorphisms associated with beta-lactam resistance within pneumococcal mosaic genes. PLoS Genet (2014); 10:e1004547; PMID:25101644; http://dx.doi.org/10.1371/journal.pgen.1004547
- Laabei M, Recker M, Rudkin JK, Aldeljawi M, Gulay Z, Sloan TJ, Williams P, Endres JL, Bayles KW, Fey PD, et al. Predicting the virulence of MRSA from its genome sequence. Genome Res (2014); 24:839-49; PMID:24717264; http://dx.doi.org/10.1101/gr.165415.113
- Forgetta V, Oughton MT, Marquis P, Brukner I, Blanchette R, Haub K, Magrini V, Mardis ER, Gerding DN, Loo VG, et al. Fourteen-genome comparison identifies DNA markers for severe-disease-associated strains of Clostridium difficile. J Clin Microbiol (2011); 49:2230-8; PMID:21508155; http://dx.doi.org/10.1128/JCM.00391-11
