ABSTRACT
Acinetobacter baumannii is a hospital-acquired pathogen that shows an extraordinary capacity to stay in the hospital environment. Adherence of the bacteria to eukaryotic cells or to abiotic surfaces is the first step for establishing an infection. The A. baumannii strain AbH12O-A2 showed an exceptional ability to adhere to A549 epithelial cells. The AbFhaB/FhaC 2-partner secretion (TPS) system involved in adhesion was discovered after the screening of the recently determined A. baumannii AbH12O-A2 strain genome (CP009534.1). The AbFhaB is a large exoprotein which transport to the bacterial surface is mediated by the AbFhaC protein. In the present study, the role of this TPS system in the AbH12O-A2 adherence phenotype was investigated. The functional inactivation of this 2-partner secretion system was addressed by analyzing the outer membrane vesicles (OMV) proteomic profile from the wild-type strain and its derivative mutant AbH12O-A2ΔfhaC demonstrating that AbFhaB is no longer detected in the absence of AbFhaC. Scanning electron microscopy (SEM) and adhesion experiments demonstrated that inactivation of the AbFhaB/FhaC system significantly decreases bacterial attachment to A549 alveolar epithelial cells. Moreover, it has been demonstrated that this 2-partner secretion system is involved in fibronectin-mediated adherence of the A. baumannii AbH12O-A2 isolate. Finally, we report that the AbFhaB/FhaC system is involved in virulence when tested using invertebrate and vertebrate hosts. These data suggest the potential role that this AbFhaB/FhaC secretion system could play in the pathobiology of A. baumannii.
Introduction
Acinetobacter baumannii is an opportunistic human pathogen considered a major cause of nosocomial infections worldwide. A. baumannii causes pneumonia, skin and soft tissue infections, bacteremia, meningitis, endocarditis and urinary tract infections resulting in high rates of morbidity and mortality.Citation1 The emergence of multidrug-resistant (MDR) strains has led to a number of hospital outbreaks that have become a serious health problem worldwide.Citation2-6
Many studies have been focused on the investigation of antimicrobial resistance. Although the clinical importance of A. baumannii infections has increased, the pathogenicity of this microorganism is sparsely understood, since it has been traditionally considered as a low virulent pathogen.Citation2 Several bacterial virulence factors are needed for the pathogenesis of Acinetobacter infections and only a few have been described in this specie. Clinical A. baumannii strains exhibit remarkably variations in virulence-associated phenotypes such as motility, adherence, biofilm formation, invasion, iron uptake, cell capsule development or penicillin binding proteins modifications among others.Citation2,7-14
Some studies have evidenced that Acinetobacter species may reach the human skin and mucosal membranes and then colonize and persist on the host several weeks.Citation15 A. baumannii has an innate capacity to interact with and adhere to diverse types of surfaces. Bacterial adherence constitutes an essential step in the colonization process. After adhesion, bacteria may form biofilms that are involved in the persistence of this pathogen in the hospital environment. Some components, such as the staphylococcal biofilm-associated protein (Bap), the CsuA/BABCDE usher-chaperone system or the poly-β-1–6-N-acetylglucosamine have been described as involved in the A. baumannii biofilm formation and adherence phenotypes.Citation16-20 A study made with 52 different clinical strains revealed that the aptitude to form biofilm and the ability to attach to host cells are independent abilities that do not have to be associated.Citation21 This could indicate that A. baumannii has several independent mechanisms involved in biofilm formation and adherence to different surfaces that also are independently regulated.
In the other hand, the capacity of A. baumannii to adhere to and achieve biofilm formation on biotic surfaces is barely known. The outer membrane protein OmpA, besides of being related to biofilm formation in abiotic surfaces, has been shown to promote the adherence to eukaryotic host and invasion.Citation22 Also, Bentancor et al.Citation23, have corroborated that the trimeric autotransporter protein Ata, belonging to the type V secretion system, participates in biofilm formation and intercedes in the attachment of A. baumannii cells to immobilized collagen type IV.Citation23 The type V secretion systems are classified into either autotransporters or 2-partner secretion systems (TPS). The protein Ata was the first adhesion component that belongs to the type V autotransporters secretion systems described in A. baumannii. The filamentous hemagglutinin protein (FHA) from Bordetella pertussis, HecA from Erwinia chrysanthemi and HMW1/2 from Haemophilus influenzae have been described as adhesins belonging to this group of proteins and constitute one of the major virulence factors described in these species.Citation24,25 The TPS are widespread secretion pathways of Gram-negative bacteria and involve 2 proteins: TpsA, a large exoprotein, and TpsB, the outer membrane protein. The genes coding for these 2 proteins are usually co-transcribed within the same operon. The TpsB protein forms a ß-barrel included into the outer membrane. In the other hand, the N-terminal domain of TpsA connects with the POTRA domain of TpsB proceeding to the secretion. After, TpsA may stay associated at the cell surface or be liberated to the extracellular medium.Citation26
The present work is focused on an A. baumannii strain that was responsible for one of the most important nosocomial outbreaks to date.Citation3 During more than 30 months more than 350 patients were colonized with MDR A. baumannii strains in a hospital in Madrid.Citation3 In recent studies, the molecular mechanisms involved in the spread and dissemination of the AbH12O-A2 strain have been characterized.Citation3,27,28 This strain was found as the main clone within this outbreak and constituted a target of many studies.Citation29,30 Recently, our group determined its complete genome (accession number CP009534.1).Citation31 We found that the most remarkable characteristic of the AbH12O-A2 strain is its notable ability to adhere to human cells. An analysis of its genome allowed us to focus on a genome region coding for an FhaB/FhaC-like 2-partner secretion system due to its possible implication in adherence. The main goal of the present work was to analyze the role of the FhaB/FhaC-like 2-partner secretion system in the pathogenicity of A. baumannii strain AbH12O-A2.
Results
The A. baumannii AbH12O-A2 strain harbors a genomic island involved in adherence
The A. baumannii strain AbH12O-A2 was the main clone isolated from a large nosocomial outbreak occurring in a hospital in Madrid (Spain) from 2006 to 2008. This A. baumannii isolate has the capacity to survive and persist for long periods in the hospital settings. Our group recently demonstrated the molecular mechanisms implicated in the reply to desiccation and persistence of this pathogen.Citation29 Several phenotype analyses were performed to find out the molecular strategies that may be involved in persistence and disease potential of A. baumannii strain AbH12O-A2.Citation3,27-31 Attachment assays showed that the AbH12O-A2 strain had a higher ability to adhere to A549 alveolar epithelial cells compare with the ATCC 17978 strain, being 37-fold more adherent with a P value < 0.001 (). This adherence phenotype may explain its propensity to remain in colonized patients, this being one of the keys of the prolonged nosocomial outbreak. Protein coding genes involved in the adherence phenotype shown by AbH12O-A2 strain were analyzed based on the fully sequenced and annotated genome of this clinical isolate (GenBank accession code CP009534.1).Citation31 The whole-genome analysis of the AbH12O-A2 strain revealed the presence of a ca. 18-kb chromosomal region containing genes involved in adherence, which also appears in the genomes of the A. baumannii 3207 and AB030 strains that showed 99% and 100% identity to the AbH12O-A2 chromosomal region, respectively. This ca. 18-kb region was also found in the genomes of the A. baumannii IOMTU 433, 6200 and SDF strains and, as assessed by the Blast tool of the NCBI, it was found coverage of 87, 87 and 70% with an identity of 96, 94 and 96%, respectively. It was observed that the ca. 18-Kb region was partially conserved in the genomes of the A. baumannii strains R2091, CIP70.10, AB031 and ZW85–1 where the genes flanking the island (LX00_12065 and LX00_12105: AbfhaC) are present and the genes LX00_12075, LX00_12080, LX00_12085, LX00_12090 and LX00_12095 are absent and only the ending region of the LX00_12100 gene (AbfhaB) appears. shows that this ca. 18-kb region described in the AbH12O-A2 genome is flanked by repeated DNA sequences. The LX00_12060 locus coding for a LuxR family transcriptional regulator and the LX00_12110 locus coding for a tRNA-Trp were found upstream and downstream of this region, respectively. Eight coding sequences were identified within this region: LX00_12065 coding for a TetR family transcriptional regulator (AIS07096.1), LX00_12075, LX00_12080, LX00_12085, LX00_12090, LX00_12095 coding for hypothetical proteins (AIS07097.1, AIS07098.1, AIS07099.1, AIS07100.1 and AIS07101.1, respectively), LX00_12100 coding for an adhesion protein (AKB90480), and LX00_12105 coding for a membrane protein (AIS07102.1), as shown in .
Figure 1. (A) Determination of the attachment ability of the AbH12O-A2 clinical strain, the AbH12O-A2ΔfhaC isogenic mutant derivate strain and the complemented strain to A549 epithelial cells. T-student tests were performed and 6 independent replicates were done. Bars indicate the standard deviation and asterisks indicate P values under 0.001. (B) SEM visualization of bacterial attachment to A549 human alveolar epithelial cells infected with A. baumannii. a) A. baumannii AbH12O-A2 clinical strain. b) A. baumannii mutant derivative AbH12O-A2ΔfhaC. c) Healthy uninfected A549 cells covered by surfactant as a negative control. Micrographs were taken at 10.000X magnifications. Bars indicated the scale (1 µm).

Figure 2. Scheme of the AbFhaB/FhaC 2-partner secretion system of A. baumannii AbH12O-A2 and its genomic context. (A) Genetic organization and orientation of the ca. 18-kb genomic island found in A. baumannii AbH12O-A2 strain (LX00_12065-LX00_12105) and the surrounding regions LX00_12060 and LX00_12110. The CDS are represented by arrows. Asterisks indicate repeated regions (*aaaaagctcc and *aggagcttt). (B) Representation of the AbFhaB (LX00_12100)/AbFhaC(LX00_12105) 2-partner secretion system of A. baumannii AbH12O-A2 strain. The CDS (AbfhaB, 11,172 bp and AbfhaC of 1,758 bp) are represented by arrows. Circles represent repeated residues. Asterisk represents an AbfhaB signal peptide.
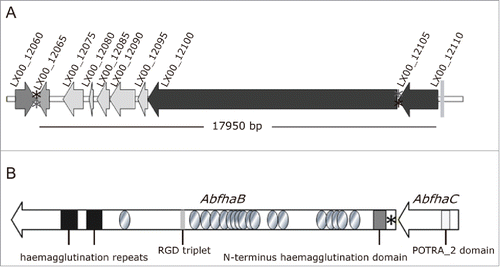
Further bioinformatic analysis revealed a TPS, named here as AbFhaB/AbFhaC, encoded by the LX00_12100 and LX00_12105 genes, respectively (). The LX00_12100 gene has been predicted to code for a 392.72-kDa exoprotein (TpsA, named here as AbFhaB) with a 25-amino acid residue signal peptide, typical of Type V secretion systems. A deeper examination of this sequence showed the appearance of an N-terminus haemagglutination activity domain (amino acids 101 to 221), highly conserved in the TPS domain of the TpsA exoproteins (IPR008638 code from InterPro database). In addition, multiple copies of a 20-amino acid residue repeat (IPR010069 code from InterPro database), also found in filamentous haemagglutinins, were detected in the AbFhaB protein, as shown in . An Arg-Gly-Asp triplet (RGD) that has been related to adhesion functions occurred once in the predicted sequence (amino acids 2029 to 2031) in the TpsA ortholog.Citation32 The LX00_12105 gene was expected to code for a 65.54-kDa outer membrane protein (TpsB, named here as AbFhaC), that belongs to the Omp85/TpsB transporter family. This protein contains a polypeptide transport-associated domain named POTRA_2 (IPR005565 code from InterPro database) localized between amino acids 87 and 166. Structure prediction analysis using Phyre2 suggested that the LX00_12105 gene may have a similar structure (100% confidence and 86% coverage) to the FhaC protein, member of the Omp85/Tpsb2 transporter family.
The AbH12O-A2ΔfhaC mutant derivative strain does not produce the AbFhaB and AbFhaC proteins
In order to investigate the role of the TPS system in the pathogenicity of the AbH12O-A2 strain, this system was inactivated by deleting the POTRA_2 domain of the fhaC gene, which is responsible for the transport and linkage of FhaB in the cell surface, obtaining the AbH12O-A2ΔfhaC mutant derivative. Briefly, the POTRA_2 domain-coding region of the LX00_12105 gene (fhaC) was deleted by double crossover recombination employing the pMo130TelR. The construction of the AbH12O-A2ΔfhaC mutant strain was verified by PCR and qRT-PCR results demonstrated the lack of LX00_12105 gene expression in this mutant strain (data not shown). The AbH12O-A2ΔfhaC isogenic deletion derivative mutant of AbH12O-A2 was constructed by deleting a region encompassing the fhaC gene without affecting the upstream and downstream surrounding genes, as assessed by RNA expression analysis, since no polar effects were observed (data not shown).
The inactivation of the TPS system was also confirmed through proteomic approaches. OMVs protein profile from AbH12O-A2 and its isogenic derivative mutant AbH12O-A2ΔfhaC were analyzed using an Information Dependent Analysis (IDA) Enhanced MS–Enhanced Resolution (EM ER) method. With this approach a total of 43 proteins from the AbH12O-A2 strain and 55 proteins from the AbH12O-A2ΔfhaC strain were identified in the OMV fraction. The FhaB protein was identified in the wild type sample using the protein database of the AbH12O-A2 strain available in GenBank (protein accession codes from A1S04698.1 to A1S8323.1), whereas the FhaC protein was not identified using this database. Besides, none of these 2 proteins were detected in the mutant sample when the same database was used. However, for the wild type strain, the 2 proteins matched several peptides when a specific database (FhaBFhaC_M.) was used and, as expected, none of these 2 proteins was identified in the mutant samples. The best proteotypic peptides were chosen for the 2 target proteins to create a MRM method where 3 peptides per protein and at least 3 transitions per peptide were considered.
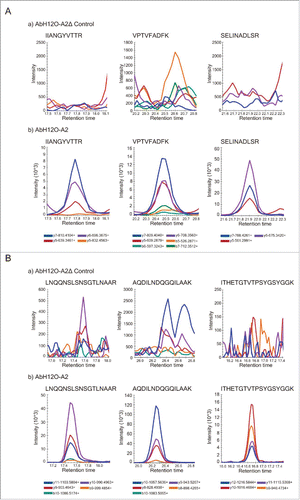
In the AbH12O-A2 samples the 2 proteins were present, the FhaB protein was readily identified by MSMS spectra but the FhaC protein, which is a minor component, was not identified with IDA under the confidence parameters of the study (Confidence > 99%). However, their peptides were identified and its spectrum of fragmentation was present with less confidence (Confidence > 95%), enough to set the conditions for the targeted analysis. In the MRM study the co-elution of at least 3 transitions for each of the 3 peptides belonging to each protein clearly demonstrates that both FhaB and FhaC are present in the AbH12O-A2 sample, as it is showed in . In the AbH12O-A2ΔfhaC samples, none of these 2 proteins were observed. In addition, no fragmentation spectrum data are available from these proteins in the AbH12O-A2ΔfhaC sample. As a conclusion, these 2 proteins were not present in the AbH12O-A2ΔfhaC samples, being identified in the AbH12O-A2 samples using targeted proteomics methodology.
Figure 3. (A) Identification of tryptic peptides using SRM/MRM. Extracted ion chromatogram (XIC) of 3 peptides from the protein AbFhaC (FhaC) in the AbH12O-A2ΔfhaC mutant strain (a) and in the AbH12O-A2 wild type strain (b) samples. The name of the co-eluted transitions for each peptide is showed in the legend. (B) Identification of tryptic peptides using SRM/MRM. Extracted ion chromatogram (XIC) of 3 peptides from the protein AbFhaB (FhaB) in the AbH12O-A2ΔfhaC mutant strain (a) and the AbH12O-A2 wild type strain (b) samples. The name of the co-eluted transitions for each peptide is showed in the legend.
The two-partner secretion system AbFhaB/AbFhaC is related to attachment to human alveolar epithelial
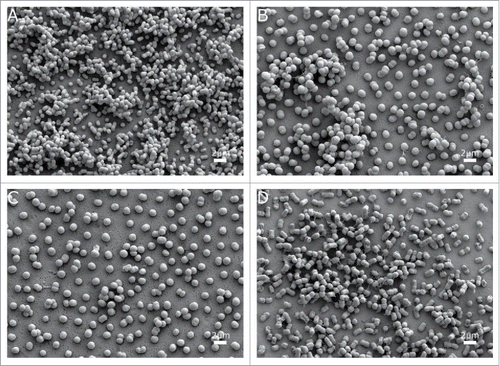
showed that the AbH12O-A2 strain is a poor biofilm former compared with the ATCC 17978 strain (micrographs A and B), being unable to form multilayered biofilm structures on abiotic surfaces. When the fhaC-deficient strain was analyzed (micrograph C) the adherence pattern showed well-separated cells. In this case, no evidence of tridimensional structures was visualized. For the complemented strain, this phenotype was somehow restored (micrograph D) showing some tridimensional cell aggregation, also observed in the wild type AbH12O-A2 strain. However, the low ability of biofilm formation of this strain does not allow a clear interpretation of the role of the AbFhaB/AbFhaC system on biofilm formation on abiotic surfaces.
Figure 4. SEM visualization of biofilm on abiotic surfaces of the A. baumannii strains (a) ATCC 17978, (b) AbH12O-A2, (c) AbH12O-A2ΔfhaC and d) AbH12O-A2ΔfhaC complemented. Micrographs were taken at 10,000x magnification. Bars indicate the scale marks (2 μm).
The AbH12O-A2ΔfhaC isogenic mutant was used to investigate the role of the AbFhaB/AbFhaC system in adherence to biotic surfaces. Thus, A549 human alveolar epithelial cells were infected with A. baumannii AbH12O-A2, the fhaC mutant derivative (AbH12O-A2ΔfhaC) and the complemented mutant (AbH12O-A2ΔfhaC complemented) strains for 3 h (). shows that adherence of AbH12O-A2 to A549 was significantly reduced (2.3-fold less, P value < 0.0001) when the fhaC gene was deleted. The adherence phenotype was restored when the fhaC gene was overexpressed in the mutant strain (). At this point, it is important to remark that the wild type strain and the mutant derivatives grow at the same rate in rich medium without selective pressure (Figure S1).
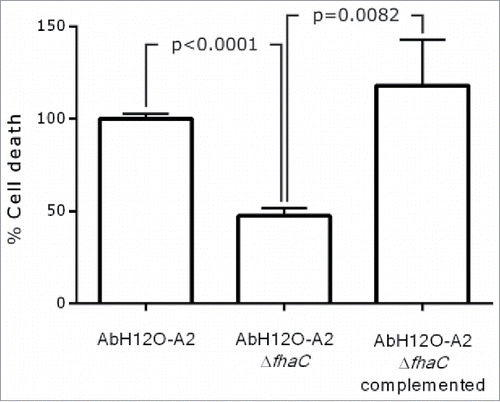
In agreement with the attachment assay, the AbH12O-A2 wild type strain showed a high attachment phenotype and appeared to form biofilm structures over the A549 cells (micrograph a, ). In addition, the fhaC-deficient AbH12O-A2 strain had reduced adherence to A549 cells and was unable to form such biofilm structures (micrograph b, ) as compared with the parental strain (micrograph a, ). Interestingly, the wild type strain (micrograph a, ) caused more damage to the cell layer than the fhac-deficient strain (micrograph b, ), demonstrated by the higher loss of the protective surfactant layer, which remained intact in the uninfected cells (micrograph c, ). In agreement with these results, the data obtained through the LIVE/DEAD assay also revealed that the fhaC-deficient strain caused less damage to the A549 cells while the complemented mutant restored the wild type phenotype ().
The two-partner secretion system AbFhaB/AbFhaC interacts with the host cell protein fibronectin
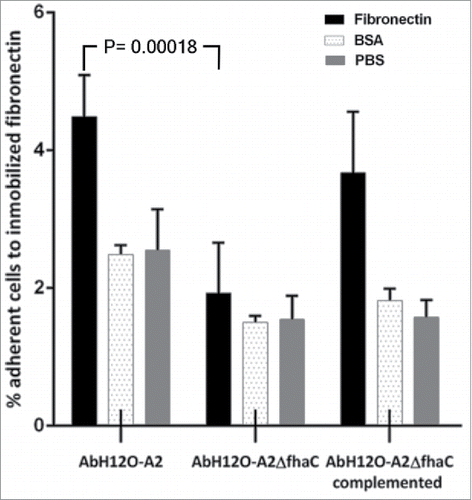
Adherence to extracellular matrix proteins, such as fibronectin, may have a role in bacterial adhesion and internalization.Citation33-36 An analysis of the interaction of the AbFhaB/AbFhaC system with fibronectin was performed incubating the AbH12O-A2 parental strain and its isogenic mutants on fibronectin-coated polystyrene wells. A. baumannii strains tested adhered more to immobilized fibronectin than to control proteins such as BSA (). FhaC was identified as a protein involved in fibronectin binding since its deletion significantly reduced (P = 0.0018) the adhesion of AbH12O-A2 to fibronectin (2.37-fold) compared with the wild-type parental strain (). These data indicate that the 2-partner secretion system AbFhaB/AbFhaC has a role on the interaction between A. baumannii AbH12O-A2 and fibronectin associated with human epithelial cells. The parental adhesion phenotype was restored in the AbH12O-A2ΔfhaC complemented strain ().
Figure 6. Binding of A. baumannii AbH12O-A2, the AbH12O-A2ΔfhaC and the AbH12O-A2ΔfhaC complemented strains to immobilized fibronectin after a 3-h incubation. Dotted bars indicate adherent bacteria to wells coated with BSA. Filled bars indicate adherent bacteria to wells coated with fibronectin. Shaded bars indicate adherent bacteria to uncoated wells. T-student test were perfomed. The standard deviation is indicated by bars. Four independent replicates were done.
The two-partner secretion system AbFhaB/AbFhaC plays a role in virulence
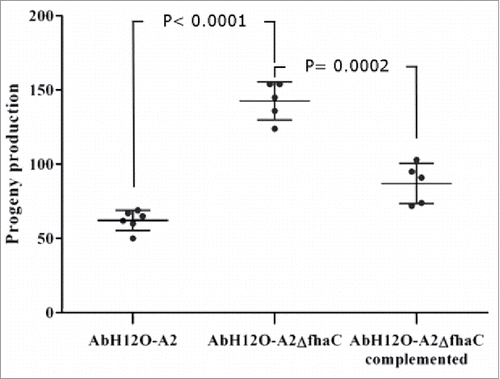
The function of the AbFhaB/AbFhaC system in the virulence of the A. baumannii AbH12O-A2 strain was assessed by a fertility assay using a C. elegans model and a survival assay using a systemic mouse infection model. The C. elegans model showed that worms infected with the A. baumannii AbH12O-A2 strain had a drop of 50% in fertility compared with those worms infected with the knockout strain AbH12O-A2ΔfhaC (). The progeny production was twice higher when C. elegans were infected the fhaC-deficient A. baumannii derivative strain compared with the parental strain AbH12O-A2, being the difference statistically significant. Complementation of the mutant strain with the parental allele restored the wild-type phenotype.
Figure 7. C. elegans fertility assays. L4 worms progeny was monitored when incubated in the presence of the wild type strain AbH12O-A2, the isogenic AbH12O-A2ΔfhaC derivative, or the corresponding AbH12O-A2ΔfhaC complemented strain. T-student test were perfomed. The standard deviation is indicated by bars. Six independent replicates were done.
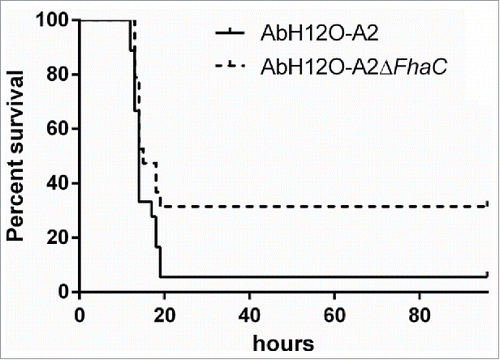
Moreover, the ability of the knockout strain to establish a systemic infection in an experimental murine model was evaluated. To assess the role of this 2-partner secretion system AbFhaB/AbFhaC on virulence, BALB/c mice were intraperitoneally infected with the AbH12O-A2 strain or its isogenic knockout strain AbH12O-A2ΔfhaC. The survival rates were monitored for several days as shown in . The results indicated a higher mortality (17 out of 18 mice) associated with wild-type strain after about 20 h of infection (). In contrast, the mortality rate was significantly reduced (12 out of 18 mice) when mice were infected with the mutant strain within the same period of time.
Discussion
The role of Acinetobacter baumannii as a nosocomial pathogen is well known, but the molecular aspects of its pathogenicity remain poorly understood. The A. baumannii clinical significance is influenced by its extraordinary ability to acquire resistance mechanisms. Besides this emerging resistance profile, A. baumannii has an amazing capacity to survive for long periods in a hospital environment, thus giving advantage to its efficiency for dissemination. Adherence is the crucial first step for colonization and therefore for the development of complex communities and it is an important virulence factor for extracellular bacteria that facilitates their persistence in the host. Previous works have analyzed the capacity of several clinical isolates of A. baumannii to adhere to surfaces and to form biofilm.Citation15,21 Here, we have investigated, using genotypic and phenotypic approaches, the characteristics that may be involved in the persistence and disease potential of the MDR A. baumannii AbH12O-A2 isolate, which caused a large outbreak for more than 30 months producing 65 bacteremia episodes.Citation3 Phenotypic analysis of the AbH12O-A2 isolate revealed its remarkable ability to adhere to human alveolar epithelial cells. However, this A. baumannii clinical strain is a non-motile isolate that does not form multilayered biofilm structures on abiotic surfaces, as assessed by SEM.
It has been previously reported that the filamentous hemagglutinin (FHA) participates in biofilm maturing by encouraging the formation of microcolonies in Bordetella pertussis.Citation37 Also, the TPS system described by Darvish et al.Citation38 was shown to be involved in biofilm formation in the strain A. baumannii 19606T. However, the role of the TPS system from the AbH12O-A2 strain in biofilm formation cannot be clearly elucidated in the present work. Although this strain showed a remarkably attachment ability, it is indeed a poor biofilm former not being able to form a mature biofilm, which difficults the detection of any biofilm formation change in its isogenic derivative mutants. In contrast, this strain showed a high attachment ability which suggests that the mechanisms of adherence of A. baumannii AbH12O-A2 strain to either abiotic or biotic surfaces could be different. This supposition is in agreement with data previously found by Eijkelkamp et al.Citation21 who reported that there is not a mandatory relationship between the capacity to form biofilms and eukaryotic cell adherence.
The genome analysis of the AbH12O-A2 strainCitation31 revealed that the AbFhaB/FhaC 2-partner secretion system was only present, with the 100% of coverage, in the case of the MDR A. baumannii strains 3207 and AB030, being also highly conserved in the case of the strains IOMTU 433, 6200 and SDF. Our genomic analysis suggests a low prevalence of this genomic island in A. baumannii spp and showed that the AbFhaB/FhaC operon was located into an 18 kb-region flanked by repetitive sequences. This fact along together with the lower GC content (ca. 38%) suggested that this genome 18 kb-sequence may have been transmitted by lateral transfer as a mobile genetic element to form what is known as a pathogenicity island.Citation39 AbFhaB/FhaC is a type V 2-partner secretion system with a TPS domain containing 2 proteins; AbFhaB (TpsA protein) and AbFhaC (TpsB protein), where AbFhaB includes the secretion domain and the cleavage site for its transporter AbFhaC. The amino acid sequence analysis of AbFhaB showed similarity to other bacterial TpsA proteins reported, such as Bordetella pertussis FHACitation37 or Erwinia chrysanthemi HecA.Citation40 Moreover, this protein has a RGD motif, which is found in many adhesive proteins associated with an integrin-mediated attachment to mammalian cells, although its role in pathogenesis of A. baumannii has not been determined yet. The predictive structure of AbFhaC protein showed that it is a transmembrane β-barrel channel protein hypothesized to serve as the AbFhaB-conducting pore through the outer membrane. It contains a conserved polypeptide transport-associated domain (POTRA_2), which is expected to participate in the TpsA recognition mediating the translocation of its TpsA partner across the membrane, or their integration into the outer membranes.Citation26,41 Overall, the data suggest that AbFhaC, which belongs to the Omp85-TpsB transporter superfamily, could mediate the secretion of AbFhaB, which ultimately would stay in the outer membrane. Our findings showed that bacteria producing the AbFhaB/FhaC system adhered to A549 cells significantly higher than the fhaC-deficient cells. The functional inactivation of this 2-partner secretion system was addressed by analyzing the OMV proteomic profile from both the wild-type and its derivative mutant AbH12O-A2ΔfhaC strains. Considering the OMVs as a representative sample of the bacterial membrane, we may conclude that AbFhaB is no longer present in the membrane in the absence of AbFhaC.
In previous works, the role of different 2-partner secretion systems from different genera was reported finding that they play an important role in attachment to biotic surfaces.Citation40,42,43,44 In the present work we demonstrated that when the AbFhaB/FhaC system of the AbH12O-A2 strain was functionally inactivated, the adherence phenotype was significantly reduced. Our results are in agreement with those found by Serra et al.Citation37 and Darvish et al.Citation38 wherein they demonstrate that the filamentous hemagglutinin protein has an important function in adherence to epithelial cells in both Bordetella pertussis and A. baumannii ATCC 19606T, respectively.
Structural and functional analyses of bacteria-host cell interactions provided insights into cellular processes implicated in the pathogenicity of A. baumannii AbH12O-A2. This strain resulted to show not only a remarkable ability to attach to but also a high capacity to cause serious damage in the A549 human alveolar epithelial cell monolayer, which was assessed both by SEM analysis and cell death assay, probably involving the surface exposure of AbFhaB.
Further analysis on the interplay of A. baumannii with host cells revealed that the AbFhaB/FhaC system could be involved in fibronectin-binding. Adhesion to extracellular matrix proteins, such as fibronectin, gives an advantage to pathogens to adhere to host cells, which is related with microorganism's virulence.Citation33 It has been reported that fibronectin is an important receptor for several bacterial species.Citation34,35 Smani et al. confirmed that A. baumannii has specific linkages for fibronectin revealing that fibronectin could act as a host receptor for A. baumannii cells.Citation36 Here, we reported that fibronectin could act as a host receptor for the MDR A. baumannii AbH12O-A2 strain since the binding to immobilized fibronectin was shown to be mediated by the AbFhaB/AbFhaC system. Indeed, its inactivation reduced significantly the attachment of the AbH12O-A2 strain to immobilized fibronectin. Thus, we demonstrated that this AbFhaB/AbFhaC TPS system plays a clear role in fibronectin-mediated adherence of the A. baumannii AbH12O-A2 isolate.
Several A. baumannii clinical isolates caused pneumonia, sepsis and soft tissue infections in murine experimental infections.Citation45 Since the AbFhaB/AbFhaC 2-partner system had a key role in adherence, we wanted to evaluate its involvement on A. baumannii virulence. To address this issue, 2 experimental infection models were performed demonstrating the importance of this 2-partner system in virulence. The fertility assay performed in C. elegans has been previously validated as a sensitive and reproducible model to determine subtle differences between isogenic derivative strains of A. baumannii ATCC 17978.Citation46,47 Data revealed that the fhaC gene deletion caused a significant increase in the fertility of the worm, showing the implication of the AbFhaB/AbFhaC 2-partner system in A. baumannii pathogenesis. The results obtained using an acute systemic infection model also supported the involvement of the AbFhaB/AbFhaC 2-partner system of the A. baumannii AbH12O-A2 strain in virulence. These results are in agreement with those found by Melvin et al.Citation25 who reported that FhaB is involved in Bordetella virulence.
The AbFhaB/AbFhaC 2-partner system of the A. baumannii AbH12O-A2 strain plays a role in virulence as confirmed by the infection models mediating the adhesion of bacteria to human epithelial cells. These findings could help to encourage the development of new therapeutic options.
Materials and methods
Bacterial strains, culture conditions, plasmids and DNA extraction
A. baumannii AbH12O-A2 was isolated at the Hospital 12 de Octubre, Madrid (Spain). A. baumannii and E. coli strains were grown in LB broth, adding 2% of agar when necessary. Strains were grown at 37°C and 180 rpm. The storage was done at −80°C in LB broth with 10% glycerol. Sodium tellurite (Sigma-Aldrich, St. Louis, MO) was used for selection of transformant strains at a concentration of 10 μg/mL for E. coli and 30 μg/mL for A. baumannii. Genomic DNA was obtained using the Wizard Genomic DNA purification Kit (Promega Corporation, Madison, WI, USA). Plasmids were extracted using the High Pure Isolation Kit (Roche Diagnostics GmbH, Mannheim, Germany). E. coli TG1 was employed for cloning approaches. A. baumannii ATCC 17978 was used as a control strain for adhesion and SEM analysis. The plasmid pMo130-TelR kindly gave up by Chua Kim Lee (University of Singapore) harbouring a tellurite resistance gene was used to generate the double crossover knockout derivatives as previously described.Citation48 The pWH1266-TelR plasmid used to genetically complement the mutant strain was obtained by cloning the tellurite resistance gene with its own promoter sequence (locus_tag ACIAD2922), amplified by PCR from A. calcoaceticus ADP1 using primers listed in into the HindIII site of the pWH1266 plasmid.Citation49
Table 1. Oligonucleotides and probes used in the present work.
Construction of knockout strain
A mutant derivative strain lacking the fhaC gene (AbH12O-A2ΔfhaC) was constructed using the suicide plasmid pM0130-TelR, which contains the genes xylE, sacB, and the kanamycin and tellurite resistance cassettes, as detailed previously.Citation48 Briefly, fragments of 608 bp and 889 bp located upstream and downstream of the POTRA_2 domain of the LX00_12105 locus (fhaC gene), respectively, were inserted into the pMo130-TelR vector using the oligonucleotides detailed in . The construction obtained was introduced in A. baumannii AbH12O-A2 by electroporation. Recombinant colonies were selected using tellurite and cathecol, following the method previously described by Hamad et al.Citation50 Colonies representing the second crossover events were obtained as described previouslyCitation48,50 and checked by PCR using the oligonucleotides detailed in .
Complementation of the mutant derivative strain AbH12O-A2ΔfhaC
The fhaC gene was PCR-amplified including its own promoter from AbH12O-A2 strain genomic DNA and then cloned into the SalI restriction site of the pWH1266-TelR using the primers listed in . This plasmid is a derivative of the pWH1266Citation51 that harbors tellurite resistance marker which was PCR amplified as described above. The new derivative pWH1266-TelR-fhaC was introduced in the AbH12O-A2ΔfhaC strain by electroporation. Transformants were selected on 30 µg/mL tellurite plates and PCR was used for checking the constructions using oligonucleotides detailed on .
RNA extraction and real-time RT-PCR
RNA isolation and expression level determination of the target genes were performed following the procedures and using the materials previously described by Álvarez-Fraga et al.Citation48 The oligonucleotides and probes used are detailed in .
Isolation and purification of OMVs
OMVs were isolated following the protocol described by Méndez et al.Citation30 Briefly, cells were centrifuged and the supernatant was filtered to remove residual bacteria. The OMV pellet was obtained by ultracentrifugation and then resuspended in PBS. The OMV suspension was filtered and processed for protein extraction. Trichloroacetic acid was added to precipitate the proteins from the OMV suspension. Then, for proteomic approaches, proteins were solubilized and their concentration was measured using the Bio-Rad protein assay (Bio-Rad, Munich, Germany).
In-solution protein digestion
In-solution digestions were performed as described by Méndez et al.Citation30 Two biological samples of the AbH12O-A2 and AbH12O-A2ΔfhaC were analyzed. In all cases the resulting tryptic peptide mixture was acidified with 1 µL of 10% of trifluoroacetic acid and desalted using home-made stage-tipsCitation52 as previously described.Citation53 Samples were aliquoted, subjected to a speed-vacuum and kept at −80°C until used.
Nanoscale liquid chromatography and mass spectrometry
Both enhanced MS (EM) and enhanced resolution (ER) methods were performed injecting 2 µg of AbH12O-A2 and AbH12O-A2ΔfhaC samples in the LC-MS/MS, using a nanoLC system (Tempo, Eksigent, Dublin, (CA), USA coupled to a 5500QTRAP instrument (ABSciex). After precolumn desalting using a C18 column, (5 μm, 300A, 100 μm, 2 cm, Acclaim PepMap, Thermo Scientific, USA) at a flow of 3 μL/min during 10 min, tryptic digests (2 μg) were separated on C18 nanocolumns (75 μm id, 15 cm, 3 μm particle size) (Acclaim PepMap 100, Thermo Scientific, USA) at a flow rate of 300 nL/min. A standard 120 min gradient from 5 to 40% of buffer B (0.1% formic acid in 95% acetonitrile) was used.
Peptide selection for multiple reaction monitoring (MRM)
The proteotypic peptides detected with the highest spectral counts and that follow the selection criteria, (fully tryptic, with no missed cleavages, unique to a particular protein, length between 8 and 30 amino acids) were chosen for MRM assay development using Skyline target proteomics Environment. We selected the top transitions for method development based on the presence of abundant y ions at m/z greater than that of the precursor. We used a standard 120 min gradient for the MRM method from 5 to 35% buffer B (0.1% formic acid in 95% acetonitrile). The QTRAP system was interfaced with nanospray sources with uncoated fused silica emitter tips (20 μm inner diameter, 10 μm tip, NewObjective, Woburn, MA). Analysis was done using the positive mode. MS source parameters were as follows: ion spray voltage (IS) was 2600 V, interface heater temperature (IHT) was 150°C, ion source gas 2 (GS2) was 0, curtain gas (CUR) was 20, ion source gas 1 (GS1) was 25 psi, and collision gas (CAD) was high. MS compound parameters were set to 10 for the entrance potential (EP) and to 15 for the collision cell exit potential (CXP). The collision energy (CE) and declusterin potential (DP) was set by Skyline software (Mac Coss lab). Q1 and Q3 were set to unit/unit resolution (0.7 Da) and the pause between mass ranges was set to 3 ms.
Data analysis
The raw data was processed using the Protein Pilot 4.0 software platform (ABSciex). Peptides were identified by comparison with the GenBank protein database from the strain AbH12O-A2 (protein accession codes from A1S04698.1 to A1S8323.1). Search parameters include: Iodoacetamide as Cys-alkylator; Trypsin digestion; Focus on biological modifications; NR as protein database; PSEP mode on and, finally, Detection Protein Threshold Unused ProtScore (Conf)> 1.3 (95.0%). The mass spectrometer switched from MRM to enhanced product ion (EPI) scanning mode automatically when the individual MRM signal exceeded 1000 counts. Each precursor was fragmented a maximum of 2 times before being excluded for 10 s and the mass was scan from 250 to 1000 Da. MRM analysis was conducted with up to 57 transitions per run (dwell time, 35 ms; cycle time or 2.5 s). Each MS/MS spectrum acquired in the 5500 QTRAP spectrometer using IDA (independent data acquisition) was searched in GenBank protein database (protein accession codes from A1S04698.1 to A1S8323.1) with a low confidence due to the lack of very accurate fragments to select the best peptides that give the best identification of the target protein using the protein Pilot software as mentioned before. For each of the 2 target proteins selected, at least 3 transitions were monitored for each peptide and charge 2 and 3 were monitored to confirm the co-elution of the 2 fragments of the same peptide. For each sample 2 biological replicates were analyzed. PSORTb, version 2.0.4 (http://www.psort.org/psortb2/index.html) was used for prediction of sub-cellular locations of the proteins.
Adhesion to A549 human alveolar cells
A549 ephitelial cells were grown in 5% CO2 at 37°C in DMEM medium (Sigma-Aldrich) with 10% heat-inactivated fetal bovine serum and 1% of penicillin-streptomycin (Gibco). Confluent monolayers were washed following the protocol previously described.Citation22,48 The multiplicity of infection (MOI) of 10 was used; in each well 104 A549 cells were infected with 105 bacterial cells. Infected cells were then incubated during 3 h at 37°C in mHBSS medium. Bacterial adhesion was determined as previously described by Álvarez-Fraga et al.Citation48 The number of colony forming units (CFUs) was established to assess the percentage of bacteria attached to eukaryotic cells in comparison with a growth control (bacteria grown in HBSS and 37°C for 3 h without eukaryotic cells). T-student tests were done to determine the statistical significance of the differences. Six biological replicates were performed.
Scanning electron microscopy (SEM)
SEM analysis of biofilms formed on polystyrene coverslips was performed following the methods described by Gaddy et al.Citation22 Briefly, A. baumannii cultures were incubated with sterile polystyrene coverslips for 48 h at 37°C without shaking and prepared for SEM.Citation22 A Zeiss Supra Gemini Series 35V scanning electron microscope was used to view each sample.
SEM analysis of bacterial attachment to A549 human alveolar epithelial cells infected with A. baumannii
A549 human alveolar cells were maintained as previously described.Citation22 Human epithelial cells were grown at a liquid-air interface following the methods described by Álvarez-Fraga et al.Citation48 Briefly, each membrane was seeded with 105 eukaryotic cells. Cells were grown and processed during 3 weeks. Then, each membrane containing the A549 cells was infected with a 1 μL suspension of 102 bacteria in HBSS, as previously described.Citation48 After 72 h of infection, the membranes were washed, fixed with 4% formaldehyde-HBSS, and prepared for SEM using the previously described methods.Citation19,48
Cell death assay on infected A549 monolayers
A549 human alveolar epithelial cells were grown with 10% CO2 at 37°C in DMEM supplemented with 100 mg/L of penicillin, 10% fetal bovine serum, and 100 mg/L streptomycin. For LIVE/DEAD assays, A549 cells were grown to a density of 1 × 105 cells per well in 24-well plates. Then, the cells were infected with 3.6 × 109 CFU/well of A. baumannii in HBSS without antibiotics and grown for 24 h at 37°C.Citation54 The cell death assay was performed as previously described by Alvarez-Fraga et al.Citation48 using the LIVE/DEAD Cell Double Staining Kit (Sigma-Aldrich) according to the manufacturer's instructions. Cell viability was measure after infection. Cell death was determined as the number of red cells related to the total cells visualized. The results were presented as a percentage. Each assay was done by triplicate and Student's t test was used to determine the statistical significance.
Fibronectin-binding assay
The fibronectin-binding assay was performed as described previously.Citation36,55 Briefly, wells were coated with fibronectin. After washing and blocking the wells bacteria were added. Controls were done with BSA and PBS as previously described.Citation36 Overnight cultures of A. baumannii AbH12O-A2 and derivative strains grown in LB at 180 rpm and 37°C were used. Grown bacteria were washed 3 times in PBS and resuspended in a double-volume of the same buffer. One-hundred µL of bacterial suspension were added to each coated or non-coated wells (controls) and incubated 3 h at room temperature. Then, non-adhered bacteria were removed by washing the plates 6 times with PBS. Adherent bacteria were then collected with PBS and Triton X-100 as previously described.Citation36 Finally, bacterial suspensions were plated onto LB agar and incubated at 37°C for 24 h in order to determine the number of CFU and the percentage of bacteria that had attached to immobilized fibronectin compared with the growth control (bacteria grown in LB at 37°C for 3 h in non-coated wells). Four independent replicates were performed. Student's t test was done and bars indicate the standard deviation.
Caenorhabditis elegans virulence assay
Fertility assays were performed as previously described.Citation47 Briefly, both the AbH12O-A2 and the mutant derivative strains were grown overnight in LB and then cultured at 37°C for 24 h in nematode growth medium (NG). The eggs of C. elegans N2 Bristol were hatched in M9 medium and L1 worms were arrested overnight at 20°C. Then, the L1 worms were incubated in NG medium plates in presence of the bacterial strains. One L4 worm was placed on a peptone-glucose-sorbitol medium (PGS) plate individually seeded with each A. baumannii strain and incubated at 25°C for 24 h. The worms were transferred to new plates seeded with the same bacterial strain and the worm progeny was counted daily for 3 d to determine their viability. Six independent replicates were done with each strain. The differences were statistically evaluated by using Student's t test. Means of the differences between strains are reported.
Virulence assays in a mouse systemic-infection model
A. baumannii strains were evaluated for virulence in terms of mortality and survival time of infected mice as previously described.Citation54 Groups of 18 BALB/c female mice, weighting 20–25 g, 6–9 weeks old, were inoculated by intra-peritoneal injection with 250 µL of bacterial suspension with 33×107 CFU/mouse of the wild type strain or 46×107 CFU/mouse of the knockout strain. Mice were monitored for signs of disease during a week. Animals that survived after this period were euthanized. Results were analyzed using the log rank test. A P value of 0.05 was considered statistically significant. Mice were maintained in the specific pathogen-free facility at the Technology Training Center of the Hospital of A Coruña (CHUAC, Spain). All experiments were done with the approval of and in accordance with regulatory guidelines and standards set by the Animal Ethics Committee (CHUAC).
Bioinformatic analysis
The MAUVE 2.3.1 software was used for multiple genome analysis.Citation56 Functional analysis of proteins were done using the The Interpro program.Citation57 The Phyre2 server was used for predicting the 3-dimensional structure of the protein.Citation58 VectorNT11 (Thermo Fisher Scientific) was used to analyze the DNA sequences and the genetic constructions. The blast tool of the National Center of Biotechnology Information was used for genetic analysis procedures.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KVIR_S_1262313.zip
Download Zip (519.2 KB)Funding
This work has been funded by Projects PI12/00552 and PI15/00860 to GB, PI11/01034 to MP, CP13/00226 to AB, and P14/000059 to MP and AB, all integrated in the National Plan for Scientific Research, Development and Technological Innovation 2013–2016 and funded by the ISCIII - General Subdirection of Assessment and Promotion of the Research – European Regional Development Fund (FEDER) “A way of making Europe.” Miami University Research Funds from the department of Microbiology supported this work. We also want to thank the Spanish Network for Research in Infectious Diseases (REIPI RD12/0015/0014 to GB), co-financed by the European Development Regional Fund (EDRF) “A Way to Achieve Europe, Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad as well as Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica (SEIMC). A.P. was financially supported by the Galician Plan for Research, Innovation and Growth (I2C Plan 2012–2016). J. A. Vallejo was financially supported by the Sara Borrell Program (ISCIII, Spain CD13/00373). S. Rumbo-Feal was financially supported by the Agustí Pumarola Grant (Societat Catalana de Malalties Infeccioses i Microbiologia Clínica, SCMIMC) and Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica (SEIMC). Finally, we thank Chua Kim Lee (University of Singapore) for providing pMo130-TelR.
References
- Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 2007; 5:939-51; PMID:18007677; http://dx.doi.org/10.1038/nrmicro1789
- McConnell MJ, Actis L, Pachon J. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev 2013; 37:130-55; PMID:22568581; http://dx.doi.org/10.1111/j.1574-6976.2012.00344.x
- Acosta J, Merino M, Viedma E, Poza M, Sanz F, Otero JR, Chaves F, Bou G. Multidrug-resistant Acinetobacter baumannii Harboring OXA-24 carbapenemase, Spain. Emerg Infect Dis 2011; 17:1064-7; PMID:21749771; http://dx.doi.org/10.3201/eid/1706.091866
- del Mar Tomas M, Cartelle M, Pertega S, Beceiro A, Llinares P, Canle D, Molina F, Villanueva R, Cisneros JM, Bou G. Hospital outbreak caused by a carbapenem-resistant strain of Acinetobacter baumannii: patient prognosis and risk-factors for colonisation and infection. Clin Microbiol Infect 2005; 11:540-6; PMID:15966971; http://dx.doi.org/10.1111/j.1469-0691.2005.01184.x
- Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 2007; 51:3471-84; PMID:17646423; http://dx.doi.org/10.1128/AAC.01464-06
- Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 2008; 21:538-82; PMID:18625687; http://dx.doi.org/10.1128/CMR.00058-07
- Eijkelkamp BA, Stroeher UH, Hassan KA, Paulsen IT, Brown MH. Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics 2014; 15:1020; PMID:25422040; http://dx.doi.org/10.1186/1471-2164-15-1020
- Antunes LC, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis 2014; 71:292-301; PMID:24376225; http://dx.doi.org/10.1111/2049-632X.12125
- Gaddy JA, Arivett BA, McConnell MJ, López-Rojas R, Pachón J, Actis LA. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun 2012; 80:1015-24; PMID:22232188; http://dx.doi.org/10.1128/IAI.06279-11
- Jacobs AC, Hood I, Boyd KL, Olson PD, Morrison JM, Carson S, Sayood K, Iwen PC, Skaar EP, Dunman PM. Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect Immun 2010; 78:1952-62; PMID:20194595; http://dx.doi.org/10.1128/IAI.00889-09
- Russo TA, Luke NR, Beanan JM, Olson R, Sauberan SL, MacDonald U, Schultz LW, Umland TC, Campagnari AA. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect Immun 2010; 78:3993-4000; PMID:20643860; http://dx.doi.org/10.1128/IAI.00366-10
- Russo TA, MacDonald U, Beanan JM, Olson R, MacDonald IJ, Sauberan SL, Luke NR, Schultz LW, Umland TC. Penicillin-binding protein 7/8 contributes to the survival of Acinetobacter baumannii in vitro and in vivo. J Infect Dis 2009; 199:513-21; PMID:19143563; http://dx.doi.org/10.1086/596317
- Harding CM, Tracy EN, Carruthers MD, Rather PN, Actis LA, Munson RS, Jr. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. MBio 2013; 4: e00360-13; PMID:23919995; http://dx.doi.org/10.1128/mBio.00360-13
- Smani Y, Dominguez-Herrera J, Pachón J. Association of the outer membrane protein Omp33 with fitness and virulence of Acinetobacter baumannii. J Infect Dis 2013; 208:1561-70; PMID:23908480; http://dx.doi.org/10.1093/infdis/jit386
- Lee JC, Koerten H, van den Broek P, Beekhuizen H, Wolterbeek R, van den Barselaar M, van der Reijden T, van der Meer J, van de Gevel J, Dijkshoorn L. Adherence of Acinetobacter baumannii strains to human bronchial epithelial cells. Res Microbiol 2006; 157:360-6; PMID:16326077; http://dx.doi.org/10.1016/j.resmic.2005.09.011
- Loehfelm TW, Luke NR, Campagnari AA. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J Bacteriol 2008; 190:1036-44; PMID:18024522; http://dx.doi.org/10.1128/JB.01416-07
- Choi AH, Slamti L, Avci FY, Pier GB, Maira-Litran T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol 2009; 191:5953-63; PMID:19633088; http://dx.doi.org/10.1128/JB.00647-09
- Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 2008; 154:3398-409; PMID:18957593; http://dx.doi.org/10.1099/mic.0.2008/019471-0
- Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 2003; 149:3473-84; PMID:14663080; http://dx.doi.org/10.1099/mic.0.26541-0
- de Breij A, Gaddy J, van der Meer J, Koning R, Koster A, van den Broek P, Actis L, Nibbering P, Dijkshoorn L. CsuA/BABCDE-dependent pili are not involved in the adherence of Acinetobacter baumannii ATCC19606(T) to human airway epithelial cells and their inflammatory response. Res Microbiol 2009; 160:213-8; PMID:19530313; http://dx.doi.org/10.1016/j.resmic.2009.01.002
- Eijkelkamp BA, Stroeher UH, Hassan KA, Papadimitrious MS, Paulsen IT, Brown MH. Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol Lett 2011; 323:44-51; PMID:22092679; http://dx.doi.org/10.1111/j.1574-6968.2011.02362.x
- Gaddy JA, Tomaras AP, Actis LA. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 2009; 77:3150-60; PMID:19470746; http://dx.doi.org/10.1128/IAI.00096-09
- Bentancor LV, Camacho-Peiro A, Bozkurt-Guzel C, Pier GB, Maira-Litran T. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J Bacteriol 2012; 194:3950-60; PMID:22609912; http://dx.doi.org/10.1128/JB.06769-11
- Jacob-Dubuisson F, Fernandez R, Coutte L. Protein secretion through autotransporter and two-partner pathways. Biochim Biophys Acta 2004; 1694:235-57; PMID:15546669; http://dx.doi.org/10.1016/j.bbamcr.2004.03.008
- Melvin JA, Scheller EV, Noel CR, Cotter PA. New Insight into Filamentous Hemagglutinin Secretion Reveals a Role for Full-Length FhaB in Bordetella Virulence. MBio 2015; 6: e01189-15; http://dx.doi.org/10.1128/mBio.01189-15
- Thanassi DG, Stathopoulos C, Karkal A, Li H. Protein secretion in the absence of ATP: the autotransporter, two-partner secretion and chaperone/usher pathways of gram-negative bacteria (review). Mol Membr Biol 2005; 22:63-72; PMID:16092525; http://dx.doi.org/10.1080/09687860500063290
- Merino M, Acosta J, Poza M, Sanz F, Beceiro A, Chaves F, Bou G. OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob Agents Chemother 2010; 54:2724-7; PMID:20385865; http://dx.doi.org/10.1128/AAC.01674-09
- Rumbo C, Fernandez-Moreira E, Merino M, Poza M, Méndez JA, Soares NC, Mosquera A, Chaves F, Bou G. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother 2011; 55:3084-90; PMID:21518847; http://dx.doi.org/10.1128/AAC.00929-10
- Gayoso CM, Mateos J, Méndez JA, Fernández-Puente P, Rumbo C, Tomás M, Martínez de Ilarduya O, Bou G. Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J Proteome Res 2014; 13:460-76; PMID:24299215; http://dx.doi.org/10.1021/pr400603f
- Mendez JA, Soares NC, Mateos J, Gayoso C, Rumbo C, Aranda J, Tomas M, Bou G. Extracellular proteome of a highly invasive multidrug-resistant clinical strain of Acinetobacter baumannii. J Proteome Res 2012; 11:5678-94; PMID:22966805.
- Merino M, Alvarez-Fraga L, Gomez MJ, Aransay AM, Lavin JL, Chaves F, Bou G, Poza M. Complete Genome Sequence of the Multiresistant Acinetobacter baumannii Strain AbH12O-A2, Isolated during a Large Outbreak in Spain. Genome Announc 2014; 2: e01182-14; PMID:25395646; http://dx.doi.org/10.1128/genomeA.01182-14
- D'Souza SE, Ginsberg MH, Plow EF. Arginyl-glycyl-aspartic acid (RGD): a cell adhesion motif. Trends Biochem Sci 1991; 16:246-50; PMID:1926332; http://dx.doi.org/10.1016/0968-0004(91)90096-E
- Henderson B, Nair S, Pallas J, Williams MA. Fibronectin: a multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS Microbiol Rev 2011; 35:147-200; PMID:20695902; http://dx.doi.org/10.1111/j.1574-6976.2010.00243.x
- van der Flier M, Chhun N, Wizemann TM, Min J, McCarthy JB, Tuomanen EI. Adherence of Streptococcus pneumoniae to immobilized fibronectin. Infect Immun 1995; 63:4317-22; PMID:7591065.
- Tsang TM, Felek S, Krukonis ES. Ail binding to fibronectin facilitates Yersinia pestis binding to host cells and Yop delivery. Infect Immun 2010; 78:3358-68; PMID:20498264; http://dx.doi.org/10.1128/IAI.00238-10
- Smani Y, McConnell MJ, Pachón J. Role of fibronectin in the adhesion of Acinetobacter baumannii to host cells. PLoS One 2012; 7:e33073; PMID:22514602; http://dx.doi.org/10.1371/journal.pone.0033073
- Serra DO, Conover MS, Arnal L, Sloan GP, Rodriguez ME, Yantorno OM, Deora R. FHA-mediated cell-substrate and cell-cell adhesions are critical for Bordetella pertussis biofilm formation on abiotic surfaces and in the mouse nose and the trachea. PLoS One 2011; 6:e28811; PMID:22216115; http://dx.doi.org/10.1371/journal.pone.0028811
- Darvish Alipour Astaneh S, Rasooli I, Mousavi Gargari SL. The role of filamentous hemagglutinin adhesin in adherence and biofilm formation in Acinetobacter baumannii ATCC19606(T). Microb Pathog 2014; 74:42-9; PMID:25086432; http://dx.doi.org/10.1016/j.micpath.2014.07.007
- Hayek N. Lateral transfer and GC content of bacterial resistant genes. Front Microbiol 2013; 4:41; PMID:23487592; http://dx.doi.org/10.3389/fmicb.2013.00041
- Rojas CM, Ham JH, Deng WL, Doyle JJ, Collmer A. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc Natl Acad Sci U S A 2002; 99:13142-7; PMID:12271135; http://dx.doi.org/10.1073/pnas.202358699
- Clantin B, Hodak H, Willery E, Locht C, Jacob-Dubuisson F, Villeret V. The crystal structure of filamentous hemagglutinin secretion domain and its implications for the two-partner secretion pathway. Proc Natl Acad Sci U S A 2004; 101:6194-9; PMID:15079085; http://dx.doi.org/10.1073/pnas.0400291101
- Tala A, Progida C, De Stefano M, Cogli L, Spinosa MR, Bucci C, Alifano P. The HrpB-HrpA two-partner secretion system is essential for intracellular survival of Neisseria meningitidis. Cell Microbiol 2008; 10:2461-82; PMID:18680551; http://dx.doi.org/10.1111/j.1462-5822.2008.01222.x
- Di Venanzio G, Stepanenko TM, Garcia Vescovi E. Serratia marcescens ShlA pore-forming toxin is responsible for early induction of autophagy in host cells and is transcriptionally regulated by RcsB. Infect Immun 2014; 82:3542-54; PMID:24914224; http://dx.doi.org/10.1128/IAI.01682-14
- van den Berg BM, Beekhuizen H, Willems RJ, Mooi FR, van Furth R. Role of Bordetella pertussis virulence factors in adherence to epithelial cell lines derived from the human respiratory tract. Infect Immun 1999; 67:1056-62; PMID:10024543.
- Parra Millan R, Jimenez-Mejias ME, Sanchez Encinales V, Ayerbe Algaba R, Gutierrez Valencia A, Pachon-Ibanez ME, Diaz C, Perez Del Palacio J, Lopez Cortes LF, Pachon J, et al. Efficacy of lysophosphatidylcholine (LPC) in combination with antimicrobial agents against Acinetobacter baumannii in experimental murine peritoneal sepsis and pneumonia models. Antimicrob Agents Chemother 2016; 60(8): 4464-70; PMID:27161639.
- Begun J, Gaiani JM, Rohde H, Mack D, Calderwood SB, Ausubel FM, Sifri CD. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog 2007; 3:e57; PMID:17447841; http://dx.doi.org/10.1371/journal.ppat.0030057
- Vallejo JA, Beceiro A, Rumbo-Feal S, Rodríguez-Palero MJ, Russo TA, Bou G. Optimisation of the Caenorhabditis elegans model for studiying the pathogenesis of opportunistic Acinetobacter baumannii. Int J Antimicrob Agents 2016; S0924-8579 (15) 00241-1
- Alvarez-Fraga L, Perez A, Rumbo-Feal S, Merino M, Vallejo JA, Ohneck EJ, Edelmann RE, Beceiro A, Vazquez-Ucha JC, Valle J, et al. Analysis of the role of the LH92_11085 gene of a biofilm hyper-producing Acinetobacter baumannii strain on biofilm formation and attachment to eukaryotic cells. Virulence 2016; 7(4): 443-55
- Nowak J, Seifert H, Higgins P. The tellurite-resistance determinant Tpm of the Acinetobacter baylyi strain ADP1 as a useful non-antibiotic selection marker for genetic manipulation in Acinetobacter baumannii. In: ESMIC, ed. 23rd European Congress of Clinical Microbiology and Infectious Diseases. Berlin, German, 2013.
- Hamad MA, Zajdowicz SL, Holmes RK, Voskuil MI. An allelic exchange system for compliant genetic manipulation of the select agents Burkholderia pseudomallei and Burkholderia mallei. Gene 2009; 430:123-31.; PMID:19010402; http://dx.doi.org/10.1016/j.gene.2008.10.011
- Hunger M, Schmucker R, Kishan V, Hillen W. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 1990; 87:45-51; PMID:2185139; http://dx.doi.org/10.1016/0378-1119(90)90494-C
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2007; 2:1896-906; PMID:17703201; http://dx.doi.org/10.1038/nprot.2007.261
- Mateos J, Pintor-Iglesias A, Fernandez-Puente P, Garcia-Camba M, Ruiz-Romero C, Domenech N, Blanco FJ. Cryoconservation of peptide extracts from trypsin digestion of proteins for proteomic analysis in a hospital biobank facility. J Proteome Res 2014; 13:1930-7; PMID:24521361; http://dx.doi.org/10.1021/pr401046u
- Beceiro A, Moreno A, Fernandez N, Vallejo JA, Aranda J, Adler B, Harper M, Boyce JD, Bou G. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother 2014; 58:518-26; PMID:24189257; http://dx.doi.org/10.1128/AAC.01597-13
- Zimbler DL, Arivett BA, Beckett AC, Menke SM, Actis LA. Functional features of TonB energy transduction systems of Acinetobacter baumannii. Infect Immun 2013; 81:3382-94; PMID:23817614; http://dx.doi.org/10.1128/IAI.00540-13
- Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 2004; 14:1394-403; PMID:15231754; http://dx.doi.org/10.1101/gr.2289704
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 2014; 30:1236-40; PMID:24451626; http://dx.doi.org/10.1093/bioinformatics/btu031
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 2015; 10:845-58; PMID:25950237; http://dx.doi.org/10.1038/nprot.2015.053