ABSTRACT
The type VI secretion system (T6SS) is a widespread molecular weapon deployed by many bacterial species to target eukaryotic host cells or rival bacteria. Using a dynamic injection mechanism, diverse effectors can be delivered by T6SS directly into recipient cells. Here, we report a new family of T6SS effectors encoded by extended Hcps carrying diverse toxin domains. Bioinformatic analyses revealed that these Hcps with C-terminal extension toxins, designated as Hcp-ET, exist widely in the Enterobacteriaceae. To verify our findings, Hcp-ET1 was tested for its antibacterial effect, and showed effective inhibition of target cell growth via the predicted HNH-DNase activity by T6SS-dependent delivery. Further studies showed that Hcp-ET2 mediated interbacterial antagonism via a Tle1 phospholipase (encoded by DUF2235 domain) activity. Notably, comprehensive analyses of protein homology and genomic neighborhoods revealed that Hcp-ET3–4 is fused with 2 toxin domains (Pyocin S3 and Colicin-DNase) C-terminally, and its encoding gene is followed 3 duplications of the cognate immunity genes. However, some bacteria encode a separated hcp-et3 and an orphan et4 (et4O1) genes caused by a termination-codon mutation in the fusion region between Pyocin S3 and Colicin-DNase encoding fragments. Our results demonstrated that both of these toxins had antibacterial effects. Further, all duplications of the cognate immunity protein contributed to neutralize the DNase toxicity of Pyocin S3 and Colicin, which has not been reported previously. In conclusion, we propose that Hcp-ET proteins are polymorphic T6SS effectors, and thus present a novel encoding pattern of T6SS effectors.
Introduction
Bacteria are often under heavy competition pressure for limited resources, and have to export diverse toxins to establish and defend their niche.Citation1,2 The type VI secretion system (T6SS) plays a critical role in this process,Citation3 and delivers diverse antagonistic effectors to adjacent cells by one-step progress in a contact-dependent manner.Citation4,5 In fact, anti-bacterial T6SSs also confer competitive persistence in mixed-species biofilms and even co-surgical,Citation6-8 self-recognition behavior.Citation9 The self-protection mechanism for predator cells uses cognate immunity proteins to neutralize the specific toxic effectors, thus preventing self-intoxication or sibling-intoxication.Citation5,10 Toxic antibacterial effectors are always encoded adjacent to their cognate immunity proteins, either within or outside T6SS gene clusters.
In recent years, bioinformatic and proteomic approaches have been applied successfully to identify T6SS-exported effectors in several organisms.Citation11-13 Three T6SS effector families for interbacterial competition have been well characterized: amidases,Citation11 phospholipases,Citation12,14 and peptidoglycan hydrolases.Citation12,15,16 Additionally, some other antibacterial effectors, such as nucleases,Citation17 NAD(P)+ Glycohydrolase,Citation18 and pore-forming proteins,Citation19 have been characterized. However, with the identification of T6SS in more than 25% of gram-negative bacteria,Citation20 the characterization of these antibacterial effectors has lagged behind. Indeed, for many T6SS-possessing bacteria, their toxic effectors have not been identified and their antibacterial mechanisms are unclear.
The T6SS apparatus comprises a membrane-bound baseplate, an Hcp inner tube, a VipAB outer sheath, and a spike complex consisting of VgrG and PAAR proteins.Citation21 The Hcp protein is both a core component of the T6SS tail tube and the exported receptor/chaperone of the effectors.Citation22,23 As an abundant protein of T6SS, Hcp forms a ring-shaped hexamer that is essential for the assembly of the T6SS apparatus.Citation22,24,25 In T6SS sci-1 of enteroaggregative Escherichia coli, the Hcp1 hexamers assemble tubes in an ordered manner with head-to-tail stacking, and this process was controlled by VgrG.Citation26 In fact, 3 distinct phylogenetic T6SSs (classified into T6SS1/Sci-1, T6SS2 and T6SS3/Sci-2 by different gene organizations and homologies) have been reported in E. coli.Citation27,28 The T6SS2 clusters encoded by the intestinal pathogenic E. coli (including enterohemorrhagic E. coli, enterotoxigenic E. coli, enteropathogenic E. coli and so on) and extraintestinal pathogenic E. coli, show significant difference in their sequence identity, while share similar gene organizations.Citation27,29 Notably, the T6SS2 from E. coli has never been verified to mediate interbacterial antagonism.
Interestingly, bacteria often possess multiple copies of PAAR, hcp and vgrG genes located outside of the major T6SS cluster. Commonly, these ‘orphan’ hcp and vgrG genes are always encoded in the proximity of putative effector genes.Citation30,31 Further studies demonstrated that in T6SS-dependent delivery, the antibacterial effectors are loaded onto the secretion complex by interaction with PAAR/VgrG or by binding to the inner surface of the Hcp tube.Citation3 Notably, some VgrG and PAAR proteins can carry antibacterial extension domains, thus acting as secreted T6SS effectors.Citation16,32 In fact, the interaction with the Hcp pore is a general requirement for the secretion of non-VgrG/PAAR related effectors.Citation26 A previous study has described the “evolved” Hcps with C-terminal extensions,Citation33 while extended Hcps fused with toxin domains have never been validated to mediate interbacterial antagonism experimentally.
In this study, we report a novel encoding pattern of antibacterial effectors using Hcp proteins to carry diverse C-terminal extension toxins (ET domains). Bioinformatic analyses revealed that this type of T6SS effectors, designated as Hcp-ETs, is widespread in the Enterobacteriaceae. Five effector types, Hcp-ET1 to -ET5, and their immunity proteins were identified and characterized. Further studies demonstrated that the Hcp/DUF796 domain was required for their delivery and interaction with the T6SS2 apparatus. Thus, we propose that extended Hcps containing C-terminal toxin domains are polymorphic T6SS effectors.
Results
A C-terminal toxin domain carried by an extended Hcp exhibited antibacterial effect
Previous studies demonstrated that each T6SS can possess more than one cognate Hcp, VgrG, or PAAR-repeat protein.Citation31,32 The VgrG and PAAR proteins have been demonstrated to carry diverse C-terminal extension domains functioning as T6SS antibacterial effectors,Citation6,16,32 while the toxic domains fused to Hcp proteins have never been verified to mediate interbacterial antagonism. Our comparative genomics analysis identified an extended Hcp with a C-terminal HNH-DNase toxin domain (ET-toxin) in human O104 Shiga toxin-producing E. coli (STEC) strain C227–11 and piglet diarrhea isolate STEC004 (). Further NCBI database searching revealed that the T6SS2 cluster and this extended Hcp with the C-terminal ET-toxin domain are widely prevalent in O104:H4 STEC strains. Thus, we designated this extended Hcp as Hcp-ET1, and managed to verify its predicted antibacterial function in strain STEC004.
Figure 1. A C-terminal toxin domain carried by an extended Hcp exhibited antibacterial activity in E. coli. (A) Schematic diagram of the genetic organization of the T6SS locus and Hcp-ET1 effector-immunity pair in strain STEC004. The related sequences have been submitted to GenBank under the accession nos. KX118291 and KX118294, and exhibit high sequence identity to that of sequenced O104:H7 strain C227–11. (B) Growth competition assays between indicated E. coli donor and recipient strains. Experiments were initiated by mixing the donor and recipient bacteria at a CFU ratio of 5:1. The Log10 CFU values of the surviving recipient cells are shown on the y-axis. The pGEN MCS with the target gene inserted under the control of Pcm promoter was used as the complementary plasmid. Asterisks indicate significant differences in the outcome of competition assay for the same donor strain against the indicated recipient strains (**, P < 0.05). Error bars indicate standard deviations (SDs) of 3 independent experiments. (C) Hcp-ET1 degraded plasmid-DNA pUC19 completely in vitro. Purified Hcp-ET1 was incubated with linear pUC19 DNA in the presence and absence of cognate ETI1 or non-cognate Crp (cAMP receptor protein). Crp is a stable protein that exists generally in the cytoplasm of E. coli, and was used as a control for cytoplasmic toxin.Citation60 Reactions were analyzed by native agarose-gel electrophoresis and Goldview (Vazyme, China) staining. All samples were also analyzed by SDS-PAGE and Coomassie blue staining to confirm the proteins' stabilities.
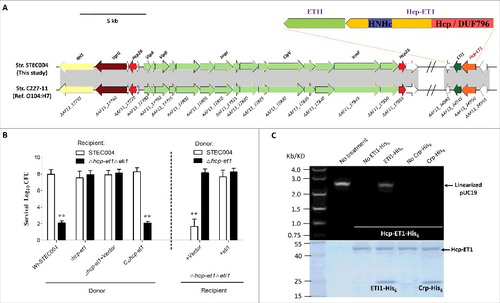
We deleted the hcp-et1/eti1 pair in E. coli STEC004 as the recipient strain, reasoning that cells lacking eti1 immunity genes should be susceptible killing by hcp-et1+ donor cells. Indeed, the hcp-et1 mutant displayed a marked attenuation in its capacity to kill Δhcp-et1Δeti1 recipient cells (). Immunity proteins can protect cells effectively from attack by the cognate toxic effector produced by donors. Thus, we reintroduced the eti1 gene carried by the pGEN-MCS vector into Δhcp-et1Δeti1 recipient cells, and demonstrated effective protection provided by ETI1 against growth inhibition (). We next asked how Hcp-ET1 inhibit target cell growth. The extension domain of Hcp-ET1 from E. coli STEC004 shares ∼35% identity with the HNH-DNase domain of the RhsB C-terminal region from Dickeya dadantii 3937.Citation34 HNH-DNase is cytotoxic by virtue of its DNase activity,Citation34,35 suggesting that Hcp-ET1 might also use the nuclease activity to inhibit target cell growth. Further studies confirmed that purified Hcp-ET1 could degrade linear plasmid DNA completely (), while its immunity protein ETI1 neutralized this DNase toxin (). These results, coupled with the above findings, indicated that E. coli STEC004 cells deploy Hcp-ET1 to kill recipient cells and protect themselves via the immunity protein ETI1.
The Hcp-ET1 is delivered by T6SS2 to degrade target cell DNA
Hcp is a central component of the bacteriophage-like model of T6S-dependent intercellular effector transport,Citation22,36 suggesting that the Hcp-ET1 might be delivered by T6SS via its Hcp/DUF796 domain. To confirm this prediction, we tested whether the Hcp conserved domain (DUF796) and T6SS2 are required for the antibacterial activity of Hcp-ET1 against Δhcp-et1Δeti1 cells. Similar to Δhcp-et1, the DUF796 or T6SS2 mutant shown a significant defect in killing recipient cells (). Furthermore, we confirmed the HNH-DNase toxicity of Hcp-ET1 according to the degradation of plasmid-DNA in vivo. Using the Δhcp-et1Δeti1 with plasmid pUC19 as recipient cells, we found that the donor strains lacking any one of hcp-et1, DUF796 or T6SS2 completely lost their capacity to degrade plasmid-DNA (). Taken together, these observations indicated that Hcp-ET1 is delivered by T6SS2, relying on its Hcp/DUF796 domain, to mediate interbacterial antagonism.
Figure 2. Hcp-ET1 is a T6S-dependent effector that degrades target cell DNA. (A) The toxicity of Hcp-ET1 was dependent on the DUF796 domain and a functional T6SS. The donor and recipient strains were mixed together at a ratio of 3:1, and then incubated for 6 h at 30 °C. The bacterial plaques (NalR recipients) from left to right were increasing serial 10-fold dilutions on NalR eosin-methylene blue agar. The pGEN MCS with the hcp-et1 gene inserted under the control of Pcm promoter was used for complementation (strain CΔhcp-et1). (B) The Hcp-ET1 degraded the plasmid DNA of target cells. The recipient (Δhcp-et1Δeti1 with pUC19) was mixed with the indicated donor strains at a ratio of 3:1, and then collected after a 6 h incubation at 30°C. Equal cell mass was collected from each group, and plasmid DNA was extracted. The linear plasmid DNA from each preparation was analyzed by agarose-gel electrophoresis and Goldview (Vazyme, China) staining. The “untreated STEC004” means only STEC004 cells that were not mixed with recipient cells for the bacterial competition assay. (C) Effects of Hcp proteins on Hcp-ET1 delivery. Donor cells were STEC004 and its derivative hcp and T6SS mutants, as indicated. Experiments were initiated by mixing the donor and recipient bacteria at a CFU ratio of 5:1. The Log10 CFU values of the surviving recipient cells are shown on the y-axis. Error bars indicate standard deviations (SDs) of 3 independent experiments. (D) Fluorescence microscopy for E. coli STEC004 co-incubation. The GFP-labeled donor cells (containing plasmid pGEN-pcm-GFP) were co-incubated with the unlabeled target cells lacking eti1 on an LB plate at a 1:1 ratio. Samples from 0 and 12 h were stained with DAPI to visualize genomic DNA. White arrows indicated anucleate target cells. GFP, green fluorescent protein.
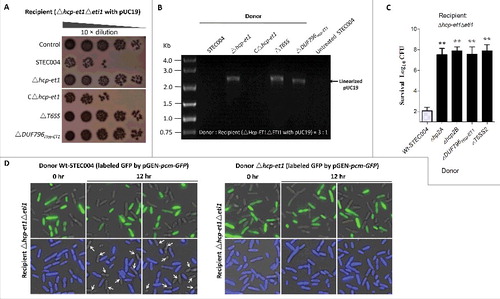
We then tested whether the 2 hcp genes encoded in the T6SS2 cluster were required for the antibacterial activity of Hcp-ET1. The results showed that Wt-STEC004 killed the recipient cells efficiently, whereas the Δhcp2A and Δhcp2B mutants did not (). These observations suggested that Hcp2A and Hcp2B likely formed a heterohexamer with the Hcp/DUF796 domain to control the translocation of the ET1 toxin via the T6SS2 complex. We next asked whether the genomic DNA is degraded in Δhcp-et1Δeti1 recipient cells during co-incubation with hcp-et1+ donors. We labeled STEC004 and Δhcp-et1 donor cells with GFP to differentiate them from unlabeled recipient cells under fluorescence microscopy. Staining with 4′,6-diamidino-2-phenylindole (DAPI) staining revealed that many recipient cells had lost DNA during co-incubation with STEC004 donors (). However, the recipient cells co-incubated with Δhcp-et1 donors retained their DNA (). These findings demonstrated that the Hcp-ET1 toxin acts by degrading target cell DNA.
The extended Hcps with diverse ET-toxins are widespread in Enterobacteriaceae
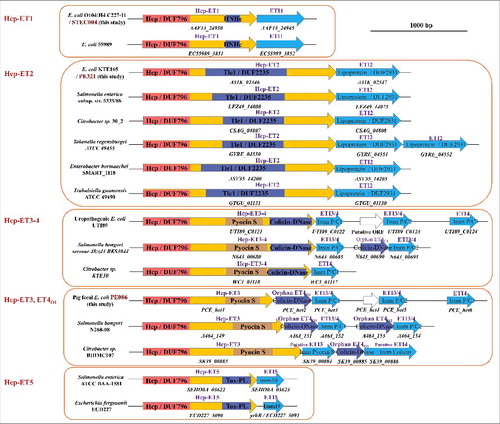
Hcp proteins are distributed widely in Gram-negative bacteria, and have been reported to fuse with C-terminal extensions to exert unknown functions.Citation33 We speculated that a few of these extended Hcps might carry diverse toxins in their C-termini to serve as T6SS effectors. Thus, the template, N-terminal Hcp/DUF796 domain plus the C-terminal toxin domain, was used for domain-architecture retrieval and a large-scale search for T6S-dependent Hcp-ETs in Gram-negative bacteria. Subsequently, 17 bacterial species from the Enterobacteriaceae were found to encode more than 350 Hcp-ETs (Table S1). To verify these search results, we performed a neighborhood analysis, and no distinct conserved or T6SS-related proteins were found. Next, we examined whether their C-terminal domains have putative antibacterial activity. Applying a protein homology detection program, HHpred (43), we found that most Hcp-ETs encoded predicted toxic enzymes targeting cell walls, membranes or nucleic acids, with the exception of a few domains of unknown function (Table S1). In fact, these toxic Hcp-ETs could be classified into 5 clans (designated as Hcp-ET1 to -ET5), namely HNH-DNase, DUF2235, Pyocin S3, Colicin-DNase and Papain-like peptidase, respectively (). Such a wide distribution of the C-terminal domains fused to Hcp/DUF796 highlights its ability to acquire potential functions, acting as a new T6SS effector.
Figure 3. The extended Hcps harbored diverse ET-toxins in multiple bacterial species from Enterobacteriaceae. The genomic organization of the Hcp-ET modules was shown. Red boxes indicate the DUF796 domains. The dark-blue and brown boxes indicate the toxic domains, and the sky-blue boxes indicate the putative immunity genes. All of the predicted Hcp-ET effector-immunity pairs are listed in Table S1, and some representative Hcp-ET pairs from 5 clans are presented here.
Hcp-ET2 functioned as a Tle1 family phospholipase
To functionally validate the predicted Hcp-ET modules, we managed to characterize the Hcp-ET2, which contains a C-terminal DUF2235 domain in enterotoxigenic E. coli (ETEC) strain PE321 (). In a comprehensive analysis of polymorphic toxin systems, DUF2235 was identified as an AB hydrolase1 that acts on lipids.Citation35 The Tle phospholipases are well-defined T6SS effectors that target both prokaryotic and eukaryotic host cells,Citation14,37,38 and family members always contain a DUF2235 domain. In fact, our phylogenetic analyses demonstrated that ET2 toxins exhibited a close evolutionary relationship with the known Tle1 family effectors from E. coli KD1 and Salmonella enterica ATCC BAA-1581 (). Furthermore, the ET2 domains encode a conserved GxSxG sequence, which has been identified as the catalytic motif of Tle1 proteins.Citation14 These results suggested that Hcp-ET2s might be the putative T6SS effectors of the Tle1 family.
Figure 4. Hcp-ET2 functions as Tle1 family phospholipase for interbacterial competition. The sequence of the Hcp-ET2 effector-immunity pair from ETEC PE321 has been submitted to GenBank under the accession no. KX118290. (A) Hcp-ET2 is a T6S-dependent Tle1 phospholipase. The phylogenetic tree of ET2 and Tle1 phospholipase effectors was constructed based on an alignment of catalytic motifs, as described previously.Citation14 ET2 proteins (labeled red) were clustered into the same branch with 2 known Tle1 members of E. coli KD1 and Salmonella enterica ATCC BAA-1581. Sequence logos analysis indicated that ET2 proteins also contained a GxSxG catalytic motif, which has been identified as the conserved catalytic motif of Tle1–4 families.Citation14 (B) Hcp-ET2 provided a competitive growth advantage under cell contact-promoting conditions. Donor and recipient strains were mixed at a 5:1 ratio, incubated for 6 h on solid media and differentiated using blue/white screening. The pGEN MCS with the hcp-et2 gene inserted under the control of Pcm promoter was used for complementation (CΔhcp-et2). The Log10 CFU values of the surviving recipient cells are shown on the y-axis. Asterisks denote significant difference in the outcome of the competition assay for the same donor strain against the indicated recipient strains (**, P < 0.05). Error bars represent the SDs for 3 independent experiments. (C) ETI2 could bind to its cognate Hcp-ET2 directly. Purified Hcp-ET2 and ETI2-His6 proteins were mixed at equimolar ratios, and then purified by Ni2+-affinity chromatography. Crp (cAMP receptor protein) is a stable protein in the cytoplasm of E. coli generally, and was used as a control in this assay. Input samples represent the protein mixtures before chromatography. All fractions were analyzed by SDS-PAGE and Coomassie blue staining.
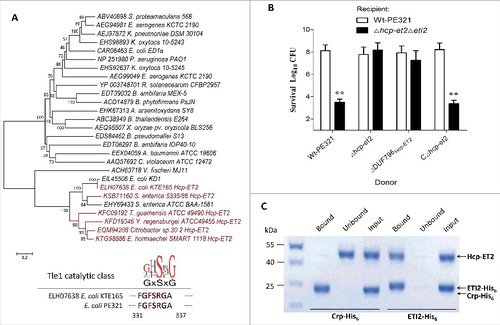
Using a bacterial competition assay to test the antibacterial-effect of the DUF2235 domain, we found that the Δhcp-et2Δeti2 mutant was outcompeted by hcp-et2+ strains during co-incubation on solid medium (). Further analyses demonstrated that the deletion of DUF796Hcp-ET2 attenuated their capacity to inhibit growth of eti2− cells severely (), suggesting that the DUF796 domain is required for the T6SS-dependent delivery of Hcp-ET2. To explore the mechanism of immunity proteins protecting against cognate Hcp-ETs, we speculated that specific binding between ETI2 and DUF2235/Tle1 might prevent ET2-mediated autoinhibition. Therefore, the Ni2+-affinity pull-down experiments were performed to examine the interaction between Hcp-ET2 and ETI2-His6 proteins. The Ni2+-NTA resin retained Hcp-ET2 that was pre-incubated with ET2-His6, but not that pre-incubated with non-cognate Crp-His6 (), which confirmed the toxin/immunity binding between Hcp-ET2 and ETI2.
The domains-fusion of Pyocin S3 and Colicin-DNase toxins, and multiple duplications of immunity proteins in the Hcp-ET3–4 module
Interestingly, the neighborhood analyses for extended Hcp proteins revealed 2 specific Hcp-ET modules, namely Hcp-ET3–4 and Hcp-ET3_ET4O1 (). In the first module, the Pyocin S3 and Colicin-DNase domains are fused to the C-terminus of the extended Hcp to form the 3 domains architecture of Hcp-ET3–4. However, some Hcp-ET3–4s were disrupted by a mutation of the termination-codon to generate Hcp-ET3 and an orphan ET4 (ET4O1), which was designed as the Hcp-ET3_ETO4 module (). The putative immunity protein (pfam01320) was annotated to antagonize both pyocin and colicin toxins in pfam database; therefore, we designated it as ETI3/4. To further analyze its genomic neighborhood in ETEC strain PE086, we found 3 et4-eti3/4 duplications located downstream of hcp-et3 (). However, the second and third orphan et4 open reading frames (ORFs) were destroyed by a series of point mutations, and only the 3 eti3/4 ORFs were retained (). These observations suggested that excessive copies of ET4O1 might lead to self-destruction of Hcp-ET3-possessing cells. To rescue themselves, cells have to invoke self-protection measures to destroy the excessive toxic genes and preserve more immunity genes.
Figure 5. The ET3 domain is involved in interbacterial antagonism. The sequence of the Hcp-ET3_ET4O1 module from ETEC PE086 has been submitted to GenBank under the accession number KX118289. (A) Artemis Comparison Tool (ACT) comparison of the Hcp-ET3–4 module from uropathogenic E. coli UTI89 and the Hcp-ET3_ET4O1 module from ETEC PE086. The PEC_het4–6 is located downstream of PEC_het1–3 in the genome of PE086. The highly conserved genomic region (red blocks denote nucleotide conservation) included these 2 loci from 5’ to 3’, and showed 3 duplications of the et4-eti3/4 fragment. (B, C) Hcp-ET3 provided a competitive growth advantage under cell contact-promoting conditions between the indicated E. coli donor and recipient strains. Experiments were initiated by mixing the donor and recipient bacteria at a CFU ratio of 5:1. The Log10 CFU values of the surviving recipient cells are shown on the y-axis. Asterisks denote significant difference in the outcome of the competition assay of the same donor strain against the indicated recipient strains (**, P < 0.05). Error bars represent the SDs for 3 independent experiments. (D) Growth curves of E. coli cells producing Hcp-ET3 or co-producing Hcp-ET3 and ETI3/4. The cytoplasmic expression of et3 and eti3/4 was induced by L-Arabinose at the indicated time (arrow). A native ribosome binding sites of E. coli located upstream of eti3 gene contributed to achieve its co-expression (blue curve). The experiments were run in triplicate.
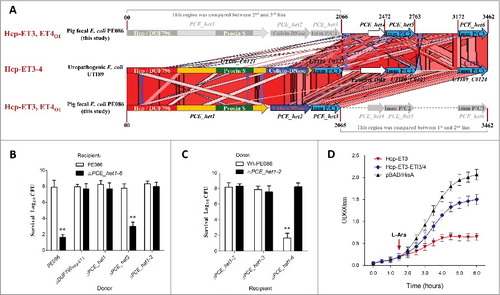
Hcp-ET3 carrying a pyocin S3 domain could inhibit the growth of target cells
To test our observations, we first characterized the Hcp-ET3 with a C-terminal pyocin S3 toxin in ETEC strain PE086. We found that the deletion of hcp-et3 in strain PE086 severely impaired its capacity to kill eti3/4− cells (). To explore whether all the eti3/4 duplications are functional for pyocin S3 antagonism, multiple eti3/4 mutants were used in competition assays. The results showed that only the tripartite deletion of eti3/4 completely abrogated self-protection against pyocin S3 toxin (), indicating that all the eti3/4 copies were functional in vivo. Pyocin S3 is cytotoxic by virtue of its DNase activity,Citation39 suggesting that Hcp-ET3 might utilize cytoplasmic nuclease activity to inhibit target cell growth. Indeed, a cytoplasmic expression assay of hcp-et3 in E. coli LMG194 abruptly inhibited cells growth after L-arabinose induction (). However, the co-expression of hcp-et3 and eti3/4 could partially eliminate the toxic inhibition of Pyocin S3 (), further indicating that the downstream eti3/4 was the cognate immunity gene of hcp-et3.
The orphan ET4 toxin was also involved in bacterial competition
We next asked whether the orphan ET4 is also functional for interbacterial antagonism via growth inhibition by its colicin-DNase toxin. To determine the expression of orphan et4 in strain PE086, a quantitative real-time reverse transcription PCR (qRT-PCR) assay was performed to amplify the potential et4O1 and hcp-et3 transcripts (). The results revealed that orphan et4 was expressed in Wt-PE086 cells, and had a similar transcript level to hcp-et3 (). Additionally, the et4O1 was transcribed at similar levels in the Δhcp-et3 mutant (PCE_het1 deletion) and Wt-PE086 (), suggesting that the promoter driving the orphan region transcription lay upstream of hcp-et3. These results implied that the orphan et4 might be co-transcribed with the upstream hcp-et3 gene.
Figure 6. Orphan ET4 toxin is functional for interbacterial competition. (A) The orphan et4 might be co-transcribed with the hcp-et3 by the same promoter. The data were normalized to the transcription level of the housekeeping gene tus. The relative expression levels represent the mean ± SD for 3 independently isolated RNA samples. (B) The toxic activity of ET4O1 was dependent on the retained DUF796 domain and functional T6SS. The recipient (screened NalR MG1655, a reference strain for T6SS killingCitation40) and indicated donor strains were mixed together at a ratio of 3:1, and then incubated for 6 h at 30°C. The bacterial plaques from left to right were increasing serial 10-fold dilutions on NalR eosin-methylene blue agar. The pGEN MCS with the target gene inserted under the control of the Pcm promoter was used as the complementary plasmid. The presence and absence of Colicin-DNase (ET4) in donor strains were indicated by blue “+” and “−,” respectively. Similarly, red icons indicate the presence and absence of DUF796. (C) Growth competition assays between the indicated donor and recipient strains. Experiments were initiated by mixing the donor and recipient bacteria at a CFU ratio of 5:1. The Log10 CFU values of the survivaing recipient cells are shown on the y-axis. Asterisks denote significant difference in the outcome of the competition assay for the same donor strain against the indicated recipient strains (**, P < 0.05). Error bars represent the SDs for 3 independent experiments.
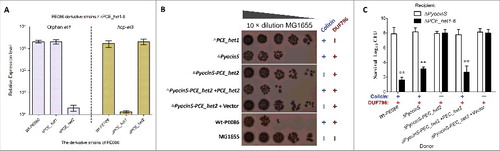
To further validate the predicted function of ET4, a series of et4− or et4+ strains were constructed. In MG1655 (reference strain for T6SS killing Citation40) survival assay, the strain lacking only the pyocin S3 domain significantly inhibited MG1655 cells growth by more than 100-fold compared with the Δhcp-et3 mutants (), suggesting that the retained Hcp/DUF796 plus orphan ET4 could confer an effective antibacterial activity. Indeed, the ΔPCE_het1–6 survival assay also showed similar results when Hcp/DUF796 and orphan ET4 coexisted in the donor cells (). Nevertheless, the antibacterial activity of colicin-possessing strains was completely abolished by the lack of the Hcp/DUF796 domain ( and ), suggesting that Hcp/DUF796 was essential for orphan ET4 delivery. Thus, we speculated that some unknown interaction between orphan ET4 and the retained DUF796 containing C-terminal residual fragment might contribute to the export of the colicin-DNase toxin.
Discussion
Previous research has highlighted that numerous bacterial pathogens take advantage of the T6SS organelles through the spike/tube VgrG/Hcp complex to deliver toxic effectors to their bacterial competitors or eukaryotic hosts.,Citation3,41 Current research further underlined the importance of T6SS in dictating bacterial dynamics in complex communities, such as the microbiota in humans, animals and plants.Citation10,17,42 Nevertheless, the systematic identification and assignment of enzymatic function to T6SS effectors remains a challenge. Recently, an N-terminal sequence marker, the MIX motif, was applied successfully to identify several antibacterial effectors with diverse C-terminal toxin domains,Citation13 suggesting a novel method for effector screening by using specifically conserved domain. Similarly, we found that the extended Hcp proteins with diverse C-terminal toxin domains (Hcp-ETs) were widespread in Enterobacteriaceae. Although “evolved” Hcp proteins with C-terminal extensions have been reported by a previous study, Citation33 they have never been verified as antibacterial effectors of T6SS. Hcp proteins are required for both the assembly of a functional T6SS and the export of its effectors.Citation22,23,26,43 Crystallographic data of Hcp homologs showed that the protein forms hexameric rings, which can stack into a tube.Citation26,44,45 The T6SS effectors are injected into target cells by binding to the inner surface of the Hcp ring-shaped hexamer.Citation23
Given that VgrG proteins have been reported to form a heterotrimer for effectors' delivery,Citation36,40 we speculated that Hcp proteins have the potential to form a heterohexamer with the DUF796 domain of Hcp-ETs, thereby facilitating the delivery of fused toxins. Indeed, our results showed that all 3 Hcps (Hcp2A, Hcp2B and Hcp/DUF796 domain of Hcp-ET1) were required for the delivery of the ET1 toxin, which partially proved our speculation. A previous study has reported that Hcp proteins are also required for the stability of posttranslational effector proteins in cytoplasm.Citation23 Therefore, the defect in antibacterial capacity of 3 hcp mutants (Δhcp2A, Δhcp2B and ΔDUF796/hcp) might be caused by 3 reasons: (i) the failure of secretion of ET1 toxin, (ii) the degradation of posttranslational ET1 toxin in cytoplasm, and (iii) losing the synergy among Hcps and ET1 which is required for the complete antibacterial activity of ET1 toxin. These potential mechanisms need to be further clarified in future work.
To expand our findings in Hcp-ET antibacterial effectors, a systematic search for extended Hcps was performed. According to the enzymatic activity of C-terminal toxins, more than 350 Hcp-ETs from 17 bacterial species of Enterobacteriaceae (Table S1) could be classified into 5 clans, namely HNH-DNase, DUF2235, Pyocin S3, Colicin-DNase and Papain-like peptidase (). The HNH-endonuclease domain has been demonstrated to function in interbacterial antagonism by degrading target cell DNA in the RhsB-CT of Dickeya dadantii 3937,Citation34 which accorded with our results about Hcp-ET1 delivered by T6SS2 in Shiga toxin-producing E. coli. Similarly, the homolog of Pyocin S3 toxin in Hcp-ET3 also inhibited the growth of competitors in the contact-dependent growth inhibition (CDI) system of Dickeya dadantii 3937.Citation39 In fact, the Colicin-DNase and DUF2235 toxin domains are distributed wildly in the MIX family and DUF4123-related effectors,1340 and mediate interbacterial competition via T6SS delivery.
It should be noted that the extended Hcps with toxin domains are so prevalent in the Enterobacteriaceae but not so in other bacteria (Vibrio, Aeromonas, or Pseudomonas) even though related T6SS core islands have been reported. We speculated that the mobile genetic elements related to these Hcp-fusions may be transferred among the bacterial species of Enterobacteriaceae specifically,Citation46 and have a very low probability to be transferred into other bacteria. However, their toxin domains (HNH-DNase, DUF2235, Pyocin S3 and Colicin-DNase) exist widely in diverse bacteria species by being fused to or associating with other T6SS-related proteins (such as PAAR, Mix, Rhs and DUF4123),Citation12,13,32,34 suggesting that these toxins are not the specific weapons for Enterobacteriaceae species to antagonize unrelated bacteria.
Multiple duplication events for immunity genes have been observed in the Tle/Tli superfamily and Tae4/Tai4 pairs of T6SS,Citation11,14 and also reported in rhs loci and CDI systems.Citation47-49 However, it is not fully understood whether each copy is functional in immunity against the cognate toxin. Although we confirmed that only the deletion of all 3 eti3/4 could completely abolish the immunity against the Hcp-ET3 toxin, it remains unclear whether dissimilar transcript regulations occur among duplications under different conditions. Remarkably, the immunity protein, ETI3/4, could neutralize the toxicity of both Pyocin S3 and Colicin-DNase specifically, which has never been reported for effector/immunity pair of T6SS. In fact, both Pyocin S3 and Colicin are cytotoxic by virtue of their DNase activity,Citation34,39 and this similar enzymatic activity might partially explain this phenomenon.
Orphan toxins have been found in rhs loci and CDI systems of multiple bacteria.Citation47,48 The orphan toxins in CDI or Rhs modules are encoded by a fragmented reading frame, and they lack translation initiation and protein export signals.Citation47,49 Nonetheless, the orphan CdiA-CTO1 of E. coli EC93 is functional in contact-dependent growth inhibition when fused to a full-length CdiA protein.Citation49 Indeed, it also has been confirmed that evolved cells undergo recombination between the main rhs gene and the rhs orphan toxin gene fragment, yielding a fusion that enables the production and delivery of the orphan toxin.Citation47 In contrast to the above orphan toxins, the orphan ET4O1 is encoded by a complete ORF, does not need a signal peptide and is transported across the envelope by the T6SS complex in a Sec-independent way. In particular, we identified 3 et4-eti3/4 duplications located downstream of hcp-et3; however, the second and third orphan et4 ORFs are destroyed by a series of point mutations (), only 3 complete eti3/4 ORFs are retained for neutralization of Pyocin S3 and Colicin-DNase toxins. It is possible that excessive copies of ET4O1 might cause serious self-destruction, which partially supports our experimental results that ET4O1 is a functional antibacterial effector.
In conclusion, the formerly well-known T6SS core component, the Hcp protein, was demonstrated to harbor diverse toxin domains in their C-termini, thus presenting a novel encoding pattern of T6SS effectors. The Hcp-ETs greatly increased the growth advantage for interbacterial competition via diverse enzymatic activities, and were bound specifically by cognate immunity proteins to prevent self-intoxication. The systematic identification of these Hcp-ET effectors in multiple species from the Enterobacteriaceae contributed to a better understanding of the complex mechanism of T6SS for interbacterial antagonism.
Materials and method
Bacterial strains and growth conditions
A list of strains and plasmids used in this study are shown in Table S2. The competitive strains were derived from intestinal pathogenic strains STEC004, PE321 or PE086. Cultures were routinely grown in LB medium containing 0.5% sodium chloride at 37°C with aeration, unless otherwise indicated. When necessary, the following antibiotics and chemicals were added to the medium: kanamycin (Kan, 50 µg/ml), ampicillin (Amp, 100 µg/ml), chloramphenicol (Clm, 25 µg/ml), and nalidixic acid (Nal, 50 µg/ml). E. coli strains TOP10 and LMG194 were used for plasmid maintenance and gene expression, respectively. LMG194 with recombinational pBAD/HisA was used for the toxicity assay of ET3, and for purification of ET2, ETI2 and Crp.
DNA manipulations and plasmids construction
All oligonucleotide primers were synthesized by Genewiz Corporation (Suzhou, China) and are listed in Table S3. DNA amplification, ligation and electroporation were performed as described previously, unless otherwise indicated.Citation29 The creation, maintenance and transformation of plasmid constructs followed standard molecular cloning procedures. DNA sequencing was performed by Sunny Biotechnology Corporation (Shanghai, China). All restriction and DNA-modifying enzymes were purchased from Thermo Fisher Scientific and used according to the suppliers' instructions.
Deletion mutants were constructed using the lambda Red recombinase system as described previously.Citation50 For complementation, a Pcm promoter region from pKD3 was inserted pGEN MCS Citation51 between the EcoRI and NdeI restriction sites, and then the target genes were cloned into the new pGEN using NdeI and BamHI restriction sites, independently. The native coding ribosome binding sites were used to achieve the co-expression of effector/immunity pairs in pBAD/HisA.
Bioinformatic identification of Hcp-ET proteins
Protein sequences were retrieved from the National Center for Biotechnology Information (NCBI) database, and the GenBank accession numbers for all Hcp-ET proteins identified in this study are listed in Table S1. The functional prediction was performed using HHpredCitation52 and Phyre2.Citation53,54 The extended Hcp protein with N-terminal Hcp/DUF796 and C-terminal toxin domains was suggested as a potential template to search for T6S-dependent effectors. A bioinformatics screen for this specific domain-architecture was performed using the CDART tool as described previously.Citation55 Using these screened Hcp-ETs, BLASTP analyses were performed against the non-redundant protein database (ftp://ftp.ncbi.nih.gov/blast/db/) to identify their homologs. The phospholipases Tle1 family members were aligned using the ClustalW method,Citation14 and a phylogenetic tree was generated using the MEGA 7.0 Neighbor-joining method with bootstrap analysis of 1,000 replicates. Visual representation of the comparative alignments between the Hcp-ET3/ET4 module of E. coli UTI89 and the ET3_ET4O1 module of E. coli PE086 was performed using the Artemis Comparison Tool (ACT) by nucleotide similarities (TBLASTX). Sequence logos were generated based on the manual alignment of conserved catalytic motifs with the Geneious software.
Interbacterial competition assays
The interbacterial competition experiment was performed as described previously with minor modifications.Citation29 Briefly, fresh cultures of donor and recipient bacteria were adjusted to an OD600nm of 0.5 and mixed at a 5:1 ratio (donor to recipient) after 50 times concentration.Citation5 Then, 10 µl of the mixture was spotted on a 0.2-µm nitrocellulose membrane placed on low-salt LB agar. After 6 h of incubation at 30°C, the mixed inoculum was used to obtain the final CFU of the surviving recipient bacteria by plate counting. The activity of a native β-galactosidase reporter was used to distinguish donor (lacZ+) and recipient (lacZ−) colonies via blue/white screening on plates containing 40 µg/ml X-gal and 1 mM IPTG, as described previously.Citation14 All assays were performed in triplicate. Statistical analyses were performed with a 2-tailed Student's t-test. Error bars represent the standard deviations of 3 independent experiments.
Recipient survival assays for visualization on plates
Briefly, the spontaneous nalidixic acid-resistant (NalR) mutants of the recipient strains were screened by plating 109 CFU onto LB containing 50 µg/ ml Nal and confirmed to retain full growth properties.Citation56 The donor and recipient strains (NalR) were mixed together at a ratio of 3:1 after the cultures were normalized to an OD600nm of 0.5. The mixture was then spotted on the nitrocellulose membrane placed on low-salt LB agar for 6 h incubation at 30°C. The bacterial spots were then resuspended, serially diluted in PBS and visualized on plates (eosin-methylene blue medium) containing 50 µg/ ml Nal to compare the survivals of the recipient cells co-incubated with different donor strains.
Fluorescence microscopy
Recipient and GFP-labeled donor cells were mixed at a 1:1 cell ratio on a 0.2−µm nitrocellulose membrane placed on low-salt LB agar. After 12 h of incubation at 30°C, the mixed inoculum was resuspended by 1×PBS and fixed in 4% (vol/vol) formaldehyde for 12 h, quenched with 125 mM glycine (pH 7.5) for 15 min and washed twice with 1×PBS. Diluted bacteria were plated onto glass slides coated with poly-D-lysine and stained with DAPI supplemented with 0.1% Triton X-100. Images were collected and processed under a confocal laser scanning microscope (CLSM) (LSM 710; Zeiss, Germany). Cells were scored as anucleate if they lacked visible DAPI staining.
Protein purification
All Hcp-ET/ETI-His6 complexes were overproduced and purified under non-denaturing conditions in reaction buffer (20 mM sodium phosphate at pH 7.0, 150 mM NaCl, 10 mM β- ME), as described previously.Citation39 Untagged Hcp-ET was isolated from ETI-His6 by Ni2+-affinity chromatography with denaturing wash buffer (20 mM sodium phosphate at pH 7.0, 6 M guanidine-HCl, 10 mM β- ME). ETI-His6 was then eluted from Ni2+-NTA using denaturing elution buffer (20 mM sodium phosphate at pH 7.0, 6 M guanidine-HCl, 10 mM β-ME; 250 mM imidazole). Purified Hcp-ET and ETI-His6 proteins were refolded by dialysis into reaction buffer and quantified by their absorbance at 280 nm.
Plasmid DNA degradation activity of Hcp-ET1
The in vitro DNA degradation experiments were performed as described previously.Citation34,39 The HNH-DNase activity of Hcp-ET1 was assayed using 100 ng of EcoRI-digested pUC19 DNA as the substrate in the presence and absence of cognate ETI1 or non-cognate Crp. The Hcp-ET1 was mixed with ETI1 or Crp, and incubated for 30 min at room temperature before the assays. Plasmid DNA stability in the recipient strains derived from E. coli STEC004 with pUC19 was assessed. Donor and recipients were cultured in low-salt LB at 30°C overnight. Equivalent cell masses (OD600nm, 0.5) were collected from each culture and co-incubated for 6 h as described above. Equal cell masses were collected from each group, and the plasmid DNA was extracted. The linear plasmid DNA from each preparation was analyzed by 1% agarose-gel electrophoresis and Goldview (Vazyme, China) staining.
The Ni2+-affinity pull-down of Hcp-ET2 and ETI2
Binding interactions between Hcp-ET2 and ETI2 proteins were assayed following previously published procedures.Citation39,57 Briefly, the purified Hcp-ET2 (untagged) was mixed with ETI2-His6 or Crp-His6 at a final concentration of 10 μM in reaction buffer, and incubated for 30 min at room temperature. An aliquot was removed for subsequent SDS-PAGE analysis, and the mixtures were then incubated with Ni2+-NTA for 1.5 h at 4°C. The reactions were centrifuged, and the supernatant containing unbound proteins was collected for SDS-PAGE analysis. The Ni2+-NTA was washed 3 times and loaded onto Poly-Prep column. Bound complexes were eluted with native elution buffer supplemented with 250 mM imidazole for SDS-PAGE analysis.
Growth curves for the ET3 toxicity assay
The vector pBAD/HisA (Invitrogen) was used to express hcp-et3 and for co-expression of hcp-et3 and eti3. A native ribosome binding sites of E. coli, located upstream of the eti3 gene, aided their co-expression. Both vectors added a C-terminal hexahistidine tag to the expressed proteins, allowing for western blot analysis of their expressions. For E. coli growth curves, LMG194 cells harboring the above recombinat plasmids were grown overnight and sub-inoculated to a starting OD600nm of 0.05 in LB at 37°C with shaking for 1.5 h. The target genes then were induced with 0.25% L-arabinose. Cell growth was tracked by measuring the OD600nm every 30 min. The results represented the mean plus standard deviations (error bars) of 3 independent experiments.
Measurement of gene expression by qRT-PCR
The bacteria were cultured overnight in low-salt LB with shaking in 30°C. Total RNA was then extrated using an E.Z.N.A. bacterial RNA isolation kit (Omega), followed by treatment with RNase-free DNase I (Roche) to remove residual genomic DNA. The cDNA was synthesized using a PrimeScript RT reagent kit (Takara) instantly. Quantitative real-time PCR was conducted on an ABI StepOne RT-PCR detection system using SYBR green supermix (Takara) and gene-specific primers (Table S3). The data were normalized to the transcript levels of the housekeeping gene tus.Citation58 The relative fold change was calculated by using the threshold cycle (2−△△CT) methodCitation59 The reported values represented the mean plus standard deviations (error bars) of 3 independent RNA extractions.
Statistical analysis
The data were analyzed using SPSS version 17.0 (SPSS Inc.). Two-way ANOVA was performed for the qRT-PCR results, and a 2-tailed Student's t-test was used for the interbacterial competition assays. P values of less than 0.05 were defined as significant and indicated with a double asterisk (“**”).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KVIR_S_1279374.zip
Download Zip (55.9 KB)Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 31372455) and the project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
References
- Hayes CS, Aoki SK, Low DA. Bacterial contact-dependent delivery systems. Ann Rev Genet 2010; 44:71-90; PMID:21047256; http://dx.doi.org/10.1146/annurev.genet.42.110807.091449
- Konovalova A, Sogaard-Andersen L. Close encounters: contact-dependent interactions in bacteria. Mol Microbiol 2011; 81:297-301; PMID:21651624; http://dx.doi.org/10.1111/j.1365-2958.2011.07711.x
- Ho BT, Dong TG, Mekalanos JJ. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 2014; 15:9-21; http://dx.doi.org/10.1016/j.chom.2013.11.008
- LeRoux M, Kirkpatrick RL, Montauti EI, Tran BQ, Peterson SB, Harding BN, Whitney JC, Russell AB, Traxler B, Goo YA, et al. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. eLife 2015; 4:e05701; PMID:25643398; http://dx.doi.org/10.7554/eLife.05701
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 2011; 475:343-7; PMID:21776080; http://dx.doi.org/10.1038/nature10244
- Schwarz S, Singh P, Robertson JD, LeRoux M, Skerrett SJ, Goodlett DR, West TE, Mougous JD. VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect Immun 2014; 82:1445-52; PMID:24452686; http://dx.doi.org/10.1128/IAI.01368-13
- Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, Durand E, Journet L, Cascales E, Monack DM. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A 2016; 113:E5044-51; PMID:27503894; http://dx.doi.org/10.1073/pnas.1608858113
- Hecht AL, Casterline BW, Earley ZM, Goo YA, Goodlett DR, Bubeck Wardenburg J. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep 2016; 17:1281-91; PMID:27432285; http://dx.doi.org/10.15252/embr.201642282
- Wenren LM, Sullivan NL, Cardarelli L, Septer AN, Gibbs KA. Two independent pathways for self-recognition in Proteus mirabilis are linked by type VI-dependent export. mBio 2013; 4; PMID:23882014; http://dx.doi.org/10.1128/mBio.00374-13
- Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, Provenzano D, Pukatzki S. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nate Commun 2014; 5:3549; PMID:24686479; http://dx.doi.org/10.1038/ncomms4549
- Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, et al. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 2012; 11:538-49; http://dx.doi.org/10.1016/j.chom.2012.04.007
- Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci U S A 2013; 110:2623-8; PMID:23362380; http://dx.doi.org/10.1073/pnas.1222783110
- Salomon D, Kinch LN, Trudgian DC, Guo X, Klimko JA, Grishin NV, Mirzaei H, Orth K. Marker for type VI secretion system effectors. Proc Natl Acad Sci U S A 2014; 111:9271-6; PMID:24927539; http://dx.doi.org/10.1073/pnas.1406110111
- Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 2013; 496:508-12; PMID:23552891; http://dx.doi.org/10.1038/nature12074
- Whitney JC, Chou S, Russell AB, Biboy J, Gardiner TE, Ferrin MA, Brittnacher M, Vollmer W, Mougous JD. Identification, structure, and function of a novel type VI secretion peptidoglycan glycoside hydrolase effector-immunity pair. J Biol Chem 2013; 288:26616-24; PMID:23878199; http://dx.doi.org/10.1074/jbc.M113.488320
- Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S. Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J Biol Chem 2013; 288:7618-25; PMID:23341465; http://dx.doi.org/10.1074/jbc.M112.436725
- Ma LS, Hachani A, Lin JS, Filloux A, Lai EM. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 2014; 16:94-104; http://dx.doi.org/10.1016/j.chom.2014.06.002
- Whitney JC, Quentin D, Sawai S, LeRoux M, Harding BN, Ledvina HE, Tran BQ, Robinson H, Goo YA, Goodlett DR, et al. An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell 2015; 163:607-19; PMID:26456113; http://dx.doi.org/10.1016/j.cell.2015.09.027
- Miyata ST, Unterweger D, Rudko SP, Pukatzki S. Dual expression profile of type VI secretion system immunity genes protects pandemic Vibrio cholerae. PLoS Pathog 2013; 9:e1003752; PMID:24348240; http://dx.doi.org/10.1371/journal.ppat.1003752
- Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner's guide. Curr Opin Microbiol 2008; 11:3-8; PMID:18289922; http://dx.doi.org/10.1016/j.mib.2008.01.006
- Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 2015; 13:343-59; PMID:25978706; http://dx.doi.org/10.1038/nrmicro3456
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 2006; 312:1526-30; PMID:16763151; http://dx.doi.org/10.1126/science.1128393
- Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, Catalano CE, Gonen T, Mougous JD. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell 2013; 51:584-93; PMID:23954347; http://dx.doi.org/10.1016/j.molcel.2013.07.025
- Osipiuk J, Xu X, Cui H, Savchenko A, Edwards A, Joachimiak A. Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa. J Struct Funct Genomics 2011; 12:21-6; PMID:21476004; http://dx.doi.org/10.1007/s10969-011-9107-1
- Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci U S A 2009; 106:4160-5; PMID:19251647; http://dx.doi.org/10.1073/pnas.0900044106
- Brunet YR, Henin J, Celia H, Cascales E. Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep 2014; 15:315-21; PMID:24488256; http://dx.doi.org/10.1002/embr.201337936
- Journet L, Cascales E. The type VI secretion system in escherichia coli and related species. EcoSal Plus 2016; 7:1-20; PMID:27223818; http://dx.doi.org/10.1128/ecosalplus.ESP-0009-2015
- Ma J, Sun M, Bao Y, Pan Z, Zhang W, Lu C, Yao H. Genetic diversity and features analysis of type VI secretion systems loci in avian pathogenic Escherichia coli by wide genomic scanning. Infect Genet Evol 2013; 20:454-64; PMID:24120694; http://dx.doi.org/10.1016/j.meegid.2013.09.031
- Ma J, Bao Y, Sun M, Dong W, Pan Z, Zhang W, Lu C, Yao H. Two functional type VI secretion systems in avian pathogenic Escherichia coli are involved in different pathogenic pathways. Infect Immun 2014; 82:3867-79; PMID:24980972; http://dx.doi.org/10.1128/IAI.01769-14
- Barret M, Egan F, Fargier E, Morrissey JP, O'Gara F. Genomic analysis of the type VI secretion systems in Pseudomonas spp.: novel clusters and putative effectors uncovered. Microbiology 2011; 157:1726-39; PMID:21474537; http://dx.doi.org/10.1099/mic.0.048645-0
- De Maayer P, Venter SN, Kamber T, Duffy B, Coutinho TA, Smits TH. Comparative genomics of the Type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins. BMC Genomics 2011; 12:576; PMID:22115407; http://dx.doi.org/10.1186/1471-2164-12-576
- Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 2013; 500:350-3; PMID:23925114; http://dx.doi.org/10.1038/nature12453
- Blondel CJ, Jimenez JC, Contreras I, Santiviago CA. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics 2009; 10:354; PMID:19653904; http://dx.doi.org/10.1186/1471-2164-10-354
- Koskiniemi S, Lamoureux JG, Nikolakakis KC, t'Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 2013; 110:7032-7; PMID:23572593; http://dx.doi.org/10.1073/pnas.1300627110
- Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 2012; 7:18; PMID:22731697; http://dx.doi.org/10.1186/1745-6150-7-18
- Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A 2007; 104:15508-13; PMID:17873062; http://dx.doi.org/10.1073/pnas.0706532104
- Jiang F, Wang X, Wang B, Chen L, Zhao Z, Waterfield NR, Yang G, Jin Q. The pseudomonas aeruginosa Type VI secretion PGAP1-like effector induces host autophagy by activating endoplasmic reticulum stress. Cell reports 2016; 16:1502-9; PMID:27477276; http://dx.doi.org/10.1016/j.celrep.2016.07.012
- Jiang F, Waterfield NR, Yang J, Yang G, Jin Q. A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe 2014; 15:600-10; http://dx.doi.org/10.1016/j.chom.2014.04.010
- Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 2010; 468:439-42; PMID:21085179; http://dx.doi.org/10.1038/nature09490
- Liang X, Moore R, Wilton M, Wong MJ, Lam L, Dong TG. Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc Natl Acad Sci U S A 2015; 112:9106-11; PMID:26150500; http://dx.doi.org/10.1073/pnas.1505317112
- Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 2014; 12:137-48; PMID:24384601; http://dx.doi.org/10.1038/nrmicro3185
- Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 2014; 16:227-36; http://dx.doi.org/10.1016/j.chom.2014.07.007
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 2010; 7:25-37; http://dx.doi.org/10.1016/j.chom.2009.12.007
- Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci U S A 2008; 105:3733-8; PMID:18310321; http://dx.doi.org/10.1073/pnas.0712247105
- Douzi B, Spinelli S, Blangy S, Roussel A, Durand E, Brunet YR, Cascales E, Cambillau C. Crystal structure and self-interaction of the type VI secretion tail-tube protein from enteroaggregative Escherichia coli. PloS one 2014; 9:e86918; PMID:24551044; http://dx.doi.org/10.1371/journal.pone.0086918
- Frohlich KS, Papenfort K. Interplay of regulatory RNAs and mobile genetic elements in enteric pathogens. Mol Microbiol 2016; 101:701-13; PMID:27232692
- Koskiniemi S, Garza-Sanchez F, Sandegren L, Webb JS, Braaten BA, Poole SJ, Andersson DI, Hayes CS, Low DA. Selection of orphan Rhs toxin expression in evolved Salmonella enterica serovar Typhimurium. PLoS Genet 2014; 10:e1004255; PMID:24675981
- Hayes CS, Koskiniemi S, Ruhe ZC, Poole SJ, Low DA. Mechanisms and biological roles of contact-dependent growth inhibition systems. Cold Spring Harbor perspectives in medicine 2014; a010025; PMID:24492845; http://dx.doi.org/10.1101/cshperspect.a010025
- Poole SJ, Diner EJ, Aoki SK, Braaten BA, t'Kint de Roodenbeke C, Low DA, Hayes CS. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet 2011; 7:e1002217; PMID:21829394
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 2000; 97:6640-5; PMID:10829079
- Alteri CJ, Smith SN, Mobley HL. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS pathog 2009; 5:e1000448; PMID:19478872
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 2005; 33:W244-8; PMID:15980461
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 2015; 10:845-58; PMID:25950237
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 2009; 4:363-71; PMID:19247286
- Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome Res 2002; 12:1619-23; PMID:12368255
- Li G, Laturnus C, Ewers C, Wieler LH. Identification of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect Immun 2005; 73:2818-27; PMID:15845486
- Diner EJ, Beck CM, Webb JS, Low DA, Hayes CS. Identification of a target cell permissive factor required for contact-dependent growth inhibition (CDI). Genes Dev 2012; 26:515-25
- Skyberg JA, Johnson TJ, Nolan LK. Mutational and transcriptional analyses of an avian pathogenic Escherichia coli ColV plasmid. BMC Microbiol 2008; 8:24; PMID:18230176
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001; 25:402-8; PMID:11846609
- Zhou Y, Tao J, Yu H, Ni J, Zeng L, Teng Q, Kim KS, Zhao GP, Guo X, Yao Y. Hcp family proteins secreted via the type VI secretion system coordinately regulate Escherichia coli K1 interaction with human brain microvascular endothelial cells. Infect Immun 2012; 80:1243-51; PMID:22184413
