To the Editor
Nontyphoidal Salmonella enterica (NTS) is a leading cause of bacterial foodborne disease throughout the world, and it is estimated to cause 93.8 million cases of gastroenteritis and 155,000 deaths each year.Citation1 Although NTS generally causes self-limiting gastroenteritis, severe invasive infections can occur, requiring appropriate antimicrobial treatment.Citation2 In this regard, the emergence of strains resistant to third-generation cephalosporins raises particular concern, since they are frequently chosen for the treatment of salmonellosis, mainly for invasive NTS infections.Citation3 This resistance profile has been mostly attributed to the large dissemination of plasmids carrying genes encoding extended-spectrum β-lactamases (ESBLs) and plasmid-mediated AmpC β-lactamases (pAmpCs).Citation4
Besides antibiotic resistance, virulence plays an important role in NTS infections, and the association of ESBL/pAmpC and a virulent profile in NTS strains represents a serious public health issue, once it makes these strains more harmful, contributing to the increase of morbidity and mortality rates.Citation5 Moreover, it causes important economic impact due to medical costs, and losses on productivity and marketing of foods of animal origin.Citation6,7
As the most populous country in South America, with more than 200 million inhabitants, Brazil has faced problems in controlling foodborne diseases. In this regard, between 2000 and 2015, according to data available by the Brazilian Ministry of Health, Salmonella was considered the major causative agent of foodborne outbreaks (http://www.saude.gov.br/svs). Because Salmonella is typically found in poultry, this type of meat has been an important vehicle of foodborne diseases. In fact, in Brazil, chicken meat is widely consumed, representing a potential risk to public health. So, the aim of the present study was to determine the genetic relatedness, the plasmid profile, and the virulence potential of ESBL/pAmpC-producing NTS strains from different serovars, sources and geographic locations in Brazil.
During a Brazilian multicentric antimicrobial resistance surveillance study, broad-spectrum cephalosporin resistance was investigated in 283 nontyphoidal Salmonella (NTS) isolates recovered from human (n = 4), farm animals (n = 2), food samples (n = 225) and other sources (n = 52), collected from 2008 to 2015. The isolates were identified by conventional biochemical methodology and, further, serotyped according to the Kauffmann-Le Minor scheme.Citation8 Antimicrobial drug susceptibility was evaluated by disc diffusion method, according to the guidelines of the Clinical Laboratory Standards Institute.Citation9
In this regard, from the 283 NTS investigated, ten isolates belonging to serovars Minnesota (n = 3, chicken meat), Typhimurium (n = 1, chicken meat), Infantis (n = 1, chicken meat), Heidelberg (n = 1, turkey meat), Agona (n = 1, turkey meat), Schwarzengrund (n = 1, drag swab), Muenchen (n = 1, human cerebrospinal fluid), and S. enterica subsp. enterica 4,5,12:i:- (n = 1, swine feces), exhibited resistance to cephalosporins, and were evaluated for the presence of ESBL, by the double-disc synergy test, and pAmpC, by resistance to cefoxitin.Citation9 Further, individual polymerase chain reactions (PCR) were carried out to evaluate the presence of ESBLCitation10 and pAmpCCitation11 genes, and then, blaCTX-M-8 (n = 5), blaCTX-M-2 (n = 4), and blaCMY-2 (n = 1) genes were confirmed by sequencing, among different NTS serovars ().
Table 1. Epidemiologic and molecular characterization of ESBL/pAmpC-producing non-typhoidal Salmonella strains recovered from livestock, food and human infection, Brazil (2008–2015).
Multilocus sequence typing (MLST) was performed according to the Salmonella enterica MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Senterica), and seven sequence types (STs) () were identified, including a new ST. Most STs match with their respective serovars, with the exception of ST19, which was shared by S. Typhimurium, S. Heidelberg, and Salmonella enterica subsp enterica 4,5,12:i:-, suggesting that the expression of cell-surface antigens can be altered by horizontal transfer of genes and genetic recombination of chromosome, without interfering in the seven housekeeping genes used in the MLST scheme.Citation12 The novel ST (ST3088) was detected in a S. Minnesota strain recovered from a chicken meat sample. According to data available in the Salmonella enterica MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Senterica/GetTableInfo_html), most STs reported in this study correspond to the more common ones to their respective serovars, with a wide host range distribution around the world. However, with exception of S. Infantis ST32, the other six STs (i.e., ST13, ST19, ST96, ST112, ST548 and ST3088) have never before been reported in Brazil.Citation13 Moreover, the human strain identified in this study corresponds to the first description (to our knowledge) of a CTX-M-2-producing S. Muenchen ST112 isolated from a cerebrospinal fluid sample of an infected newborn ().
Most plasmids carrying ESBL/pAmpC genes were successfully transferred by conjugation or transformation to E. coli J53, E. coli HB101 or E. coli TOP10 recipient strains. The size of plasmids carrying CTX-M- or CMY-2-type genes was estimated by S1 nuclease digestion following pulsed-field gel electrophoresis (S1-PFGE),Citation10 ranging from ~97 to 291-kb. Moreover, PCR-based replicon typing (PBRT)Citation14 revealed that most plasmids harboring blaESBL and blaCMY-2 genes belonged to the IncI1 incompatibility group. IncI1 plasmids were submitted to plasmid multilocus sequence typing (pMLST) (http://pubmlst.org/plasmid/), most of them being assigned to ST113, with the exception of the blaCMY-2-carrying plasmid identified in the S. Minnesota strain from chicken meat, which belonged to ST12 ().
IncI1/ST113 plasmids harbouring blaCTX-M-8 gene have been previously reported in Enterobacteriaceae isolated from humans and food, in Germany.Citation15 In Brazil, IncI1/ST113 plasmids have been associated with the transfer of blaCTX-M-8 gene in E. coli strains isolated from poultry.Citation16 On the other hand, IncI1/ST12 plasmids have been disseminated worldwide, being linked to the spread of blaCMY–type pAmpC genes among members of the Enterobacteriaceae family from different sources and clinical contexts.Citation17,18 Interestingly, in a recent study conducted in the Netherlands, IncI1/ST12 plasmids encoding blaCMY-2 were found in S. Heidelberg strains isolated from poultry meat imported from Brazil.Citation19 So, our results support that IncI1/ST113 and IncI1/ST12 plasmids might be key vectors responsible for the dissemination of ESBL and pAmpC genes among NTS isolates through the Brazilian food production chain, which is worrisome, since Brazil is the third largest producer of chicken meat (only after the United States and China) and is the largest exporter of this product.Citation20
Investigation of the virulence behavior of extended-spectrum cephalosporin-resistant NTS was initially performed using PCR for the detection of the chromosomal virulence genes aceK,Citation21 h-1i,Citation21 invA,Citation22 slyA,Citation23 and sopB,Citation21 and the plasmidial virulence gene spvC,Citation22 revealing the presence of aceK, invA, slyA, and sopB genes in all strains. In addition, S. Typhimurium and S. enterica subsp enterica 4,5,12:i:- strains, isolated from poultry meat and commercial swine, carried the h-1i gene (). The high similarity of virulence profile among different serovars of NTS from unrelated sources and geographic area, denote that important virulence genes might be highly conserved in NTS strains.Citation24
In order to overcome the limitation related to the few virulence genes screened by PCR and the lack of their expression, which is necessary to demonstrate the virulence potential of strains, in vivo experiments were carried out with the Galleria mellonella infection model,Citation25,26 using ESBL-negative S. Typhimurium ATCC® 14028™, E. coli ATCC® 25922™, and S. Typhimurium IAL 1431, as comparative strains. In this regard, S. Typhimurium IAL 1431, a drug-susceptible strain belonging to the culture collection of the National Reference Center Instituto Adolfo Lutz (São Paulo, Brazil), was negative for aceK, invA and spvC virulence genes. G. mellonella larvae, of nearly 250 to 350 mg, were inoculated with 105 CFU of each strain and survival analysis was evaluated every hour, during 48 hours. For each strain, groups of G. mellonella containing five larvae were evaluated in two separate experiments.
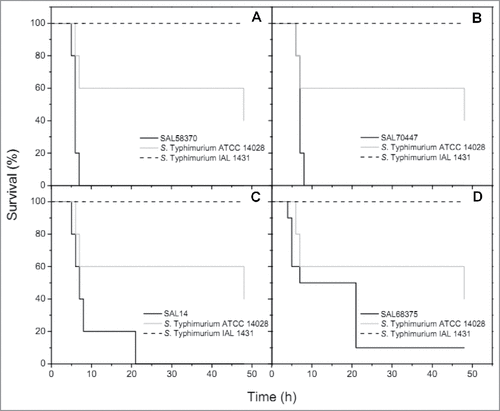
Groups infected with S. Typhimurium SAL58370 and S. enterica subsp enterica 4,5,12:i:- SAL219 achieved 100% of mortality at 7 h post-infection, while a group infected with the S. Minnesota strain SAL70447, killed all the larvae at 8 h post-infection. For the G. mellonella groups infected with the other two S. Minnesota strains (SAL65505 and SAL77088) or with S. Schwarzengrund SAL785, S. Agona SAL769, S. Muenchen SAL14, or S. Infantis SAL64, 100% of mortality was achieved at 21 h post-infection. On the other hand, in the group infected with S. Heidelberg SAL68375, 90% of mortality was observed at 7 h post-infection (). No mortality was observed in larvae infected with E. coli ATCC 25922 and S. Typhimurium IAL 1431. Otherwise, S. Typhimurium ATCC 14028 killed 40% and 60% of the larvae at 7 h and 48 h post-infection, respectively. The summarizes the in vivo evaluation of the virulence of four representative NTS strains in comparison to S. Typhimurium ATCC 14028 and S. Typhimurium IAL 1431. Survival curves were plotted using the Kaplan-Meier method, and data were analyzed by the log rank test, with P<0.05 indicating statistical significance (Graph Pad Software, San Diego, CA, USA). In this study, the low survival rates of G. mellonella suggest a high virulent background of NTS producing CTX-M and CMY-2 β-lactamases. In fact, mortality (%) of G. mellonella larvae infected with these strains was higher than in the ones infected with S. Typhimurium ATCC 14028, which is known to be highly virulent.Citation27,28 Despite G. mellonella not being a natural host of S. enterica, it has been successfully utilized as an infection model to assess the pathogenic potential of NTS strains, since it displays many similarities with vertebrates, such as the innate immune system.Citation24,28,29 Therefore, responses to bacterial infections observed in this model could closely mimics responses displayed by mammalian models.Citation30-32 However, since G. mellonella infection model is not yet an established approach for the study of NTS, it not discards the need of using other models.
Figure 1. Kaplan-Meier survival curves of Galleria mellonella infected with 105 CFU/larva of NTS strains. Cephalosporin-sensitive (ESBL-negative) S. Typhimurium ATCC 14028 and S. Typhimurium IAL 1431 were utilized as control strains. (A) Strain SAL58370, which had 100% mortality at 7 h post-infection. (B) Strain SAL70447, which had 100% mortality at 8 h post-infection. (C) Strain SAL14, which had 100% mortality at 21 h post-infection. (D) Strain SAL68375, which had 90% mortality at 7 h post-infection. Injection with the wild-type strains resulted in significantly higher mortality rate compared to injection with the control strains (P<0.05, log rank test).
In summary, this study reports the emergence of virulent ESBL/pAmpC-producing NTS strains in food, food-producing animals and human over a 6-year period, in Brazil. The identification of IncI1/ST113 and IncI1/ST12 plasmids highlights the important role of these vectors in the spreading of ESBL and pAmpC genes among NTS belonging to different clinically significant serovars. Furthermore, the association of an extended-spectrum cephalosporin-resistant profile with a high virulence background deserves special attention, since NTS are a leading cause of food-borne zoonoses, constituting a major public health concern worldwide. So, meticulous investigation of strains of this sort is necessary to prevent their dissemination.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Cefar Diagnóstica Ltda. (São Paulo, Brazil) for kindly supplying antibiotic discs for susceptibility testing, E.M.F. Reis (FIOCRUZ) and M.R.T. Casas (Instituto Adolfo Lutz) for assistance with serotyping, and Dr. Caetano P. Sabino for guidance and help with statistical analysis.
Funding
This work was supported by research grants from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, process number: 2016/08593-9), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). N.L. is a research grant fellow of CNPq.
References
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin Infect Dis 2010; 15(50):882-9; http://doi.org/10.1086/650733
- Mandomando I, Bassat Q, Sigaúque B, Massora S, Quintó L, Ácacio S, Nhampossa T, Vubil D, Garrine M, Macete E, et al. Invasive Salmonella infections among children from Rural Mozambique, 2001–2014. Clin Infect Dis 2015; 61(Suppl 4):S339-45; PMID:26449950; http://doi.org/10.1093/cid/civ712
- Burke L, Hopkins KL, Meunier D, De Pinna E, Fitzgerald-Hughes D, Humphreys H, Woodford N. Resistance to third-generation cephalosporins in human non-typhoidal Salmonella enterica isolates from England and Wales, 2010–12. J Antimicrob Chemother 2014; 69(4):977-81; PMID:24288030; http://doi.org/10.1093/jac/dkt469
- Smith H, Bossers A, Harders F, Wu G, Woodford N, Schwarz S, Guerra B, Rodríguez I, van Essen-Zandbergen A, Brouwer MSM, et al. Characterization of epidemic IncI1-Iγ plasmids harbouring Amber class A and C genes in Escherichia coli and Salmonella enterica from animals and humans. Antimicrob Agents Chemother 2015; 59(9):5357-65; PMID:26100710; http://doi.org/10.1128/AAC.05006-14
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Dopfer D, Fazil A, Fischer-Walker CL, Hald T, et al. World health organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 2015; 12(12):e1001921; PMID:26633831; http://doi.org/10.1371/journal.pmed.1001921
- Scharff RL, Besser J, Sharp DJ, Jones TF, Peter G-S, Hedberg CW. An economic evaluation of PulseNet: a network for foodborne disease surveillance. Am J Prev Med 2016; 50(5 Suppl 1):S66-73; PMID:26993535; http://doi.org/10.1016/j.amepre.2015.09.018
- Antunes P, Mourão J, Campos J, Peixe L. Salmonellosis: The role of poultry meat. Clin Microbiol Infect 2016; 22(2):110-21; PMID:26708671; http://doi.org/10.1016/j.cmi.2015.12.004
- Grimont P, Weill F-X. Antigenic formulae of the Salmonella servovars. WHO Collab Cent Ref Res Salmonella 2007; 1-167. Available from: http://www.pasteur.fr/ip/portal/action/WebdriveActionEvent/oid/01s-000036-089/npapers2://publication/uuid/CA3447A0-61BF-4D62-9181-C9BA78AF0312.
- Clinical and Laboratory Standard Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. Wayne, PA: CLSI; 2015. CLSI document M100-S25.
- Dropa M, Balsalobre LC, Lincopan N, Matté GR, Matté MH. Complex class 1 integrons harboring CTX-M-2-encoding genes in clinical Enterobacteriaceae from a hospital in Brazil. J Infect Dev Ctries 2015; 9(8):890-7; PMID:26322883; http://doi.org/10.3855/jidc.6241
- Poppe C, Martin LC, Gyles CL, Reid-Smith R, Boerlin P, McEwen SA, Prescott JF, Forward KR. Acquisition of resistance to extended-spectrum cephalosporins by Salmonella enterica subsp. enterica serovar Newport and Escherichia coli in the turkey poult intestinal tract. Appl Environ Microbiol 2005; 71(3):1184-92; PMID:15746317; http://doi.org/10.1128/AEM.71.3.1184-1192.2005
- Liu W bing, Liu B, Zhu X na, Yu S jing, Shi X ming. Diversity of Salmonella isolates using serotyping and multilocus sequence typing. Food Microbiol 2011; 28(6):1182-9; PMID:21645818; http://doi.org/10.1016/j.fm.2011.04.001
- Almeida F, Pitondo-Silva A, Oliveira MA, Falcão JP. Molecular epidemiology and virulence markers of Salmonella Infantis isolated over 25 years in São Paulo State, Brazil. Infect Genet Evol 2013; 19:145-51; PMID:23860124; http://doi.org/10.1016/j.meegid.2013.07.004
- Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 2005; 63(3):219-28; PMID:15935499; http://doi.org/10.1016/j.mimet.2005.03.018
- Eller C, Leistner R, Guerra B, Fischer J, Wendt C, Rabsch W, Werner G, Pfeifer Y. Emergence of extended-spectrum beta-lactamase (ESBL) CTX-M-8 in Germany. J. Antimicrob. Chemother. 2014; 69:562-4; PMID:24072171; http://doi.org/10.1093/jac/dkt387
- Ferreira JC, Penha Filho RAC, Andrade LN, Berchieri AJ, Darini ALC. IncI1/ST113 and IncI1/ST114 conjugative plasmids carrying blaCTX-M-8 in Escherichia coli isolated from poultry in Brazil. Diagn Microbiol Infect Dis 2014; 80(4):304-6; PMID:25284375; http://doi.org/10.1016/j.diagmicrobio.2014.09.012
- Hansen KH, Bortolaia V, Nielsen CA, Nielsen JB, Schonning K, Agerso Y, Guardabassi L. Host-specific patterns of genetic diversity among IncI1-Iγ and IncK plasmids encoding CMY-2 beta-lactamase in Escherichia coli isolates from humans, poultry meat, poultry, and dogs in Denmark. Appl Environ Microbiol 2016; 82(15):4705-14; PMID:27235431; http://doi.org/10.1128/AEM.00495-16
- Alonso N, Miró E, Pascual V, Rivera A, Simó M, Garcia MC, Xercavins M, Morera MA, Espejo E, Gurguí M, et al. Molecular characterisation of acquired and overproduced chromosomal blaAmpC in Escherichia coli clinical isolates. Int J Antimicrob Agents 2016; 47(1):62-8; PMID:26607336; http://doi.org/10.1016/j.ijantimicag.2015.10.007
- Liakopoulos A, Geurts Y, Dierikx CM, Brouwer MSM, Kant A, Wit B, Heymans R, van Pelt W, Mevius DJ. Extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg strains, the Netherlands. Emerg Infect Dis 2016; 22(7):1257-61; PMID:27314180; http://doi.org/10.3201/eid2207.151377
- United States International Trade Commission. Brazil: Competitive Factors in Brazil Affecting U.S. and Brazilian Agricultural Sales in Selected Third Country Markets. Publication 4310. Washington, DC; 2012. Available online: https://www.usitc.gov/publications/332/pub4310.pdf
- Nayak R, Stewart T, Wang RF, Lin J, Cerniglia CE, Kenney PB. Genetic diversity and virulence gene determinants of antibiotic-resistant Salmonella isolated from preharvest turkey production sources. Int J Food Microbiol 2004; 91(1):51-62; PMID:14967560; http://doi.org/10.1016/S0168-1605(03)00330-1
- Chiu CH, Ou JT. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J Clin Microbiol 1996; 34(10):2619-22; PMID:8880536
- Soto SM, Martínez N, Guerra B, González-Hevia M a, Mendoza MC. Usefulness of genetic typing methods to trace epidemiologically Salmonella serotype Ohio. Epidemiol Infect 2000; 125(3):481-9; PMID:11218198; http://doi.org/10.1017/S0950268800004921
- Figueiredo R, Card R, Nunes C, Abuoun M, Bagnall MC, Nunez J, Mendonça N, Anjum MF, Da Silva GJ. Virulence characterization of Salmonella enterica by a new microarray: Detection and evaluation of the cytolethal distending toxin gene activity in the unusual host S. Typhimurium. PLoS One 2015; 10(8):e0135010; PMID:26244504; http://doi.org/10.1371/journal.pone.0135010
- Insua JL, Llobet E, Moranta D, Pérez-Gutiérrez C, Tomás A, Garmendia J, Bengoechea JA. Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth Galleria mellonella. Infect Immun 2013; 81(10):3552-65; PMID:23836821; http://doi.org/10.1128/IAI.00391-13
- Junqueira JC. Galleria mellonella as a model host for human pathogens: recent studies and new perspectives. Virulence 2012; 3(6):474-6; PMID:23211681; http://doi.org/10.4161/viru.22493
- Swearingen MC, Porwollik S, Desai PT, McClelland M, Ahmer BMM. Virulence of 32 Salmonella strains in mice. PLoS One 2012; 7(4):e36043; PMID:22558320; http://doi.org/10.1371/journal.pone.0036043
- Bender JK, Wille T, Blank K, Lange A, Gerlach RG. LPS structure and PhoQ activity are important for Salmonella Typhimurium virulence in the Galleria mellonella infection model [corrected]. PLoS One 2013; 8(8):e73287; PMID:23951347; http://doi.org/10.1371/journal.pone.0073287
- Viegas SC, Mil-Homens D, Fialho AM, Arraiano CM. The virulence of Salmonella enterica serovar Typhimurium in the insect model Galleria mellonella is impaired by mutations in RNase E and RNase III. Appl Environ Microbiol 2013; 79(19):6124-33; PMID:23913419; http://doi.org/10.1128/AEM.02044-13
- Jander G, Rahme LG, Ausubel FM. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol 2000; 182(13):3843-5; PMID:10851003; http://doi.org/10.1128/JB.182.13.3843-3845.2000
- Kavanagh K, Reeves EP. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol Rev 2004; 28(1):101-12; PMID:14975532; http://doi.org/10.1016/j.femsre.2003.09.002
- Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, Ausubel FM, Diener A. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun 2005; 73(7):3842-50; PMID:15972469; http://doi.org/10.1128/IAI.73.7.3842-3850.2005
