ABSTRACT
In eukaryotic organisms, myosin proteins are the major ring components that are involved in cytokinesis. To date, little is known about the biologic functions of myosin proteins in Magnaporthe oryzae. In this study, insertional mutagenesis conducted in M. oryzae led to identification of Momyo2, a pathogenicity gene predicted to encode a class-II myosin protein homologous to Saccharomyces cerevisiae Myo1. According to qRT-PCR, Momyo2 is highly expressed during early infectious stage. When this gene was disrupted, the resultant mutant isolates were attenuated in virulence on rice and barley. These were likely caused by defective mycelial growth and frequent emergence of branch hyphae and septum. The Momyo2 mutants were also defective in conidial and appressorial development, characterized by abnormal conidia and appressoria. These consequently resulted in plant tissue penetration defects that the wild type strain lacked, and mutants being less pathogenic. Cytorrhysis assay, CFW staining of appressorium and monitoring of protoplast release suggested that appressorial wall was altered, presumably affecting the level of turgor pressure within appressorium. Furthermore, impairments in conidial germination, glycogen metabolites, tolerance to exogenous stresses and scavenging of host-derived reactive oxygen species were associated with defects on appressorium mediated penetration, and therefore attenuated the virulence of Momyo2 mutants. Taken together, these results suggest that Momyo2 plays pleiotropic roles in fungal development, and is required for the full pathogenicity of M. oryzae.
Introduction
In eukaryotic organisms, cell division is a complex process required for cell proliferation and differentiation. Cytokinesis is essential during cell division. However, during development of certain organisms, cytokinesis depends on the formation of actomyosin contractile ring. Myosins are a type of actin-dependent ATPase motors and major ring components involved cytokinesis, intracellular transport, cell polarization, transcriptional regulation, and signal transduction.Citation1-3 In fungi, 3 types of myosin proteins were identified, with each having distinguished cellular roles during development.Citation4-6 The class-I myosins, such as MyoA in Aspergillus nidulans and Myo5 in Candida albicans, are involved in endocytosis and hyphal morphogenesis.Citation4,7,8 The class-V myosins are major organelle transporters and required for mediating secretory vesicle delivery and morphogenesis during fungi development and pathogenicity.Citation5,9,10 In contrast to the roles on endocytosis and exocytosis by class-I and class-V myosins, the class-II myosins are known to play important roles in cytokinesis during fungi development.Citation6,11,12 In the budding yeast, Saccharomyces cerevisiae, cytokinesis has been well studied, with type II myosin known to play an important role in this process.Citation6 During cytokinesis, invagination of the plasma membrane at the mother-daughter cell junction is the first essential process. It is followed by polysaccharide chitin extrusion into the invagination to develop chitin disk, which acts as a primary septum for cell division.Citation6,13 In these processes, molecular motor myosin-II encoded by MYO1 provides the force to constrict the actomyosin ring in an F-actin and septin-dependent manner.Citation14,15 In Schizosaccharomyces pombe, Myo2p, together with components of Cdc15p, Cdc12p, Rng2p, and Mid1p, are assembled at the division site to form cytokinesis nodes,Citation16 in which myosin II pulls adjacent actin filaments together to form actomyosin ring, and initiates septum formation.Citation17-19
In contrast to unicellular yeast cells, filamentous fungi cells consist of a continuum of cell wall-coated tubular cells called hyphae. Unlike yeast cells, hyphae do not necessarily undergo cell separation during proliferative growth, and its multicellularity is achieved by insertion of unplugged chitin-rich septa, which divide the adjacent hyphal compartments.Citation20 In fungi, type II myosin has diverse roles in different fungal pathogens such as A. nidulans, Penicillium marneffei and Fusarium graminearum.Citation11,12,21 Mutants of this gene display multiple defects, which range from abolishment of septation, aberrant chitin accumulation to nuclear division defects. However, what seems to be clear from findings in these studies is the fact that certain type-II myosin functions, such as septation, may be conserved across filamentous fungal pathogens, and these may be crucial for pathogenicity. Therefore, understanding the regulatory mechanisms of Momyo2 in septum development may be the key to provide novel strategies for disease control of rice blast fungus.
Magnaporthe oryzae, a filamentous phytopathogenic fungus, served as an excellent model organism for investigating plant-fungal interactions due to its economic and scientific importance.Citation22-24 It causes rice blast disease on many cereal crops, including rice and barley, and results in devastating yield loss of cultivated rice worldwide.Citation25 Previous studies revealed that abolishment of Woronin body, which function to seal septal pores in response to cellular damage, alters pathogenicity of M. oryzae.Citation26 This suggests that intact septum is required for normal development and pathogenicity in M. oryzae. However, there is currently limited investigation toward understanding mechanisms of septum development during M. oryzae cytokinesis. To fill this knowledge gap, this study screened the M. oryzae mutant library for mutants with defects in septation and pathogenicity. A type II myosin gene, Momyo2, was identified to be essential for fungal septation, appressorium development, invasive hyphae growth and full virulence on host plants in M. oryzae.
Results
Identification of Momyo2 in M. oryzae
To identify genes involved in plant infection by M. oryzae, more than 1,500 transformants from a T-DNA insertional mutagenesis library were screened, and a mutant (A115) was identified to show reduced virulence on 14-day-old rice seedlings (). Southern blot analysis revealed that A115 mutant contained a single-copy (> 10 kb) insertion of hygromycin cassettes in the genome (). TAIL-PCR was used to identify flanking sequences of T-DNA insertion in A115 mutant. Sequence analysis revealed an insertion of T-DNA into the second exon of MGG_03060 (GenBank XP_003720621) (), which was based on data from the Broad Institute (http://www.broadinstitute.org).
Figure 1. Isolation and characterization of Momyo2 in M. oryzae. (A) Conidial suspensions (1 × 105 spores mL−1) of wild-type Guy11, ATMT mutant A115 were inoculated on 2-week-old rice seedlings (Co-39). The infected leaves were photographed at 5 dpi. (B) Southern blot assay. Genomic DNA of wild-type Guy11 and ATMT mutant A115 were digested with EcoRV and hybridized with HPH probe. (C) Genomic location of T-DNA insertion in A115 mutants. T-DNA was integrated into second exon of Momyo2 gene (MGG_03060). The gray boxes represent exons and the black line indicates the introns. (D) Domain architectural analysis of myo2. Comparison of the domain structures of Myo2 from S. cerevisiae, S. pombe, and Homo sapiens. Myo2 contains all predicted domains that are typical for class-II myosins. AA indicated amino acids. (E) Phylogenetic tree of Momyo2 homologues was created by the distance based minimum evolution method, based on 1000 bootstraps. M. oryzae sequences characterized in this study are highlighted in red box.
A further BLAST search revealed that the gene comprises 3 exons and 2 introns spanning 7384 bp and encodes a 2388 amino acids protein (). Amino acid sequence analysis of MGG_03060 revealed several conserved domains, including an N-terminal SH3-like fold, N-terminal myosin motor domain (ATPase activity), IQ motif (light chain and calmodulin binding site) and a coiled-coil tail (cellular localization), which are characteristic structures identified in class II myosins in most fungi ().Citation12,21,27-30 Alignment of MGG_03060 with these class-II myosins demonstrates that MGG_03060 shares high identity on domain structures, with 51 to 88% identity in its motor region and 17 to 73% identity in its neck and tail regions (Fig. S1). Phylogenetic tree analysis using homologous myosin proteins from different families grouped MGG_03060 into myosin II subfamily. In class II subfamily, the myosins in filamentous fungi are distant from those of unicellular yeasts and mammals, with M. oryzae Momyo2 being most similar to sequences from filamentous fungi, including Colletotrichum gloeosporioides (Accession No. XP_007274402), Neurospora crassa (Accession No. XP_964712) and F. graminearum (Accession No. EYB29885) (). Thus, consistent with conserved sequence motifs and phylogenetic analysis, this protein can be provisionally designated as class II myosin in M. oryzae.
Expression and deletion of Momyo2 gene in M. oryzae
To understand the role of Momyo2 in development and pathogenicity of M. oryzae, we used qRT-PCR to examine its expression patterns. The results showed that Momyo2 gene is expressed during axenic growth and fungal infection of rice plants, with much higher induction at pre-penetration stage (8 hrs, 3.57-fold, 24hrs, 1.60-fold) and infectious stage (48 hrs, 1.54-fold; 72hrs, 1.66-fold)(Fig. S2A), compared with the stable expression of β-Tubulin gene.Citation31
To characterize functional roles of Momyo2, as well as to further confirm that the phenotype of the insertion mutant resulted from the disruption of Momyo2, a gene knock out vector pMDT-Momyo2-HPH was constructed using targeted disruption strategy (Fig. S2B). Two independent transformants of Momyo2 (Momyo2–1 and Momyo2–7) were selected using hygromycin B and verified by diagnostic PCR (Fig. S2C). They were further confirmed by Southern blot and semiquantitative RT-PCR (RT-PCR) analysis (Fig. S2D, S2E, S2F). To ensure that the defects observed in Momyo2 mutants were ascribed to deletion of Momyo2, a complemented strain Momyo2c was generated by introducing a genomic copy of Momyo2 into Momyo2–1 mutant, and verified by RT-PCR (Fig. S2F). As expected, all defects reported in this study were restored in the Momyo2c ().
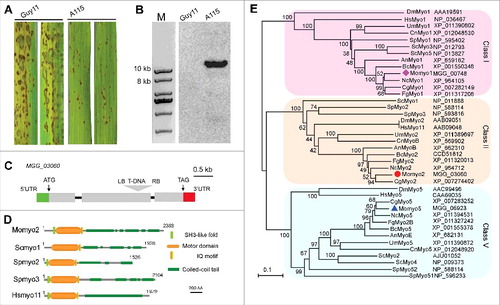
Figure 2. Septa development in Momyo2 mutants. (A) Septa of Guy11, Momyo2 mutants and Momyo2c were stained with CFW and observed under UV or DIC microscopy. White asterisk indicated septum in mycelium. (B) Measurement of average length of sub-apical cell of vegetative hyphae. The mean and standard deviations were calculated based on 3 independent experiments by measuring at least 49 sub-apical cells in each replicate. Error bar represents standard deviation, and asterisks indicate significant differences from the control (P < 0.01). (C) Septa ultrastructure in the hyphae of Guy11 and Momyo2 mutants. Wild-type Guy11 showed a typical septum while the mutant showed incomplete and aberrant septum. CW, cell wall; WB, Woronin body; S, septum; La: lacunas.
Momyo2 gene is required for normal vegetative growth and hyphal morphogenesis
To investigate the role of Momyo2 on vegetative growth, mycelial growth of wild type (Guy11), and mutant strain (Momyo2–1 and Momyo2–7) as well as the complement strain (Momyo2c) was compared on 5 artificial media. The Momyo2 mutants showed a significant reduction in radial growth on the 5 media, compared with Guy11 and Momyo2c (Fig. S3A, S3B). In liquid CM, mycelial growth of Momyo2 mutants was more compact than that of Guy11 and Momyo2c (Fig. S3C). Mycelial biomass was also determined for these strains following their inoculation into liquid CM for 72 hrs and 120 hrs. Biomass determination revealed significantly less harvested fungal mycelia for Momyo2 mutants than Guy11 and Momyo2c 72 hrs and 120 hrs post inoculation, respectively (Fig. S3D).
In view of defects on radial growth, we set out to examine mycelia cultures from the wild type strain and Momyo2 mutants by visualization using light and epifluorescence microscope. Contrary to normal vegetative mycelial of the wild type strain, Momoyo2 mutants developed aberrant vegetative mycelia characterized by thickened, collapsed and bumpy cells with excessive branches (). Mycelial septa from the mutants and wild type strain were compared using an epifluorescence microscope following staining with CFW. These results show that septa in mycelial of Momyo2 mutants were more than that observed for the wild type strain (). The average length of sub-apical hyphal cells in the Momyo2 mutants was 21.4 μm (Momyo2–1) and 20.51 μm (Momyo2–7), respectively, which was significantly shorter than the 77.66 μm of wild-type Guy11 and 73.74 μm of Momyo2c (). However, intact septa can be detected in wild type Guy11 by both DIC and epifluorescence microscopy. Regardless of their abundance in mycelia, these septa were nonetheless abnormal and narrowly spanned hyphal lumen in Momyo2 mutants (). This suggests that Momyo2 is required for normal septal development in M. oryzae.
Momyo2 is required for development of aerial hyphae, conidiation and conidium morphology
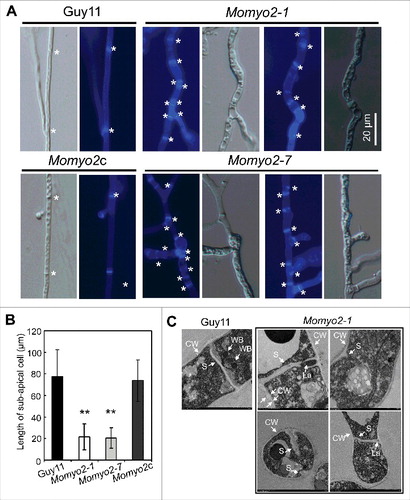
Given that quantity of conidia produced determines severity of disease epidemic in a growing season,Citation32 we assessed the role of Momyo2 in conidiation. On RDC media, hyphal growth of Momyo2 mutant was somewhat flat, exhibiting a severe defect on aerial hyphae formation, compared with wild type Guy11 and Momyo2c strain (). Microscopic examination revealed that, in most cases, only a single conidium was differentiated from the conidiophores of Momyo2 mutants, in contrast to 7 to 10 conidia in a sympodial pattern on conidiophore of Guy11 (; Fig. S4). Quantitative measurement of conidia reconfirmed that conidiation was dramatically reduced by approximately 13-fold in Momyo2 mutants (Momyo2–1, 13.1-fold; Momyo2–7, 13.3-fold), compared with Guy11 and Momyo2c (). Approximately 60% of conidia formed by Momyo2–1 and Momyo2–7 exhibited abnormal morphology. By contrast, only a few conidia formed by wild type Guy11 (3.4%) and Momyo2c (3.9%) were abnormal (, E). These data suggest Momyo2 is essential for normal conidia development in M. oryzae.
Figure 3. Effects of Momyo2 on conidiation and conidial morphology of M. oryzae. (A) Comparison of aerial hyphae growth. The indicated strains of M. oryzae were grown in constant dark condition on RDC agar plates for 7 d and then photographed. (B) Momyo2 disruption results in defects of conidiation. Conidia developed on conidiophores were examined by light microscope using strains grown on RDC medium for 7 d. Scale bar = 50 μm. (C) Analysis of conidial production. The numbers of conidia produced by strains grown on RDC medium for 14 d were statistically analyzed (p < 0.01). (D) Comparison of conidia morphology. Conidia of Guy11, Momyo2 mutants and Momyo2c were collected from 14-day-old cultures, and then imaged under light microscope. Bars = 30 μm. (E) The measurement of percentage of abnormal conidia. Conidia of Guy11, Momyo2 mutants and Momyo2c were collected from 14-day-old cultures, and the ratio of aberrant conidia was measured and statistically analyzed, respectively. Asterisks in , indicate significant differences. Error bar represents standard deviation.
Momyo2 is required for conidial germination and glycogen metabolism
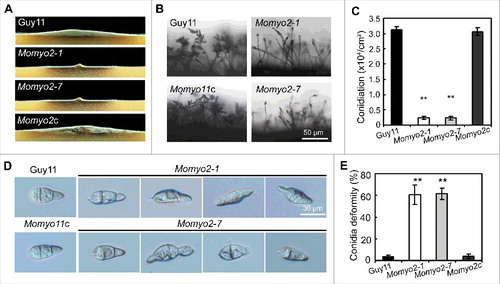
To ascertain whether abnormal conidia of the Momyo2 mutants are viable, we evaluated their germination using hydrophobic coverslips (). The results showed that conidial germination was dramatically reduced in the Momyo2 mutants, compared with that of Guy11 and Momyo2c strain. At 2 hours post inoculation (hpi), ∼10% of conidia of Momyo2 mutants germinated (Momyo2–1, 10.8%; Momyo2–7, 10.4%), compared with 98% of Guy11 and 96.3% of Momyo2c (). Conidial germination gradually increased to 71%, 72% in Momyo2–1 and Momyo2–7 mutant, respectively, at 4 hpi. Nonetheless, it was still significantly less than that observed for Guy11 and Momyo2c strain (, B). To determine whether the ability to germinate was associated with conidial age, 20-day-old conidia harvested from Momyo2 mutants were compared. Conidial germination was dramatically decreased when compared with conidia harvested from 10-day-old RDC plates, with 0% at 2 hpi and average 50% (Momyo2–1, 49.5%; Momyo2–7, 50.8%) at 4 hpi, respectively ().
Figure 4. Conidium germination and glycogen deposition. (A) Conidial germination is delayed in the Momyo2 mutants. Droplets of conidial suspension (1 × 105 spores ml−1) were inoculated on the hydrophobic coverslips for indicated time, and then photographed. Bars = 10 μm. (B) Statistical analysis of germinated conidia. Germinated conidia were examined at each indicated time under a light microscope, and then statistically analyzed (P < 0.01). (C) Impaired glycogen deposits in conidia of the Momyo2 mutants. Conidia (10-day-old) suspension stained with a glycogen staining solution were visualized with brightfield optics of a Nikon inverted Ti-S epifluorescence microscope. Bars = 20 μm. (D) Impairment of glycogen deposition in age-dependent conidium. Impaired glycogen deposits in conidia (> 99) of Guy11 and Momyo2 mutants were counted and analyzed. (P < 0.01). (E) Impaired glycogen degrading during appressorium formation in the Momyo2 mutants. Conidia (10-day-old) drops (1 × 105 spores ml−1) were inoculated on the hydrophobic coverslips for 24 hrs, and then stained with glycogen staining solution. (F) Statistical analysis of germinated conidia containing glycogen deposit at 24 hpi. The percentage of germinated conidia containing glycogen deposit was counted and statistically analyzed (p < 0.01) Asterisks in , , indicate significant differences between wild type and Momyo2 mutants.
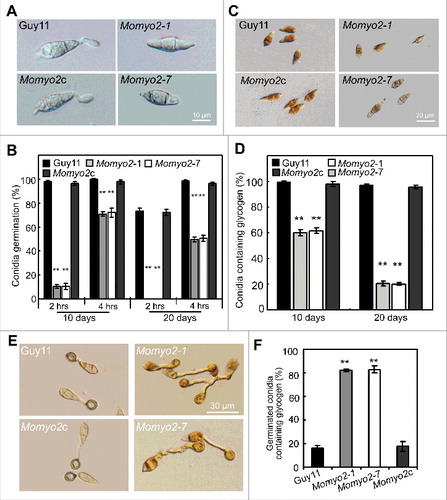
Since utilization of metabolites from internal storage compounds is an essential event during conidial germination, we thus examined cellular distribution of glycogen in conidia (). Iodine staining of 10-day-old conidia revealed 99.7% of Guy11 conidia had densely distributed glycogen, compared with 60% of Momyo2 mutants (Momyo2–1, 59.9%; Momyo2–7, 61.6%) (). During appressorium development, large amounts of glycogen were present in conidia (), even after 24 hr in Momyo2 mutants than in control strains (). Interestingly, glycogen accumulated in more than 82% of germinating conidia in Momyo2 strains compared with accumulation in about 16% of conidia in wild type Guy11 and Momyo2c (). To determine whether distribution of glycogen is age-dependent in Momyo2 mutants, 20-day-old conidia were further stained with iodine solution, and the results showed that 97% of conidia from Guy11 were visible of glycogen deposits, in contrast to average 20% of conidia of the mutants (Momyo2–1, 20.5%; Momyo2–7, 20.2%), indicating a dramatic decrease in the maintenance of glycogen deposits in the Momyo2 mutants ().
Momyo2 is required for exogenous stress tolerance
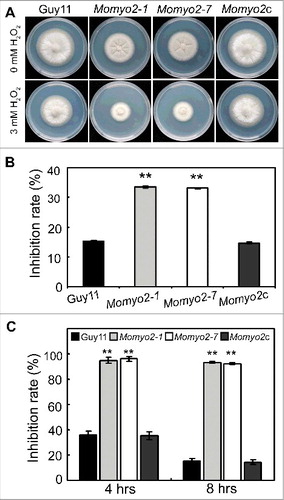
To establish the stress response of the Momyo2 mutants, we subjected strains to compounds that stimulate oxidative (H2O2), cell wall (CR and CFW) and osmotic stress (SDS, sorbitol and NaCl). Except in the presence of CFW, Momyo2 strains were highly sensitive to all tested stressful conditions, compared with Guy11 and Momyo2c strain (Fig. S5A, S5B, S6A, S6B). Similarly, mycelial growth of Momyo2 mutants was dramatically reduced on a hydrogen peroxide-containing CM medium, with high inhibition rates compared with Guy11 and Momyo2c, with relatively less inhibition rates (, B). Meanwhile, conidia of Momyo2 mutants displayed sensitivity to H2O2. Exposure to 1 mM H2O2 resulted in severely decreased conidial germination in Momyo2 mutant, which was 60% less than that of Guy11 and Momyo2c (). Furthermore, about 92% of Momyo2 conidia failed to germinate even 8 hrs post inoculation. By contrast, respectively 15.2% and 14.4% conidia of wild type and complement strain did not germinate under this condition post 8 hrs (). These findings suggest Momyo2 plays an essential role during growth under oxidative stress inducing conditions in M. oryzae.
Figure 5. The Momyo2 mutants are hypersensitive to H2O2. (A) Mycelia growth of the Momyo2 mutants under oxidative stress. The indicated strains were inoculated on CM with or without 3 mM H2O2 and cultured at 28 °C for 5 d. (B) Statistical analysis of the inhibition rate of the tested strains under oxidative stress. The colony diameters of the testing strains were measured and subjected to statistical analysis. (C) Statistical analysis of germinated conidia under oxidative stress. Conidia germination rates were measured and analyzed after treatment with or without 3 mM H2O2. Three repeats were performed and similar results obtained. Error bars represent the standard deviations and asterisks represent significant differences (p < 0.01).
Momyo2 is required for pathogenic development in M. oryzae
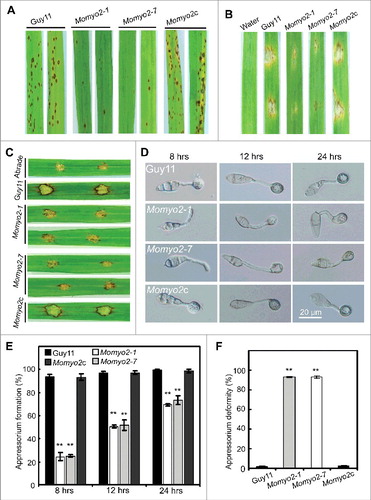
To investigate the role of Momyo2 in pathogenesis, conidial suspensions (1 × 105 spores mL−1) were inoculated on 2-week-old rice seedlings (Oryza sativa cv CO-39) by spraying and/or one-week-old barley leaves by conidial drop. At 5 d after inoculation, the Momyo2 mutant was found to be less pathogenic than Guy11 and Momyo2c. On rice seedlings, the Momyo2 mutants developed less and small restricted growth lesions on rice plants (), in contrast to numerous typical necrotic lesions caused by wild type Guy11 and Momyo2c. Similar results were observed on Momyo2 infected barley leaves, compared with that of by Guy11 (). To further investigate the pathogenic role of Momyo2 in M. oryzae, drops (20 μL) of conidial suspension from Guy11, Momyo2 mutants and Momyo2c were inoculated on wounded rice leaves. The wild type Guy11 were fully pathogenic, with extendible and necrotic lesions, whereas the Momyo2 mutants causes small necrotic lesions on the inoculation sites (). These findings suggest a potential role for Momyo2 in plant invasion and colonization.
Figure 6. Pathogenicity assays and appressorium development in Momyo2 mutants. (A) Spray assay for disease development on rice leaves. The Momyo2 deletion reduced virulence on rice leaves. Diseased leaves were collected at 5 dpi. (B) Pathogenic assays on barley leaves. Droplets of conidial suspension of indicated strains were inoculated on barley leaves and diseased leaves were harvested at 5 dpi. (C) Pathogenic assays on abraded rice leaves. Droplets of conidial suspension of indicated strains were inoculated on abraded rice leaves and diseased leaves were harvested at 5 dpi. (D) Appressorium development is delayed in the Momyo2 mutant. Conidial drops (1 × 105 spores ml−1) were inoculated on the hydrophobic coverslips for 8, 12, 24 hrs, and then imaged with light microscope with DIC equipments. (E) Analysis of appressorium formation. Appressorium formed at the germ tube were counted and statistically analyzed. P < 0.01. (F) Statistical analysis of abnormal appressorium. The appressorium deformity rate of indicated strains was measured and statistically analyzed. P < 0.01. Asterisks in , indicate significant differences between the wild type and Momyo2 mutants.
Momyo2 is essential for appressorium development, penetration and invasive growth
In M. oryzae, differentiation of mature appressorium (with turgor pressure up to 8MPa) at the apex of germ tube enables the blast fungus to invade into host cells.Citation33 Thus, the ability to develop appressorium at the tip of germ tube was first compared among Guy11, Momyo2 mutants and Momyo2c strain. The results revealed that appressoria developed by the Momyo2 mutants were dramatically decreased, with 69% by 8 hrs, 47% by 12 hrs and 28% by 24 hrs less than that of by Guy11 and Momyo2c (, E). Furthermore, of those appressoria developed by the mutants at 24 hpi, average 93.3% were not melanized and reduced in size, compared with 1.7% and 2.7% of that by Guy11 and Momyo2c, respectively (, F). Based on the above, our results suggest that Momyo2 is associated with the development of appressorium and cell morphology in M. oryzae.
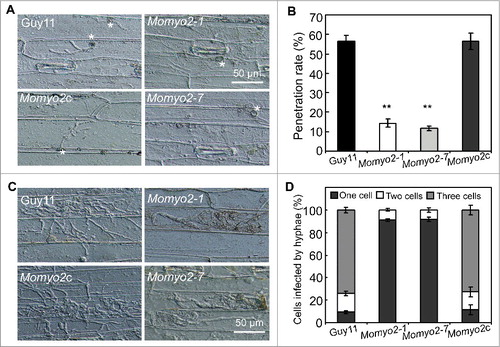
To understand the role of Momyo2 in disease development, barley leaves were used to determine the causes of attenuated virulence. As less necrotic lesions were observed on the leaves infected by Momyo2 mutants (), appressoria-mediated penetration was examined, and the results revealed that Momyo2 mutants could less effectively penetrate into the epidermal cells of barley leaves at 36 hpi, compared with Guy11 (, B). At 72 hpi, most invasive hyphae of Momyo2 mutants were still delayed in their in planta growth and colonization of the neighboring cells, with most of primary infection hyphae being restricted to the cellular compartment they initially penetrated, whereas, wild type Guy11 could freely expand and colonize the neighboring cells from the initially invaded cell (, D). The above defects were rescued when Momyo2 gene was reintroduced into the mutant, indicating that Momyo2 is required for both appressorium-mediated penetration and invasive hyphae growth in host cells.
Figure 7. Plant penetration assays. (A) Appressorium-mediated penetration was impaired in Momyo2 mutants. The Momyo2 mutantss showed defects on penetration into barley leaves after 36 hrs, compared with control strains. (B) Analysis of appressorium penetrated in barley cells. The appressoria penetrated into barley epidermal cells were measured at 36 hpi, and the data were statistically analyzed. (C) Invasive hyphae growth was impaired in Momyo2 mutants. Infectious hyphae growth in barley epidermal cells was observed at 72 hpi. (D) Quantification of hyphae infection on barley epidermal cells. The percentage of infected cells occupied by invasive hyphae of indicated strains at 72 hpi were measured and statistically analyzed. Asterisks in indicate significant differences between the wild type and Momyo2 mutants.
Momyo2 is required for turgor generation and cell wall integrity
During appressorial development, glycogen within the appressorium is rapidly degraded at the onset of melanization and required for turgor pressure in M. oryzae.Citation34 In view of defective glycogen metabolism in Momyo2 mutants, we presumed that appressorial turgor pressure might be abnormal in Momyo2 mutants. Therefore, an incipient cytorrhysis assayCitation35 was used to measure turgor pressure, and the results showed that appressoria of Momyo2 mutant were less collapsed than those of Guy11 and Momyo2c (), suggesting an increased turgor pressure in the mutant. This findings seemed further confirmed by lytic enzymes treatments, with less collection of protoplast by Momyo2 mutants than that of Guy11 and Momyo2c at given time point (Fig. S7A). However, the above results were contradictory to the findings that appressorium-mediated penetration was less effectively in Momyo2 mutants than in Guy11. To investigate the possible reason, strains expressed cytoplasmic eGFP were generated using the Momyo2 mutant and Guy11 as the recipients, respectively. Microscope examination revealed that most area of the mycelia by Momyo2 mutant is absent from green fluorescent signal compared with Guy11 (Fig. S7B), making us presume that the less harvest of protoplast might not be ascribed from the enhancement of cell wall, but from the loss of cytoplasm in the mycelia of Momyo2 mutant.
Figure 8. Assessment of turgor pressure and cell wall integrity. (A) Measurement of appressorium turgor pressure. Appressorium of the indicated strains was induced on hydrophobic surface of coverslip and treated with given concentration of glycerol solution. The collapsed appressoria (> 100) were counted and statistically analyzed. (B) Staining of appressorium of indicated strains by the CFW. Appressoria induced on the hydrophobic surface were stained by CFW and the fluorescence signal from mature appressoria was imaged by epifluorescence microscope. (C) The defective appressoria of Momyo2 mutants are permeable to glycerol. Appressoria induced on the hydrophobic surface were treated with 4 molar glycerol solutions and then the rate of cytorrhysis and the proportion of appressoria that had recovered from cytorrhysis were determined at each time point. (D) Detection of cell wall porosity. Appressoria induced on the hydrophobic surface were treated with different Stokes' radii of PEG. Cell wall porosity was measured by comparing the ratio of plasmolysis to cytorrhysis. Asterisks in and indicate significant differences among the strains (p < 0.01). Error bar represents standard deviation.
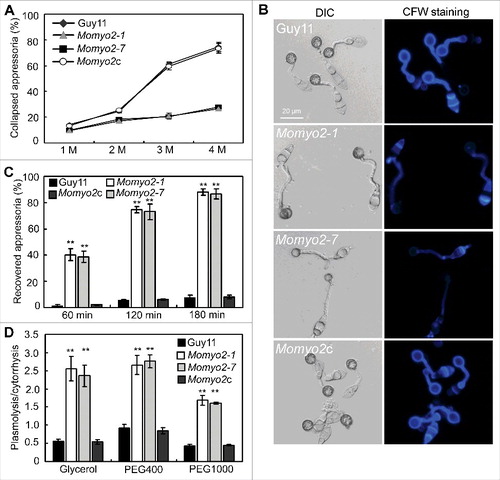
In M. oryzae, non-melanized appressorial wall is permeable to glycerol and could quickly recovered from cytorrhysis when glycerol diffuses through the cell wall.Citation36 In Momyo2 mutants, most of the appressoria were not melanized and reduced in size, making us presume that cell wall integrity (CWI) is defective in Momyo2 mutants. The staining of appressorium with CFW revealed that fluorescent signal was much stronger around appressorium of Guy11 and Momyo2c than that of Momyo2 mutants (), indicating less accumulation of chitin on appressorial wall in Momyo2 mutant than that of Guy11. The subsequent evaluation of appressoria recovered from cytorrhysis showed that average 39%, 73% and 87% of appressoria could quickly recover from cytorrhysis when incubated for 60, 90 and 120min, respectively, compared with 1.9%, 5.7% and 6.4% of that by Guy11, further suggesting that appressoria cell wall of Momyo2 mutants is much thinner and permeable to glycerol during cytorrhysis assay (). In spite of this, porosity assay also revealed that the ratios of plasmolysis to cytorrhysis by wild type Guy11 were 0.55, 0.91 and 0.44 in glycerol, PEG 400 and PEG1000, respectively, in contrast to average ratios of 2.43, 2.70 and 1.64 in glycerol, PEG400 and PEG1000 by the Momyo2 mutants (), indicating an increased porosity of cell wall in Momyo2 mutants, which make the appressorial wall permeable to glycerol and not collapse during cytorrhysis assay.
Momyo2 is essential for suppressing ROS production in plant cells
The invasive hyphae of Momyo2 mutants displayed restrained growth during plant cell infection (). This is reminiscent with restricted growth observed in Moatf1 mutant, disrupted for ATF1, which mediates oxidative stress responses.Citation37 Based on functional and phenotypic similarities of Momyo2 and Moatf1 mutants, ROS production was evaluated using DAB staining of barley leaves infected by Guy11, Momyo2 mutants and Momyo2c strain. Increased accumulation of ROS was detectable in barley epidermal cells infected by Momyo2 mutants, but not in cells penetrated by Guy11 and Momyo2c (, B). When diphenyleneiodonium (DPI), an inhibitor of NADPH oxidases,Citation38 was applied to barley leaves, the infected cells stained by DAB were dramatically decreased in Momyo2 mutants (), suggesting that the Momyo2 mutants might be unable to suppress ROS production in plant cells during infection.
Figure 9. Scavenging of ROS production in plant cells by Momyo2 mutant. (A) DAB staining of plant cells infected by Momyo2 mutant. Conidial suspensions (1 × 105 spores mL−1) of the Guy11, Momyo2 mutants, and complemental strain Momyo2c were inoculated on barley leaves for 24 hrs and then stained with DAB solution. Arrows indicate appressoria Bar = 40 μm. (B) Percentages of the DAB-stained plant cells without or with DPI treatment. Asterisks indicate significant differences among the strains (p < 0.01). Error bar represents standard deviation. (C) DPI treatment partially restored invasive hyphae growth. Barley leaves were treated with or without DPI (0.5 mM) solution. Invasive growth was observed at 30 hpi. AP, appressoria; BH, branched IH; PH, primary IH. Bar = 30 μm. (D) Percentages of distinct types of IH developed by indicated strains. Each of the indicated strains was inoculated on barley leaves pretreated with or without DPI for 30 hrs and then the percentage of distinct types of IH was statistically compared.
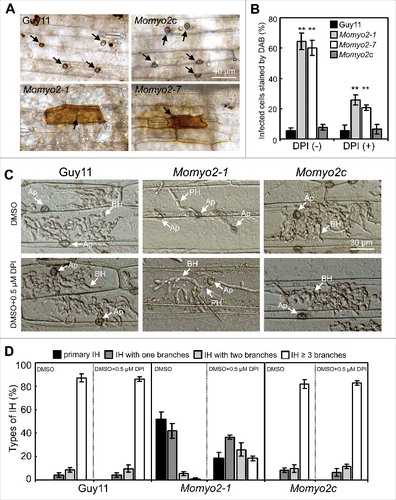
To determine whether impairment of plant colonization by Momyo2 mutants is associated with increased ROS accumulation in plant tissues, we also evaluated the effect of pretreatment with DPI on plant infection. The wild type strain formed 100% appressoria with invasive hyphae (IH), of which 95.7% had more than 2 branches. However, 48.4% of Momyo2 appressoria formed IH, with 6.5% of IH showing more than 2 branches (, D). After pretreatment with DPI, Guy11 formed 100% appressoria with IH, of which 95.8% showed more than 2 branches at 48 hpi. By contrast, extensive bulbous IH were observed in 81.7% of Momyo2 appressoria, of which ∼44.1% formed more than 2 branches at 48 hpi (, D). These results indicate that suppression of ROS production in host cells significantly increased the ability of Momyo2 mutants to penetrate and colonize in plant cells.
Momyo2 is required for secretion of extracellular peroxidases
Under cell wall stress conditions, the degradation halo of CR by the Momyo2 mutants is much smaller than that of Guy11 (Fig. S6A), indicating that Momyo2 plays an important role in degrading CR. In M. oryzae, secreted enzymes, such as peroxidases, are presumed to be responsible for degrading of CR and required for degrading host-derived ROS.Citation37, 39 Therefore, activity of extracellular enzymes was compared using peroxidase and laccase. Activity of both enzymes was severely reduced in culture filtrate of Momyo2 mutants relative to Guy11 and Momyo2c strain (Fig. S6C). To determine the possible reason, several reported peroxidase and laccase enzymes with a signal peptideCitation37,40 were compared at transcriptional level in Momyo2 mutant and wild-type Guy11. These results showed that expression of genes coding for laccase showed severely reduced expression in the mutants compared with Guy11. However, of peroxidase encoding genes, 4 revealed increased expressions while the rest were comparable in the mutants and wild type Guy11 (Fig. S6D). This expression patter suggests that secretion of extracellular peroxidases might be defective in the Momyo2 mutant.
Localization of Momyo2 and nuclear distribution in Momyo2 mutants
To investigate the function of Momyo2 during hyphal development in M. oryzae, we labeled the protein with a green fluorescent tag in the wild type Guy11 and observed its dynamic behavior during septation. As expected, the Momyo2-eGFP strain was viable, and its phenotypes such as growth, conidiation and pathogenicity were the same as wild type Guy11 (Fig. S8), confirming the fusion of a C-terminal eGFP did not affect Momyo2 activity. We next used time-lapse imaging to examine the dynamic behavior of the florescent protein during the hyphal development, and found that florescence was clearly detected on the inner cell walls on both sides of the hyphae, revealing the position of contractile ring at the site of septal initiation (; Video S1). As the septum developed, Momyo2-eGFP protein has been recruited to form a constricted contractile ring toward the middle of the septum. When the septum completely developed, the florescent ring was contracted to a point at the center of the septum, with a strong band of fluorescence at the peak of Momyo2-eGFP protein accumulation (, 40′, 50′). After that, the Momyo2-eGFP protein appeared to disperse, as indicated by the declining fluorescence signal in (70′–90′).
Figure 10. Localization of Momyo2 and nuclear distribution in Momyo2 mutants. (A) Momyo2-eGFP dynamics during septation processes in hyphae. Mycelia of the Momyo2-eGFP strain were overnight cultured, and then Momyo2-eGFP dynamics during septation were visualized by a confocal fluorescence microscope. (B, C) Nuclear distribution in Momyo2 mutants. Nuclear signal in conidia and appressorium of both Guy11 and Momyo2 mutants were visualized by a confocal fluorescence microscope. (D) Nuclear distribution in the hyphae of Momyo2 mutants. Both Guy11 and Momyo2 mutants expressing H1-RFP fusion protein were overnight cultured in liquid CM, stained with CFW and then visualized by a confocal fluorescence microscope. Arrows indicated the hyphae without cytoplasm.
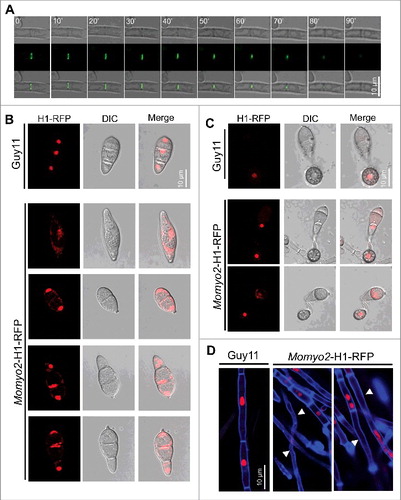
To determine whether deletion of Momyo2 affects cytokinesis, the histone H1-RFP fusion construct were transformed into Guy11 and Momyo2–1 mutant. In Guy11, septation occurred regularly in hyphae and during appressorium development, with each hyphal apartments and appressorium had only one globular nucleus (, C, D). However, in Momyo2 mutant, septa were unevenly distributed in the hyphae (; ), with some of the individual hyphal apartments contained multiple nuclei while the others are absence of nuclei (). Meanwhile, nucleus signal in mutants conidia are not aggregative and prevented from degradation in mother conidia of the mutants during appressorium formation, compared with Guy11.
Discussion
Cytokinesis is essential during proliferation and development of all organisms.Citation11,14,21 In this study, we have shown that type II myosin in M. oryzae is encoded by Momyo2 gene, and has pleiotropic functions during fungal development and pathogenicity. Consistent with previous findings in other fungi, our results indicate that Momyo2 is required for formation of primary septum during cell division.Citation11,14,21,41 Meanwhile, our study also revealed that Momyo2 is required for conidiation, conidia morphology, CWI, appressorium mediated penetration and full pathogenicity. However, we found in this study additional roles of Momyo2 that may be unique to M. oryzae. These include roles of nuclei degradation during appressorium formation, glycogen metabolism, secretion of extracellular enzymes and scavenging of host-derived ROS during plant infection. With these functions taken into account, Momyo2 is required for fungal development and for the full virulence of M. oryzae.
In M. oryzae, the disruption of Momyo2 interfered with both mycelial growth and sexual reproduction, supporting the conserved role of type II myosins in cell proliferation and cytokinesis in yeast and filamentous fungi.Citation11,14,42 In filamentous fungi, the chitin-rich septa in hyphal cells are essential for maintenance of hyphae cytoplasmic content, and its differentiation at the hyphae apex is critical for fungal growth and pathogenicity.Citation11,20,43,44 In M. oryzae, Woronin body has been shown to seal septal pores in response to cellular damage, and its abolishment results in defects of proper development of functional appressoria and pathogenicity,Citation26 further indicating the critical roles of intact septa in development and pathogenicity of M. oryzae. In this study, our findings revealed that Momyo2 hyphae elaborated more septa. However, these were incomplete and failed to span the width of hyphae. Therefore, the intracellular septa in the Momyo2 mutants are malformed. Previous studies have confirmed that type II myosins have a conserved function in fungi of providing the force to constrict the actomyosin ring, and are thus critical for fungal septa development in fungi.Citation6,28,44 In this study, septal localization of Momyo2 indicates the involvement of Momyo2 in forming a contractile ring during septum development of M. oryzae (). Disruption of this gene blocks primary septum formation, which, as a result, affects the cytokinesis, cell proliferation and hyphae elongation of the mutants. In Momyo2 mutants, more than one nucleus was observed in each cell compartment in hyphae, providing direct clues of Momyo2 in regulating cytokinesis and cell division in M. oryzae. In addition, the loss of cytoplasm in mycelium and conidium of Momyo2 mutants (, , ; Fig. S6B) might also be ascribed from the incomplete intracellular septa in cells, which potentially reduces the size of Woronin bodies and results in open septal gaps in these structures.
In M. oryzae, asexual spores produced from the knee-like conidiophores are the major source of primary inoculum, and the ability to produce conidia determines an epidemic of the disease.Citation22,24 The Myo2 protein is required for proper conidial production in F. graminearum.Citation11 Deletion of its gene results in aberrant chains of linked macroconidial spores similar to the chain-cells produced by type II myosins mutants in S. cerevisiae and S. pombe.Citation14,17 Type II myosin also show similar roles in conidiophore development and conidiation in other fungi such as P. marneffei and A. nidulans,Citation12,21 highlighting an important role of type II myosin protein in fungi asexual spores development. In this study, Momyo2 was shown to act as a determinant for asexual reproduction. Deletion of this gene resulted in a dramatic reduction in conidium production. Conidiation of M. oryzae required an apical extension and swelling of the conidiophore in a sympodial pattern.Citation45,46 In examination of early conidial ontogeny, the Momyo2 mutants showed a much lower number of conidiophores than the wild type Guy11 (). Meanwhile, the Momyo2 mutants could not develop knee-like conidiophores, but mostly produced one conidium per conidiophore (; Fig. S4), indicating that extension of conidiophores is defective in Momyo2 mutants. As septa formation usually co-occurs with conidiophore extension and is essential for proper conidial development,Citation44 we presumed that aberrant septum formation in Momyo2 mutants will restrict the development of knee-like conidiophores in early stage, which ultimately affect the conidial production in M. oryzae.
Conidial germination in M. oryzae is a complex process that involves a cascade of biologic events. The degradation of glycogen during conidial germination is regarded as one of the principal energy sources during germ tube extension.Citation31,34,47 In this research, the Momyo2 mutants produce large amount of malformed conidia with postponed germination ability in comparison to Guy11 (). Considering the roles of glycogen metabolism in conidial germination and appressorium formation in M. oryzae,Citation31,34 we monitor its deposition in conidia. This analysis showed that glycogen is diminished in most Momyo2 conidia in an age-dependent manner. Since glycogen deposits were normal in 60% of 10-day-old conidia, abnormal glycogen breakdown identified in 20-day-old conidia in Momyo2 mutants suggests an essential role of Momyo2 in maintenance rather than synthesis of conidial glycogen in M. oryzae. In addition, previous studies revealed that both spore germination and maturation of infection structures require rapid degradation and mobilization of glycogen in M. oryzae.Citation34,48 Our results indicate that glycogen degrading ability is defective during conidial germination and appressorium development in Momyo2 mutants. This might affect appressorium formation and maturation in MoMyo2 mutant.
Appressorium-mediated penetration is a critical step for fungal pathogenicity, and an intact cell wall of appressorium guarantees successful infection by pathogenic fungus M. oryzae.Citation49-53 In this study, appressorium-mediated penetration may be defective in Momyo2 mutants due to alterations in CWI. In turgor pressure assay, the less collapsed appressoria at given glycerol solution indicate higher turgor pressure in the appressoria of Momyo2 mutants. Lytic enzymes treatments seem to support this hypothesis, as indicate by less collection of protoplast. However, this possibility was excluded by examining cytoplasm in Momyo2 mutants. Presence of empty cytoplasm in Momyo2 mutants might be the main reason for less protoplast release. Given that appressorial turgor pressure increased in Momyo2 mutants, it was also paradoxical to the facts that appressorium-mediated penetration was defective in Momyo2 mutants. In M. oryzae, non-melanized appressorial wall were much thinner and permeable to glycerol, which make more plasmolysis than cytorrhysis by treatment with hyperosmotic glycerol solutions.Citation36 In this study, non-melanized appressoria developed by Momyo2 mutants showed similar phenotypes with ALB1 mutant,Citation36 with more plasmolysis than cytorrhysis () under priority test. This confirms that cell wall integrity might be defective in Momyo2 mutants. CFW staining supported this view, with much weaker fluorescent signal on appressoria of Momyo2 mutants, which indicated less accumulation of chitin on appressorial wall and much thinner cell wall of Momyo2 mutants. In addition, appressorium recovery assay also confirmed the conserved roles of Momyo2 in involvements of the CWI of M. oryzae. Based on the above information, our results strongly suggest that cell wall structure of Momyo2 appressoria is altered such that solutes easily diffuse through cell wall pores.
In fungi, host-derived ROS is a second messenger for defense-gene induction during fungal infection. Its degrading promotes fungal infection during plant-microbe interactions.Citation37,40,54 In view of sensitivity to H2O2 and restricted growth in plant cells by Momyo2 mutant, we presumed that the ability to detoxify host-derived ROS may be impaired in Momyo2 mutants. The DAB staining supported this hypothesis and greater amounts of H2O2 accumulated at primary infection sites when challenged with Momyo2 mutant compared with Guy11. When DPI was used to prevent host-derived ROS, appressorium penetration and growth of infectious hyphae in Momyo2 mutants are significantly increased. This suggests increased accumulation of host-derived ROS might prevent infection of Momyo2 mutants. In M. oryzae, extracellular peroxidases degrade ROS at infection site.Citation39 Our measurement of secreted enzymes activity revealed reduction in acitivity of extracellular peroxidases and laccases in Momyo2 mutant. In fungi, intracellular organelles involved in exocytosis are transported by molecular motors to special growth region along the cytoskeleton.Citation55,56 Myosins act as major type of actin-dependent ATPase motors and are therefore required for transporting of cargo during fungi development.Citation1-3 Therefore, combined with conserved roles of Myo2 in fungi, we presumed that Momyo2 might be required for the delivery and exocytosis of specific proteins outside of fungal cell wall. Deletion of this gene in M. oryzae might block secretion of proteins such as extracellular peroxidases during fungal development. This will ultimately impair the degrading of host-derived ROS during plant infection. However, since laccase encoding genes were downregulated in the Momyo2 mutant, we couldn't exclude possibility that the transcriptional repression of genes, which resulted in defects on synthesis of extracellular enzymes, might in part disable detoxification of host-derived ROS by Momyo2 mutant.
In M. oryzae, appressorium development is regulated by cell cycle progression.Citation57 Generally, mitosis consistently occurs within fungal germ tube before appressorium differentiation, and breakdown of nuclei within the conidium was correlated with appressorium formation. Previous studies revealed that autophagy, which results in nuclei degradation in conidium during appressoria development, is an essential process during appressorium-mediated penetration by M. oryzae.Citation58 The blocking of autophagy could result in formation of non-functional appressoria and the loss of pathogenicity during fungal invasion.Citation58,59 In M. oryzae, mutants disrupting the PMK1 or MgATG8, which resulted in defective nuclei degradation, is unable to complete fungal invasion.Citation58 In Momyo2 mutants, similar phenotypes were identified, with most of conidial nuclei close to the appressorium undegraded even after 24 hpi, indicating defective autophagy might exist in Momyo2 mutants. Combined with the fact that autophagy and functional appressorium is coupled existing, we speculated that the defect of nuclear degradation during appressorium formation might be result in inability of developing functional appressorium, and thus attenuated the virulence on rice plants in M. oryzae.
In summary, this study reveals pleiotropic roles of Momyo2 during the development of M. oryzae, including regulation of fungal septation, appressorium development, invasive hyphae growth and attenuated virulence of M. oryzae.
Materials and methods
Fungal strains and culture conditions
M. oryzae strain Guy11 was used as wild-type strain throughout this study. Guy11 and its derivative transformants were routinely grown at 28 °C on complete medium (CM) with or without agar for 3–15 d to evaluate growth and colony characteristics. Genomic DNA, RNA and protoplasts were isolated from mycelia harvested following growth in liquid CM at 28 °C for 2 d. For conidiation, fungal strains were treated following previously established methods.Citation52 Germinated conidia were obtained by placing drops of conidial suspension on hydrophobic coverslip for 2 and 4 hrs, respectively. Appressoria were obtained by holding germinated conidia on a hydrophobic coverslip for 8, 12 and 24 hrs, respectively. Conidiophore development and conidiation observation were performed as described previously.Citation40,60 To determine fungal growth under different stressors, strains were cultured on CM with or without various chemicals, including H2O2 (3 mM), NaCl (0.7 M), Congo Red (CR, 200 μg mL−1), Calcofluor White (CFW, 200 μg mL−1) and SDS (0.2%), and grew under 28°C for 5 d. The inhibition rate was calculated as described previously.Citation51
hiTAIL-PCR, qRT-PCR and semiquantitative RT-PCR
High-efficiency thermal asymmetric interlaced polymerase chain reaction (hiTAIL-PCR) was performed as described previously.Citation61 Primers used for hiTAIL-PCR are listed in Table S1. cDNA was synthesized according to the described previously methods.Citation52 RT-PCR was performed with primer pairs GL678 / GL679 to confirm deletion and reintroduction of Momyo2 gene. The stable expression β-tubulin gene amplified by primer pairs GL622 / GL623 was used as internal control. Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) were performed using a BIO-RAD CFX96 touch q-PCR system (BIO-RAD, Hercules, California, USA), following previously established procedures.Citation52 Primers used in this section were listed in Table S1.
Nucleic acid manipulation and Southern blotting
Genomic DNA prepared by the standard method was used for both diagnostic PCR and Southern blot hybridization.Citation62 DNA hybridization probes, which are amplified by primers GL455 / GL900 and GL135 / GL136 (Table S1), respectively, were labeled with digoxigenin-11-dUTP using DIG-High prime according to manufacturer instructions (11745832910, Roche, China). Southern hybridization procedures, including restriction enzyme digestion, agarose gel separation, DNA gel blotting and hybridization were performed as described previously.Citation52 Total RNA was isolated from frozen fungal mycelia, conidia and infectious plants (8, 24, 48 and 72 hrs) using the protocol described in E.Z.N.A. Total RNA Kit I (R6834–01, Omega Bio-Tek, Norcross, USA).
Targeted gene replacement and complementation
Targeted gene disruption construct was designed following previously established procedures.Citation52 The 1.0 kb DNA fragments flanking the upstream and downstream regions of targeted gene, respectively, were amplified using primers GL455 / GL456 and GL457/ GL458. Subsequently, they were jointed together by overlap PCR with primers GL455 / GL458 to construct plasmid pMDT-Momyo2. The hygromycin B-resistance cassette that amplified with primers GL135 / GL136 by KOD-Plus-Neo (KOD-401, TOYOBO, China) was inserted into the EcoRV site in plasmid pMDT-Momyo2 to generate deletion construct pMDT-Momyo2-HPH. A ∼3.4-kb DNA fragment amplified from deletion construct with primers GL455 / GL458 was transformed into protoplasts of wild type Guy11 via PEG-mediated fungal transformation. Hygromycin B-resistant transformants were screened using primer pairs P1 / P2 and P3 / P4, respectively, and then were further validated by Southern blot and RT-PCR. For complementation, a 9.33 kb DNA fragments containing a 1.25 kb upstream sequence, a full-length Momyo2 gene coding region and a 0.69-kb downstream sequence was amplified from Guy11 genomic DNA using the primers GL693 / GL696, and then cloned into the pMoC-eGFP vectorCitation63 using yeast gap repair approach to generate pMoC::Momyo2 vector. The sequenced vector was reintroduced into Momyo2–1 mutant by Agrobacterium-mediated transformation of M. oryzae. The transformants were screened on half CM platesCitation64 with 50 μg mL−1 carboxin and validated by semiquantitative RT-PCR. To observe the localization of Momyo2, the Momyo2::eGFP fusion vector was generated by yeast gap repair approach. Three DNA fragments, including the promoter region and ORF of Momyo2 gene, as well as eGFP ORF were amplified from the wild-type Guy11 genomic DNA and pMoC-eGFP plasmid, respectively. These were mixed with the HindIII digested pMoC-eGFP and then were transformed into FY834 strain using previously established procedures.Citation63 Following transformation, the generated pMoC-Momyo2::eGFP vector was isolated and used for transforming the wild type strain Guy11. Under fluorescent microscope, a Momyo2::eGFP strain expressing the fusion protein was obtained and used for examining the dynamic behavior of Momyo2 during septation. To observe the nuclei in both Guy11 and Momyo2 mutant, 2 DNA fragments, including the promoter region and ORF of Histone H1 gene, as well as RFP ORF were subcloned into pYF11, generating pYF11-H1-RFP, then transformed into Guy11 and Momyo2 mutant, respectively, to generate corresponding strains. Primers used in this section were listed in Table S1.
Glycogen, CFW Staining and Microscopy assays
Conidia harvested from 14-day-old and 21-day-old RDC culture plates were used for glycogen staining. Conidial suspension (1 × 105 conidia mL−1) was directly mixed with staining solution (60 mg mL−1 of Kl and 10 mg ml−1 of I2 in distilled water).Citation34 Microscopic examination of stained conidia was performed using a Nikon inverted Ti-S epifluorescence microscope (Nikon Co., Tokyo, Japan) with differential interference contrast (DIC). For CFW staining, 14-day-old conidia were treated as described previously.Citation52 Both vegetative hyphae and appressorium stained by CFW was visualized using the Nikon inverted Ti-S epifluorescence microscope (Nikon Co., Tokyo, Japan), respectively.
To investigate the cellular localization of Momyo2 in M. oryzae, mycelia of the transformant expressing Momyo2::eGFP fusion protein were inoculated in liquid CM and grown overnight. Mycelia were then visualized using a confocal fluorescence microscope (Zeiss LSM710). For visualizing nuclear distribution in Momyo2 mutants, mycelia of each strain were cultured as described above, then treated with CFW and visualized by a confocal fluorescence microscope (Zeiss LSM710). To view cytoplasm in mycelia, the Momyo2 mutant and Guy11 was transformed with pMoC-eGFP vector,Citation63 and then cytoplasmic GFP signal was compared under Nikon inverted Ti-S epifluorescence microscope (Nikon Co., Tokyo, Japan).
Phenotype assays
To determine fungal growth, mycelia plugs of Guy11 and its derivative mutants were inoculated onto fresh CM, minimal agar medium (MM),Citation22 V8 agar medium (V8), oatmeal agar medium (OM),Citation50 and RDC agar medium,Citation40 respectively. Strains were incubated in dark at 28 °C for 5 d. To evaluate sporulation, conidia were harvested using 5 mL of sterilized distilled water from 14-day-old RDC agar plates. These cells were counted using a hemacytometer, and size was measured using a microscope. Conidial germination and appressorium formation were determined as described previously.Citation52 Appressorium turgor was measured by incipient cytorrhysis assay using a 1–4 molar concentration of glycerol solution as described previously.Citation65 The evaluation of porosity of appressorial wall was performed using a solute exclusion technique and screened for cytorrhysis and plasmolysis.Citation66 Cytorrhysis occurs when a solute molecule is excluded from the pores in the cell wall, whlie plasmolysis is the evidence of permeation. For evaluating the proportion of appressoria that had recovered from cytorrhysis, appressorium was treated as previously established methods and measured at 60, 120 and 180 min after incubation, respectively. All the experiments described here were independently repeated 3 times with 3 replicate each, and a representative result from one experiment is shown.
Pathogenicity assay and infectious growth observation
Pathogenicity assay were conducted following a previous established precedures by using 14-day-old rice seedlings (Oryza sativa cv CO-39) and 7-day-old barley leaves (Hordeum vulgare cv Golden Promise), respectively.Citation52 The plants were examined for disease development at 5 d after inoculation. For pathogenic test on abraded rice leaves, 20 μl drops of conidia suspension (1.0 × 105 spores mL−1) of each strain were placed on wounded rice leaves. Virulence of test strains was evaluated 5 d post inoculation.. Plant penetration assays were performed using 7-day-old barley leaves as described previously.Citation52 Invasive hyphae growth was examined after at 36 hrs and 72 hrs using a light microscopy. All experiments in this section were repeated 3 times, and representative results from one experiment are shown.
Detection of extracellular enzyme activity
The extracellular enzyme activity was measured following the previously established procedures.Citation39 The culture filtrate harvested from 2-day-old CM liquid culture was used to measure the peroxidase and laccase activity. The reaction volumes and conditions were strictly following the established procedures,Citation39 and absorbance was determined at 420 nm.
Three, 3′-Diaminobenzidine (DAB) Staining and DPI treatment
The in vivo detection of H2O2 and reactive oxygen species (ROS) was performed following previously established procedures.Citation67,68 DAB forms a reddish-brown polymer upon reaction with H2O2 and ROS produced by the plant cells. One-week-old barley leaves were inoculated with conidial suspension (1.0 × 105 spores mL−1) for 30 hrs and 48 hrs, respectively, then their leaves were cut and placed in 1 mg mL−1 DAB-HCl solution (pH 3.8). The above samples were incubated in the dark condition for additional 8 hrs to allow DAB uptake and reaction with H2O2 and ROS, then were decolorized using the ethanol-chloroform solution (4: 1) at room temperature for 2 d. Samples were mounted in 50% (v/v) glycerol and examined by a Nikon inverted Ti-S epifluorescence microscope (Nikon Co., Tokyo, Japan).
To investigate the growth of IH in ROS-suppressed barley cells, conidial drops supplemented with 0.5 mM DPI was inoculated on barley leaves.Citation37 At 30 hpi, the IH growth was evaluated by Nikon inverted Ti-S epifluorescence microscope equipped with DIC.
Domain architecture and phylogenetic analysis
The class II myosin proteins from diverse organisms were obtained from NCBI database (www.ncbi.nlm.nih.gov) using the BLAST algorithm.Citation69 Sequence alignments and phylogenetic analysis were performed using the ClustalW programCitation70 and Mega 4.0 β program,Citation71 respectively. Domain architecture was provided by the SMART online software program.Citation72
Transmission electron microscopy
For transmission electron microscopy (TEM), mycelia grown in liquid CM for 48 hour were used to fix in 2.5% (v/v) glutaraldehyde and 1% (v/v) osmium tetroxide. Sections were prepared and visualized using a H-7650 transmission electron microscope (Hitachi, Tokyo, Japan) as described previously.Citation73
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KVIR_S_1323156.zip
Download Zip (23.5 MB)Funding
This work was supported by the Foundation for the Author of National Excellent Doctoral Dissertation of PR China (Grant No: 201470 to MG), the National Natural Science Foundations of China (Grant No: 31671976 to MG), the Key Grant for Excellent Young Talents of Anhui Higher Education Institutions (gxyqZD2016037 to MG), Natural Science Foundations of Anhui province (Grant No: 1608085QC49 to MG), the Foundation for the Excellent Talents of Anhui Agricultural University (Grant No: RC2015002 to MG) and the National Science Foundation for Distinguished Young Scholars of China (Grant No. 31325022 to ZZ).
References
- Mermall V, Post PL, Mooseker MS. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science 1998; 279:527-33; PMID:9438839; https://doi.org/10.1126/science.279.5350.527
- Hartman MA, Finan D, Sivaramakrishnan S, Spudich JA. Principles of unconventional myosin function and targeting. Annu Rev Cell Dev Biol 2011; 27:133-55; PMID:21639800; https://doi.org/10.1146/annurev-cellbio-100809-151502
- Hofmann WA, Richards TA, de Lanerolle P. Ancient animal ancestry for nuclear myosin. J Cell Sci 2009; 122:636-43; PMID:19225126; https://doi.org/10.1242/jcs.030205
- McGoldrick CA, Gruver C, May GS. myoA of Aspergillus nidulans encodes an essential myosin I required for secretion and polarized growth. J Cell Biol 1995; 128:577-87; PMID:7860631; https://doi.org/10.1083/jcb.128.4.577
- Weber I, Gruber C Fau - Steinberg G, Steinberg G. A class-V myosin required for mating, hyphal growth, and pathogenicity in the dimorphic plant pathogen Ustilago maydis. Plant Cell 2003; 15:2826-42; PMID:14615599; https://doi.org/10.1105/tpc.016246
- Schmidt M, Bowers B, Varma A, Roh DH, Cabib E. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J Cell Sci 2002; 115:293-302; PMID:11839781
- Yamashita RA, May GS. Constitutive activation of endocytosis by mutation of myoA, the myosin I gene of Aspergillus nidulans. J Biol Chem 1998; 273:14644-8; PMID:9603982; https://doi.org/10.1074/jbc.273.23.14644
- Oberholzer U, Marcil A, Leberer E, Thomas DY, Whiteway M. Myosin I is required for hypha formation in Candida albicans. Eukaryotic Cell 2002; 1:213-28; PMID:12455956; https://doi.org/10.1128/EC.1.2.213-228.2002
- Motegi F, Arai R, Mabuchi I. Identification of two type V myosins in fission yeast, one of which functions in polarized cell growth and moves rapidly in the cell. Mol Biol Cell 2001; 12:1367-80; PMID:11359928; https://doi.org/10.1091/mbc.12.5.1367
- Karpova TS, Reck-Peterson SL, Elkind NB, Mooseker MS, Novick PJ, Cooper JA. Role of actin and Myo2p in polarized secretion and growth of Saccharomyces cerevisiae. Mol Biol Cell 2000; 11:1727-37; PMID:10793147; https://doi.org/10.1091/mbc.11.5.1727
- Song B, Li HP, Zhang JB, Wang JH, Gong AD, Song XS, et al. Type II myosin gene in Fusarium graminearum is required for septation, development, mycotoxin biosynthesis and pathogenicity. Fungal Genet Biol 2013; 54:60-70; PMID:23507542; https://doi.org/10.1016/j.fgb.2013.02.010
- Taheri-Talesh N, Xiong Y, Oakley BR. The functions of myosin II and myosin V homologs in tip growth and septation in Aspergillus nidulans. PLoS One 2012; 7:e31218; PMID:22359575; https://doi.org/10.1371/journal.pone.0031218
- Vallen EA, Caviston J, Bi E. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol Biol Cell 2000; 11:593-611; PMID:10679017; https://doi.org/10.1091/mbc.11.2.593
- Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, et al. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol 1998; 142:1301-12; PMID:9732290; https://doi.org/10.1083/jcb.142.5.1301
- Tolliday N, Pitcher M, Li R. Direct evidence for a critical role of myosin II in budding yeast cytokinesis and the evolvability of new cytokinetic mechanisms in the absence of myosin II. Mol Biol Cell 2003; 14:798-809; PMID:12589071; https://doi.org/10.1091/mbc.E02-09-0558
- Pollard TD, Wu JQ. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol 2000; 11:149-55; https://doi.org/10.1038/nrm2834
- Kitayama C, Sugimoto A, Yamamoto M. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J Cell Biol 1997; 137:1309-19; PMID:9182664; https://doi.org/10.1083/jcb.137.6.1309
- Pollard TD, Wu JQ. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol 2010; 11:149-55; PMID:20094054; https://doi.org/10.1038/nrm2834
- Vavylonis D, Wu JQ, Hao S, O'Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science 2008; 319:97-100; PMID:18079366; https://doi.org/10.1126/science.1151086
- Hayakawa Y, Ishikawa E, Shoji JY, Nakano H, Kitamoto K. Septum-directed secretion in the filamentous fungus Aspergillus oryzae. Mol Microbiol 2011; 81:40-55; PMID:21564341; https://doi.org/10.1111/j.1365-2958.2011.07700.x
- Canovas D, Boyce KJ, Andrianopoulos A. The fungal type II myosin in Penicillium marneffei, MyoB, is essential for chitin deposition at nascent septation sites but not actin localization. Eukaryot Cell 2011; 10:302-12; PMID:21131434; https://doi.org/10.1128/EC.00201-10
- Talbot NJ. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu Rev Microbiol 2003; 57:177-202; PMID:14527276; https://doi.org/10.1146/annurev.micro.57.030502.090957
- Xu JR, Zhao X, Dean RA. From genes to genomes: a new paradigm for studying fungal pathogenesis in Magnaporthe oryzae. Adv Genet 2007; 57:175-218; PMID:17352905
- Zhang H, Zheng X, Zhang Z. The Magnaporthe grisea species complex and plant pathogenesis. Mol Plant Pathol 2016; 17:796-804; PMID:24832137; doi: 10.1111/mpp.12342.
- Ou SH. Rice Diseases. 2nd ed. Surrey, UK: Commonwealth Mycological Institute 1985.
- Soundararajan S, Jedd G, Li X, Ramos-Pamplona M, Chua NH, Naqvi NI. Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell 2004; 16:1564-74; PMID:15155882; https://doi.org/10.1105/tpc.020677
- Shannon KB, Li R. A myosin light chain mediates the localization of the budding yeast IQGAP-like protein during contractile ring formation. Curr Biol 2000; 10:727-30; PMID:10873803; https://doi.org/10.1016/S0960-9822(00)00539-X
- Bezanilla M, Forsburg SL, Pollard TD. Identification of a second myosin-II in Schizosaccharomyces pombe: Myp2p is conditionally required for cytokinesis. Mol Biol Cell 1997; 8:2693-705; PMID:9398685; https://doi.org/10.1091/mbc.8.12.2693
- Fang X, Luo J, Nishihama R, Wloka C, Dravis C, Travaglia M, Vallen EA, Bi E. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J Cell Biol 2010; 191:1333-50; PMID:21173112; https://doi.org/10.1083/jcb.201005134
- Lister IM, Tolliday NJ, Li R. Characterization of the minimum domain required for targeting budding yeast myosin II to the site of cell division. BMC Biol 2006; 4:19; PMID:16800887; https://doi.org/10.1186/1741-7007-4-19
- Han JH, Lee HM, Shin JH, Lee YH, Kim KS. Role of the MoYAK1 protein kinase gene in Magnaporthe oryzae development and pathogenicity. Environ Microbiol 2015; 17:4672-89; PMID:26248223; https://doi.org/10.1111/1462-2920.13010
- Lee K, Singh P, Chung WC, Ash J, Kim TS, Hang L, Park S. Light regulation of asexual development in the rice blast fungus, Magnaporthe oryzae. Fungal Genet Biol 2006; 43:694-706; PMID:16765070; https://doi.org/10.1016/j.fgb.2006.04.005
- Howard RJ, Valent B. Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu Rev Microbiol 1996; 50:491-512; PMID:8905089; https://doi.org/10.1146/annurev.micro.50.1.491
- Thines E, Weber RW, Talbot NJ. MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 2000; 12:1703-18; PMID:11006342
- Howard RJ, Ferrari MA, Roach DH, Money NP. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc Natl Acad Sci U S A 1991; 88:11281-4; PMID:1837147; https://doi.org/10.1073/pnas.88.24.11281
- de Jong JC, McCormack BJ, Smirnoff N, Talbot NJ. Glycerol generates turgor in rice blast. Nature 1997; 389:244; https://doi.org/10.1038/38418
- Guo M, Guo W, Chen Y, Dong S, Zhang X, Zhang H, Song W, Wang W, Wang Q, Lv R, et al. The basic leucine zipper transcription factor Moatf1 mediates oxidative stress responses and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae. Mol Plant Microbe Interact 2010; 23:1053-68; PMID:20615116; https://doi.org/10.1094/MPMI-23-8-1053
- Cross AR, Jones OT. The effect of the inhibitor diphenylene iodonium on the superoxide-generating system of neutrophils. Specific labelling of a component polypeptide of the oxidase. Biochem J 1986; 237:111-6; PMID:3800872; https://doi.org/10.1042/bj2370111
- Chi MH, Park SY, Kim S, Lee YH. A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. Plos Pathogens 2009; 5:e1000401; PMID:19390617; https://doi.org/10.1371/journal.ppat.1000401
- Guo M, Chen Y, Du Y, Dong Y, Guo W, Zhai S, Zhang H, Dong S, Zhang Z, Wang Y, et al. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog 2011; 7:e1001302; PMID:21383978; https://doi.org/10.1371/journal.ppat.1001302
- Goodson HV, Spudich JA. Identification and molecular characterization of a yeast myosin I. Cell Motil Cytoskeleton 1995; 30:73-84; PMID:7728870; https://doi.org/10.1002/cm.970300109
- Mulvihill DP, Hyams JS. Role of the two type II myosins, Myo2 and Myp2, in cytokinetic actomyosin ring formation and function in fission yeast. Cell Motil Cytoskeleton 2003; 54:208-16; PMID:12589679; https://doi.org/10.1002/cm.10093
- Momany M, Hamer JE. Relationship of actin, microtubules, and crosswall synthesis during septation in Aspergillus nidulans. Cell Motil Cytoskeleton 1997; 38:373-84; PMID:9415379; https://doi.org/10.1002/(SICI)1097-0169(1997)38:4%3c373::AID-CM7%3e3.0.CO;2-4
- Walther A, Wendland J. Septation and cytokinesis in fungi. Fungal Genet Biol 2003; 40:187-96; PMID:14599886; https://doi.org/10.1016/j.fgb.2003.08.005
- Kim S, Park SY, Kim KS, Rho HS, Chi MH, Choi J, et al. Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet 2009; 5:e1000757; PMID:19997500; https://doi.org/10.1371/journal.pgen.1000757
- Lau GW, Hamer JE. Acropetal: a genetic locus required for conidiophore architecture and pathogenicity in the rice blast fungus. Fungal Genet Biol 1998; 24:228-39; PMID:9742203; https://doi.org/10.1006/fgbi.1998.1053
- Dixon KP, Xu JR, Smirnoff N, Talbot NJ. Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 1999; 11:2045-58; PMID:10521531; https://doi.org/10.1105/tpc.11.10.2045
- Badaruddin M, Holcombe LJ, Wilson RA, Wang ZY, Kershaw MJ, Talbot NJ. Glycogen metabolic genes are involved in trehalose-6-phosphate synthase-mediated regulation of pathogenicity by the rice blast fungus Magnaporthe oryzae. PLoS Pathog 2013; 9:e1003604; PMID:24098112; https://doi.org/10.1371/journal.ppat.1003604
- Jeon J, Goh J, Yoo S, Chi MH, Choi J, Rho HS, Park J, Han SS, Kim BR, Park SY, et al. A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Mol Plant Microbe Interact 2008; 21:525-34; PMID:18393612; https://doi.org/10.1094/MPMI-21-5-0525
- Zhang H, Liu K, Zhang X, Song W, Zhao Q, Dong Y, Guo M, Zheng X, Zhang Z. A two-component histidine kinase, MoSLN1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Curr Genet 2010; 56:517-28; PMID:20848286; https://doi.org/10.1007/s00294-010-0319-x
- Guo M, Gao F, Zhu X, Nie X, Pan Y, Gao Z. MoGrr1, a novel F-box protein, is involved in conidiogenesis and cell wall integrity and is critical for the full virulence of Magnaporthe oryzae. Appl Microbiol Biotechnol 2015; 99:8075-88; PMID:26227409; https://doi.org/10.1007/s00253-015-6820-x
- Guo M, Tan L, Nie X, Zhu X, Pan Y, Gao Z. The Pmt2p-mediated protein O-Mannosylation is required for morphogenesis, adhesive properties, cell wall integrity and full virulence of Magnaporthe oryzae. Frontiers Microbiol 2016; 7:630; PMID:27199956
- Yin Z, Tang W, Wang J, Liu X, Yang L, Gao C, Zhang J, Zhang H, Zheng X, Wang P, et al. Phosphodiesterase MoPdeH targets MoMck1 of the conserved mitogen-activated protein (MAP) kinase signalling pathway to regulate cell wall integrity in rice blast fungus Magnaporthe oryzae. Mol Plant Pathol 2016; 17:654-68; PMID:27193947; https://doi.org/10.1111/mpp.12317
- Molina L, Kahmann R. An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 2007; 19:2293-309; PMID:17616735; https://doi.org/10.1105/tpc.107.052332
- Guimaraes SC, Schuster M, Bielska E, Dagdas G, Kilaru S, Meadows BR, Schrader M, Steinberg G. Peroxisomes, lipid droplets, and endoplasmic reticulum “hitchhike” on motile early endosomes. J Cell Biol 2015; 211:945-54; PMID:26620910; https://doi.org/10.1083/jcb.201505086
- Schuster M, Treitschke S, Kilaru S, Molloy J, Harmer NJ, Steinberg G. Myosin-5, kinesin-1 and myosin-17 cooperate in secretion of fungal chitin synthase. EMBO J 2012; 31:214-27; PMID:22027862; https://doi.org/10.1038/emboj.2011.361
- Wilson RA, Talbot NJ. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol 2009; 7:185-95; PMID:19219052; https://doi.org/10.1038/nrmicro2032
- Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 2006; 312:580-3; PMID:16645096; https://doi.org/10.1126/science.1124550
- Liu XH, Lu JP, Zhang L, Dong B, Min H, Lin FC. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryotic Cell 2007; 6:997-1005; PMID:17416896; https://doi.org/10.1128/EC.00011-07
- Du Y, Zhang H, Hong L, Wang J, Zheng X, Zhang Z. Acetolactate synthases MoIlv2 and MoIlv6 are required for infection-related morphogenesis in Magnaporthe oryzae. Mol Plant Pathol 2013; 14:870-84; PMID:23782532; https://doi.org/10.1111/mpp.12053
- Liu YG, Chen Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 2007; 43:649-50, 52, 54 passim; PMID:18072594; https://doi.org/10.2144/000112601
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY:Cold Spring Harbor Laboratory Press, 1989.
- Guo M, Zhu X, Li H, Tan L, Pan Y. Development of a novel strategy for fungal transformation based on a mutant locus conferring carboxin-resistance in Magnaporthe oryzae. AMB Express 2016; 6:57; PMID:27558019
- Chen XL, Yang J, Peng YL. Large-scale insertional mutagenesis in Magnaporthe oryzae by Agrobacterium tumefaciens-mediated transformation. Methods Mol Biol 2011; 722:213-24; PMID:21590424
- Zhang H, Zhao Q, Liu K, Zhang Z, Wang Y, Zheng X. MgCRZ1, a transcription factor of Magnaporthe grisea, controls growth, development and is involved in full virulence. FEMS Microbiol Lett 2009; 293:160-9; PMID:19260966
- Money NP. Measurement of pore size in the hyphal cell wall of Achlya bisexualis. Exp Mycol 1990; 14:234-42
- Thordal-Christensen H, Zhang ZG, Wei YD, Collinge DB. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 1997; 11:1187-94
- Liu G, Greenshields DL, Sammynaiken R, Hirji RN, Selvaraj G, Wei Y. Targeted alterations in iron homeostasis underlie plant defense responses. J Cell Sci 2007; 120:596-605; PMID:17244651
- McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 2004; 32:W20-5; PMID:15215342
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22:4673-80; PMID:7984417
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24:1596-9
- Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 2014; 43:D257-60; PMID:25300481
- Xu YB, Li HP, Zhang JB, Song B, Chen FF, Duan XJ, Xu HQ, Liao YC. Disruption of the chitin synthase gene CHS1 from Fusarium asiaticum results in an altered structure of cell walls and reduced virulence. Fungal Genetics Biol 2010; 47:205-15; PMID:19941967
