ABSTRACT
Streptococcus pneumoniae is a leading cause of bacterial meningitis. Here, we investigated whether pneumococcal paralogous zinc metalloproteases contribute to meningitis onset. Findings of codon-based phylogenetic analyses indicated 3 major clusters in the Zmp family; ZmpA, ZmpC, and ZmpB, with ZmpD as a subgroup. In vitro invasion assays of human brain microvascular endothelial cells (hBMECs) showed that deletion of the zmpC gene in S. pneumoniae strain TIGR4 significantly increased bacterial invasion into hBMECs, whereas deletion of either zmpA or zmpB had no effect. In a mouse meningitis model, the zmpC deletion mutant exhibited increased invasion of the brain and was associated with increased matrix metalloproteinase-9 in plasma and mortality as compared with the wild type. We concluded that ZmpC suppresses pneumococcal virulence by inhibiting bacterial invasion of the central nervous system. Furthermore, ZmpC illustrates the evolutional theory stating that gene duplication leads to acquisition of novel function to suppress excessive mortality.
Introduction
Streptococcus pneumoniae is a leading cause of pneumonia, sepsis, and meningitis, though the organism usually colonizes the human oral cavity or nasopharynx as a commensal resident. Notably, pneumococcal meningitis has a higher rate of incidence as compared with other major meningitis pathogens, such as Neisseria meningitidis and Haemophilus influenzae.Citation1 S. pneumoniae belongs to the mitis group, based on its 16S rRNA sequence classification, and all mitis group species except S. pneumoniae are known to be non-pathogenic commensal colonizers of the oral cavity or upper respiratory tract. Kilian et al. suggested that Streptococcus mitis and S. pneumoniae evolved through 2 opposing processes occurring in parallel under selective pressure, in which non-pathogenic S. mitis stabilized a reduced genome devoid of various virulence factors, whereas S. pneumoniae imported genes from related streptococci with genomic flexibility.Citation2 Although introduction of pneumococcal conjugate vaccines has significantly reduced the incidence of pneumococcal meningitis in individuals of all ages in the United States, S. pneumoniae remains a major cause of bacterial meningitis and is responsible for 2-thirds of all meningitis cases.Citation3 In addition, administration of pneumococcal conjugate vaccines could induce selective pressure and non-vaccine pneumococcus serotypes have increased throughout the world since introduction of those vaccines.Citation4,5
During the process of meningitis development, S. pneumoniae organisms penetrate the blood-brain barrier (BBB) and invade the brain mainly via the bloodstream. Although the complete mechanism remains to be identified, NanA and CbpA have been reported as pneumococcal adhesins used by S. pneumoniae to cross the BBB. NanA localizes on bacterial cell surfaces via its cell-wall anchoring motif and contributes to pneumococcal invasion into human brain microvascular endothelial cells (hBMECs),Citation6, 7 while its LamG domain promotes pneumococcal internalization into hBMECs via inflammatory cytokine production by the cells and resulting cell activation.Citation7 CbpA also localizes on bacterial cell surface proteins via choline binding repeats and interacts with 2 host receptors, laminin and poly-immunoglobulin receptors, on host brain endothelial cells. In cases of pneumococcal meningitis, the levels of interferon-γ, monocyte chemoattractant protein-1, and matrix metalloproteinase-9 (MMP-9) are significantly elevated as compared with cases of meningitis caused by N. meningitidis or H. influenzae.Citation8,9
Gene duplication is considered to be a major force in the evolution of novel gene functions.Citation10-12 Although several theoretical models have been proposed, gene duplication is generally considered to allow for functional changes of the original and/or new copy under relaxed purifying selection. It has also been speculated that preserved paralogous genes play important roles for bacterial survival, as duplicated genes are most likely to be lost.Citation11 Since S. pneumoniae contains 2 to 4 paralogous zinc metalloproteases (IgA1 protease/ZmpA, ZmpB, ZmpC, and ZmpD), investigation of those should be helpful for revealing pneumococcal evolutional history in a host environment. Various functions of pneumococcal zinc-metalloproteases have been reported, of which ZmpA is known as an IgA1 protease, while ZmpB, ZmpC, and ZmpD do not have IgA1 protease activity. The zmpB gene is present in virtually all mitis and salivarius groups, except for Streptococcus thermophiles and some Streptococcus oralis strains. A previous minimum evolutional phylogenetic analysis of 297 zmp sequences and representative housekeeping genes indicated that zmpB is the ancestral gene predating the evolution of today's humanoid species.Citation13 zmpD sequences form a subgroup within the zmpB cluster, and it has been suggested that zmpD originated from an ancient duplication of the zmpB gene in a common ancestor of S. pneumoniae and S. mitis. Although the substrate specificities of ZmpB and ZmpD remain unknown,Citation13 various functions of ZmpC have been reported. For example, ZmpC was found to cleave to human matrix metalloproteinase 9, while intranasal infection with a zmpC-mutant in a mouse pneumonia model significantly reduced mortality as compared with infection with wild-type strains.Citation14 In addition, ZmpC is known to cause shedding of syndecan-1, a type I transmembrane heparan sulfate proteoglycan.Citation15 In other studies, TIGR4 zmpA, zmpB, and zmpC mutant strains showed reduced virulence as compared with a wild-type strain in mice following intranasal infection.Citation14,16 However, the role of the zmp family in pneumococcal meningitis remains unknown.
In this study, we investigated whether zmp family members contribute to development of pneumococcal meningitis. Our in vitro findings indicated that zmpA and zmpB have limited effects on bacterial invasion of hBMECs, whereas zmpC inhibited invasion. Furthermore, in a mouse meningitis model, a zmpC mutant strain showed a significantly greater number of colony forming units (CFUs) in the brain and greater virulence. Our findings indicate that pathogenic bacteria retain a gene that functions to suppress virulence, suggesting that a reduction in host mortality is related to their success.
Results and discussion
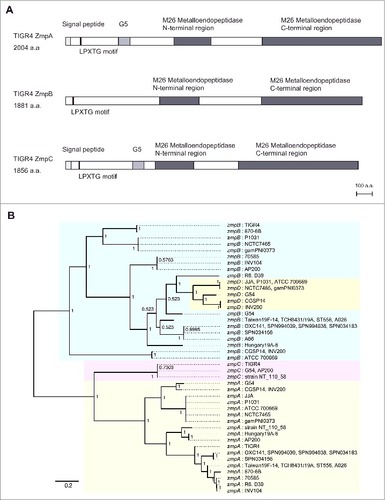
S. pneumoniae TIGR4 contains ZmpA, ZmpB, and ZmpC, and amino acid sequence analysis showed that Zmp proteins possess an LPXTG motif in the N-terminus and M26 metalloendopeptidase domains. In addition, ZmpA and ZmpC contain a deduced signal peptidase cleavage site and G5 domain, whereas ZmpB does not (). To examine the relationship among the pneumococcal Zmp sequences, we performed codon-based Bayesian and maximum likelihood phylogenetic analyses using 63 pneumococcal zmp sequences, which revealed similar patterns of genetic classification with high posterior probabilities or bootstrap values ( and Fig. S1). Both trees indicated the presence of 3 clusters, ZmpA, ZmpC, and ZmpB, while ZmpD was classified as a subgroup of ZmpB, as indicated by the results of a previously performed nucleotide-based minimum evolution algorithm analysis.Citation13 To investigate the phylogenetic relationship among pneumococcal and other bacterial zmp genes, we also performed codon-based Bayesian and maximum likelihood phylogenetic analyses using 132 streptococcal zmp sequences. Those results indicated that oral commensal bacteria contain zmpC orthologues and the zmpC gene of Streptococcus suis was found to be located at a more genetically distant position as compared with pneumococcal zmpC (Figs. S2 and S3).
Figure 1. Phylogenetic analysis of zmp family. (A) Schematic illustration of domains in S. pneumoniae TIGR4 ZmpA, ZmpB, and ZmpC. Scale bar = 100 amino acids. (B) Codon-based Bayesian phylogenetic tree of zmpA, zmpB, zmpC, and zmpD genes. Additional information regarding these bacterial strains is presented in Table S5. Strains with identical sequences are listed on the same branch. The percentage of posterior probabilities is shown near the nodes. The scale bar indicates nucleotide substitutions per site. S. pneumoniae zmpA, zmpB, zmpC, and zmpD genes are shaded in yellow, blue, red, and green, respectively. The tree is unrooted, though presented as midpoint rooted for clarity
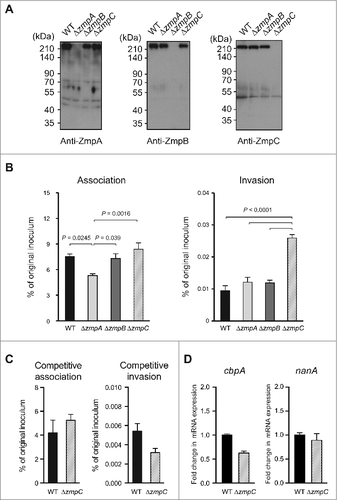
To investigate the role of Zmps in bacterial meningitis, we constructed isogenic zmpA, zmpB, and zmpC mutants of the S. pneumoniae TIGR4 strain. Zmp proteins contain an LPXTG motif in the N-terminus. However, a typical LPXTG motif functions as a cell wall anchoring motif in the C-terminus, followed by a stretch of hydrophobic and positively charged amino acid residues,Citation17 while Zmp proteins were reported to be secreted into culture supernatant.Citation18 Our Western blot analysis also indicated that ZmpA, ZmpB, and ZmpC become localized in pneumococcal supernatant. On the other hand, the zmpA, zmpB, and zmpC mutant strains lacked those protein expressions and did not have effects on expressions of the other Zmp family members (). In addition, there were no remarkable differences in regard to minimum inhibitory concentration and minimum bactericidal concentration among the strains (Table S1). In bacterial broth medium, the zmpC mutant strain showed slightly faster growth, while that of the zmpA mutant strain was slightly slower as compared with the other strains (Fig. S4). To examine the role of pneumococcal ZmpA, ZmpB, and ZmpC in invasion of BBB endothelium, we performed association/invasion assays using hBMECs (). To quantify pneumococcal invasion, hBMECs were incubated with the pneumococcal strains for 1 hour without antibiotics and then for 1 hour in medium containing antibiotics. There were no differences in regard to adherence and invasion phenotypes between the wild-type and zmpB mutant strains with hBMECs, while the association of the zmpA mutant strain was slightly decreased as compared with that of the other strains. On the other hand, the rate of invasion of hBMECs by the zmpC mutant strain was significantly higher as compared with the wild-type strain. These results indicated that pneumococcal ZmpC, but not ZmpA or ZmpB, can inhibit pneumococcal invasion of brain endothelial cells. Next, we performed competitive association and invasion assays by co-infection of the wild-type and zmpC mutant strains. In the competition assay, the zmpC mutant strain infected ZmpC-affected hBMECs because of co-infection with the wild-type strain. Our results showed no significant differences in regard to association and invasion (). Furthermore, the differences between the wild-type and zmpC mutant strains were less than 2-fold in regard to expressions of the nanA and cbpA genes (). These results indicate that the higher invasion rate of the zmpC mutant strain is not dependent on a different of bacterial cell ability, such as growth or adhesin expression pattern. In addition, there was no significant difference, but a tendency toward an increased amount of cleaved syndecan-1 in the wild-type strain-infected hBMECs (Fig. S5).
Figure 2. Rates of S. pneumoniae association with and invasion of hBMECs. (A). Culture supernatants of S. pneumoniae strain TIGR4 and its isogenic mutant strains were analyzed by Western blotting using antisera against ZmpA, ZmpB, and ZmpC. (B) S. pneumoniae strain TIGR4 and its isogenic mutant strains were examined for their association and invasion activities. Data are presented as the mean values of 6 samples, with SE values are represented by vertical lines. Differences between groups were analyzed using one-way ANOVA followed by Tukey's multiple comparisons test. (C) S. pneumoniae strain wild-type (WT) and ΔzmpC strains were examined for their competitive association and invasion activities. Data are presented as the mean values of 6 samples, with SE values are represented by vertical lines. (D) The mRNA expression of S. pneumoniae ΔzmpC strain relative to that of the WT strain was examined by quantitative PCR, with 16s rRNA was used as an internal control. SE values are represented by vertical lines. Data were pooled and normalized from 3 independent experiments, each performed in quadruplicate
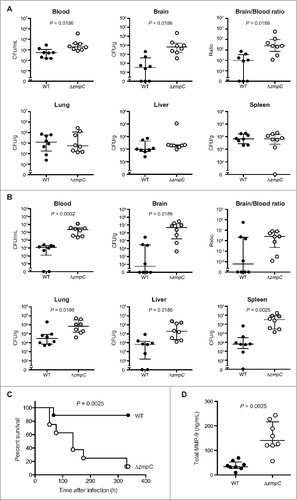
Finally, to investigate the role of ZmpC in vivo in a meningitis model, we intravenously infected mice with pneumococcal strains, then compared bacterial CFUs in blood and brain samples at 12, 24, and 36 hours after infection. In addition, we compared the CFUs of bacteria isolated from the lungs, livers, and spleens at 12 and 36 hours after infection. All infected mice showed piloerection after infection. At 12 hours after infection, the numbers of CFUs of both the wild-type and zmpC mutant strains in the lung, liver, and spleen samples were comparable. However, in the brain samples, the zmpC mutant strain-infected mice showed an approximately 100-fold greater number of CFUs as compared with the wild-type strain-infected mice, while blood samples from zmpC mutant strain-infected mice showed an approximately 8-fold greater number. Also, the mean ratio for CFUs in the brain and blood of zmpC mutant-infected mice was significantly higher as compared with the wild-type-infected mice (). At 24 hours after infection, the same tendency for mean ratio for CFU count in the brain and blood samples was seen (Fig. S6). At 36 hours after infection, 2 mice in the zmpC mutant strain-infection group had died. Furthermore, bacteria were not detected in the blood of 2 of the wild-type strain-infected mice, while the zmpC mutant strain-infected mice showed significantly higher CFU counts in blood and all examined organs (), and also showed significantly higher mortality (). One or 2 of the survived mice showed abnormal behavior, such as continuous turning to the right. We considered that our results were mainly due to the greater level of brain invasion by the zmpC mutant strain during the early infection phase, which worsened the systemic condition. Total MMP-9 ELISA findings showed a significantly greater amount in plasma of the zmpC mutant strain-infected mice at 12 hours after infection (). We concluded that ZmpC inhibits pneumococcal invasion into host brain tissues as well as virulence following infection.
Figure 3. ZmpC decreases pneumococcal invasion into the brain and pathogenesis in vivo. (A, B). Mice were intravenously infected with ∼1.2 × 106 CFUs of S. pneumoniae TIGR4 wild-type or ΔzmpC. All mice were perfused with PBS after blood collection, then organ samples were collected at 12 (A) and 36 (B) hours after infection. Gray circles indicate dead mice. Median and interquartile range are represented by lines. Differences between groups were analyzed using a Kolmogorov-Smirnov test. (C) Mice were intravenously infected with ∼1.5 × 106 CFUs of S. pneumoniae TIGR4 wild-type or ΔzmpC. Mouse survival was monitored for 14 d. Differences between groups were analyzed using a Log-rank test. (D) Mice were intravenously infected with ∼1.2 × 106 CFUs of S. pneumoniae TIGR4 wild-type or ΔzmpC. Total MMP-9 amounts were examined by ELISA using mice plasma samples collected at 12 hours after infection
The presence of the zmpC gene was found to be mainly associated with serotype 8, 4, 33A/F, and 11A/D strains in Italy and the Netherlands.Citation18,19 In the present study, we examined ZmpC protein expression in Japanese clinical strains using western blot analysis and found that 19 of 167 strains expressed ZmpC (Table S2). That expression was significantly associated with serotype 4 strains (7 of 7), while some serotype 14, 15A, 11A, 33, 6B, 23F, and 19A strains also expressed the protein. Meanwhile, there were no large differences among the strains isolated from different sites (Table S3).
Pneumococcal ZmpC directly sheds a heparan sulfate proteoglycan from syndecan-1 ectodomains.Citation15 On the other hand, interactions between serotype 19F S. pneumoniae and heparan sulfate mediate pneumococcal invasion of nasopharyngeal epithelial cells.Citation20 Heparan sulfate chains on the surface of brain capillary cells also function as a receptor for invasion by Streptococcus agalactiae into the host brain.Citation21 S. agalactiae does not contain zmpC homologs, though it is possible that ZmpC inhibits BBB penetration and CNS infection through syndecan-1 shedding. An additional speculated mechanism is possible ZmpC cleavage with or reduced expression of MMP-9, which is a risk factor for pneumococcal meningitis.Citation8,14 MMP-9 modulates opening of the BBB during inflammation, and inhibitors of matrix metalloproteinases including MMP-9 have been found to be associated with reduced mortality rates and inflammatory cytokines in experimental meningitis cerebrospinal fluid samples.Citation14,22,23 Although the actual effect of ZmpC-cleavage with MMP-9 remains unknown, the present zmpC mutant strain-infected mice showed significantly higher MMP-9 amounts in plasma. It is possible that ZmpC inhibits pneumococcal invasion into the central nervous system via interactions with host syndecan-1 and MMP-9.
Previous studies reported that a TIGR4 zmpC mutant strain showed reduced or similar virulence as compared with a wild-type strain in assays following intranasal infection.Citation14,16 The discrepancy between those previous results and our findings is likely due to the different route of infection used. The host environments encountered by bacteria in the respiratory tract and blood are largely different. In addition, some host molecules play distinct roles based on their anatomic location. For example, previous studies reported inconsistent roles of MMP-9 in pneumococcal infection, as it was reported to play a protective role in intranasal pneumococcal infection, and also found to be critical for effective bacterial phagocytosis and reactive oxygen species generation by neutrophils.Citation24 On the other hand, it has also been shown that MMP-9 contributes to disrupt the BBB and is involved in meningitis onset.Citation25 Thus, several factors including MMP-9 may have had effects on the different findings.
Gene duplications and the following selective pressure play important roles in the evolution of novel gene functions. No common substrate for paralogous Zmp proteases has been reported. IgA1 protease activity is specific for ZmpA, while ZmpC has a unique ability to cleave MMP-9 and Syndecan-1.Citation13-15 In the present study, only the zmpC mutant strain showed increased pneumococcal invasion into hBMECs. The limited sequence diversification of the zmpC gene indicates that the gene was acquired later than other zmp genes in S. pneumoniae. Our results provide an actual example of the evolution of duplicated genes to obtain different roles. Generally, excess virulence is disadvantageous for bacterial success, as a reduction in the period of infection caused by host death limits the number of bacteria reproduced. Standard predictions based on virulence theory assume that pathogens evolve to maximize their reproductive number. From a virulence evolutionary point of view, it is plausible that ZmpC suppresses host mortality following intravenous infection. The zmpC gene was previously found to be conserved among non-pathogenic mitis group streptococci, including all Streptococcus sanguinis and most S. mitis strains,Citation13 which indirectly supports our speculation. The present results suggest that zmp gene duplication contributes to the evolutional survival strategy of S. pneumoniae.
Materials and methods
Bacterial strains, cell lines, and reagents
Streptococcus pneumoniae strains were provided by the Pathogenic Microbes Repository Unit, RIMD, Osaka University, and cultured in Todd-Hewitt broth (BD Biosciences) supplemented with or without 0.2% yeast extract (BD Biosciences) (THY medium) at 37°C. An Escherichia coli strain, XL10-Gold, was used as a host for derivatives of plasmids pQE30. E. coli strains were cultured in Luria-Bertani broth at 37°C with agitation. For selection and maintenance of mutants, antibiotics were added to medium at the following concentrations: ampicillin (Wako), 100 µg/ml for E. coli; spectinomycin (Wako), 120 µg/ml for S. pneumoniae. The TIGR4 isogenic mutant strain was constructed using a double crossover recombination technique, as described previously.Citation26 All mutations were confirmed by PCR findings of genomic DNA and Western blotting. The primers used are shown in Table S4. Human brain endothelial cells (hBMECs) were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS, 10% NuSerum (BD), and 1% MEM nonessential amino acids, and incubated at 37°C in 5% CO2.
Phylogenetic analyses
Phylogenetic analyses were performed as described previously.Citation27 Details are shown in the Supplementary information section.
hBMEC association and invasion assays
Pneumococcal association with and invasion of hBMECs was quantified as described previously.Citation21,27,28 S. pneumoniae strains were grown to the mid-log phase (OD600 = 0.4) and resuspended in PBS. In each well, ∼2.0 × 106 CFUs of S. pneumoniae was added for infection of ∼2.0 × 105 hBMECs at a multiplicity of infection (MOI) of 10, then the plate was centrifuged to initiate contact. To determine bacterial association, the infected cells were incubated for 1 hour and washed 3 times with PBS, then harvested with a solution containing trypsin and 0.025% Triton X-100. The number of bacteria showing an association was quantified by serial dilution plating. To examine bacterial invasion, hBMECs were washed following 1 hour of incubation, then medium containing 100 µg/mL of gentamicin was added and the cells were incubated for an additional 1 hour. Next, the cells were washed and lysed, and the number of invaded bacteria was quantified. Bacterial association and invasion rates were calculated by dividing the number of bacteria showing association/invasion by the number of original inoculums. For competitive association and invasion assays, a 1:1 ratio was obtained by mixing equal volumes of wild-type and ΔzmpC strains resuspended in PBS. Thereafter, total and mutant strain CFUs were determined by serial dilution plating on TS blood agar with or without spectinomycin. The CFU number for the wild-type strain was calculated by subtracting that of the mutant strain from total CFUs.
ELISA
To determine the amount of cleaved human syndecan-1, supernatant from cultured hBMECs was collected at 1 hour after pneumococcal infection, which was performed as well as an hBMEC association assay. Cleaved human syndecan-1 was quantified using an sCD138 ELISA Kit (Diaclone). To measure the amount of MMP-9, blood aliquots were collected from mice following induction of general euthanasia at 12 hours after infection. After adding heparin, mice plasma was obtained by centrifuging the heparinized blood. Total MMP-9 amount in mice plasma was measured using a Mouse Total MMP-9 Quantikine ELISA Kit (R&D systems).
Western blot analysis
Recombinant proteins for mouse immunization were constructed using primers (Table S4) and a pQE30 vector. Mouse antisera against ZmpA, ZmpB, and ZmpC were raised by immunizing BALB/c mice with purified recombinant proteins, as described previously.Citation29 Details of the Western blot analysis method are shown in the supplementary material online.
Mouse infection assays
All mouse experiments were conducted in accordance with animal protocols approved by the Animal Care and Use Committee of Osaka University Graduate School of Dentistry (24–025–2). The ethical end point was defined as the point in time when mice were unable to move spontaneously or respond to gentle physical stimulation. CD-1 (ICR: IGS) mice (6 weeks, female; Oriental) were infected with 1.2–1.5 × 106 CFUs of S. pneumoniae via the tail vein. At 12, 24, and 36 hours after infection, blood aliquots were collected from mice following induction of general euthanasia. Brain/meninges, lung, liver, and spleen samples were collected following perfusion with PBS. All brain, lung, and spleen whole tissues, and the anterior segment of the liver were resected. Bacterial counts in blood as well as brain, lung, liver, and spleen homogenates were determined by separate plating of serial dilutions, with those of the organs corrected for differences in organ weight. Detection limits were 50 CFU/organ or 50 CFU/mL in blood. Mouse survival was monitored twice daily for 14 d.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KVIR_S_1328333.pdf
Download PDF (2 MB)Acknowledgments
This work was supported by GSK Japan under a GSK Japan Research Grant 2015 G-2, the Asahi Glass Foundation, and the Japan Society for the Promotion of Science (JSPS) under a Grant-in-Aid for challenging Exploratory Research 16K15787, Grant-in-Aid for Young Scientists (A) 17H05103, and Grants-in-Aid for Scientific Research (B) 15H05012 and 16H05847.
References
- Castelblanco RL, Lee M, Hasbun R. Epidemiology of bacterial meningitis in the USA from 1997 to 2010: a population-based observational study. Lancet Infect Dis 2014; 14:813-9; PMID:25104307; https://doi.org/https://doi.org/10.1016/S1473-3099(14)70805-9
- Kilian M, Riley DR, Jensen A, Bruggemann H, Tettelin H. Parallel evolution of Streptococcus pneumoniae and Streptococcus mitis to pathogenic and mutualistic lifestyles. MBio 2014; 5:e01490-14; PMID:25053789; https://doi.org/https://doi.org/10.1128/mBio.01490-14
- McIntyre PB, O'Brien KL, Greenwood B, van de Beek D. Effect of vaccines on bacterial meningitis worldwide. Lancet 2012; 380:1703-11; PMID:23141619; https://doi.org/https://doi.org/10.1016/S0140-6736(12)61187-8
- Golubchik T, Brueggemann AB, Street T, Gertz RE, Jr, Spencer CC, Ho T, Giannoulatou E, Link-Gelles R, Harding RM, Beall B, et al. Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat Genet 2012; 44:352-5; PMID:22286217; https://doi.org/https://doi.org/10.1038/ng.1072
- Flasche S, Van Hoek AJ, Sheasby E, Waight P, Andrews N, Sheppard C, George R, Miller E. Effect of pneumococcal conjugate vaccination on serotype-specific carriage and invasive disease in England: a cross-sectional study. PLoS Med 2011; 8:e1001017; PMID:21483718; https://doi.org/https://doi.org/10.1371/journal.pmed.1001017
- Uchiyama S, Carlin AF, Khosravi A, Weiman S, Banerjee A, Quach D, Hightower G, Mitchell TJ, Doran KS, Nizet V. The surface-anchored NanA protein promotes pneumococcal brain endothelial cell invasion. J Exp Med 2009; 206:1845-52; PMID:19687228; https://doi.org/https://doi.org/10.1084/jem.20090386
- Banerjee A, Van Sorge NM, Sheen TR, Uchiyama S, Mitchell TJ, Doran KS. Activation of brain endothelium by pneumococcal neuraminidase NanA promotes bacterial internalization. Cell Microbiol 2010; 12:1576-88; PMID:20557315; https://doi.org/https://doi.org/10.1111/j.1462-5822.2010.01490.x
- Grandgirard D, Gaumann R, Coulibaly B, Dangy JP, Sie A, Junghanss T, Schudel H, Pluschke G, Leib SL. The causative pathogen determines the inflammatory profile in cerebrospinal fluid and outcome in patients with bacterial meningitis. Mediators Inflamm 2013; 2013:312476; PMID:23864766; https://doi.org/https://doi.org/10.1155/2013/312476
- McGill F, Heyderman RS, Panagiotou S, Tunkel AR, Solomon T. Acute bacterial meningitis in adults. Lancet 2016; 388:3036-47; PMID:27265346; https://doi.org/https://doi.org/10.1016/S0140-6736(16)30654-7
- Zhang J. Evolution by gene duplication: an update. Trends Ecol Evolution 2003; 18:292-8; https://doi.org/https://doi.org/10.1016/S0169-5347(03)00033-8
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 2010; 11:97-108; PMID:20051986; https://doi.org/https://doi.org/10.1038/nrg2689
- Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nat Rev Genet 2015; 16:472-82; PMID:26184597; https://doi.org/https://doi.org/10.1038/nrg3962
- Bek-Thomsen M, Poulsen K, Kilian M. Occurrence and evolution of the paralogous zinc metalloproteases IgA1 protease, ZmpB, ZmpC, and ZmpD in Streptococcus pneumoniae and related commensal species. MBio 2012; 3:e00303-12; https://doi.org/https://doi.org/10.1128/mBio.00303-12
- Oggioni MR, Memmi G, Maggi T, Chiavolini D, Iannelli F, Pozzi G. Pneumococcal zinc metalloproteinase ZmpC cleaves human matrix metalloproteinase 9 and is a virulence factor in experimental pneumonia. Mol Microbiol 2003; 49:795-805; PMID:12864860; https://doi.org/https://doi.org/10.1046/j.1365-2958.2003.03596.x
- Chen Y, Hayashida A, Bennett AE, Hollingshead SK, Park PW. Streptococcus pneumoniae sheds syndecan-1 ectodomains through ZmpC, a metalloproteinase virulence factor. J Biol Chem 2007; 282:159-67; PMID:17098735; https://doi.org/https://doi.org/10.1074/jbc.M608542200
- Chiavolini D, Memmi G, Maggi T, Iannelli F, Pozzi G, Oggioni MR. The three extra-cellular zinc metalloproteinases of Streptococcus pneumoniae have a different impact on virulence in mice. BMC Microbiol 2003; 3:14; PMID:12841855; https://doi.org/https://doi.org/10.1186/1471-2180-3-14
- Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev 1999; 63:174-229; PMID:10066836
- Camilli R, Pettini E, Del Grosso M, Pozzi G, Pantosti A, Oggioni MR. Zinc metalloproteinase genes in clinical isolates of Streptococcus pneumoniae: association of the full array with a clonal cluster comprising serotypes 8 and 11A. Microbiology 2006; 152:313-21; PMID:16436419; https://doi.org/https://doi.org/10.1099/mic.0.28417-0
- Cremers AJ, Kokmeijer I, Groh L, de Jonge MI, Ferwerda G. The role of ZmpC in the clinical manifestation of invasive pneumococcal disease. Int J Med Microbiol 2014; 304:984-9; PMID:25023076; https://doi.org/https://doi.org/10.1016/j.ijmm.2014.06.005
- Tonnaer EL, Hafmans TG, Van Kuppevelt TH, Sanders EA, Verweij PE, Curfs JH. Involvement of glycosaminoglycans in the attachment of pneumococci to nasopharyngeal epithelial cells. Microbes Infect 2006; 8:316-22; PMID:16239116; https://doi.org/https://doi.org/10.1016/j.micinf.2005.06.028
- Chang YC, Wang Z, Flax LA, Xu D, Esko JD, Nizet V, Baron MJ. Glycosaminoglycan binding facilitates entry of a bacterial pathogen into central nervous systems. PLoS Pathog 2011; 7:e1002082; PMID:21731486; https://doi.org/https://doi.org/10.1371/journal.ppat.1002082
- Liechti FD, Grandgirard D, Leppert D, Leib SL. Matrix metalloproteinase inhibition lowers mortality and brain injury in experimental pneumococcal meningitis. Infect Immun 2014; 82:1710-8; PMID:24491581; https://doi.org/https://doi.org/10.1128/IAI.00073-14 https://doi.org/10.1128/IAI.01799-14
- Liechti FD, Bachtold F, Grandgirard D, Leppert D, Leib SL. The matrix metalloproteinase inhibitor RS-130830 attenuates brain injury in experimental pneumococcal meningitis. J Neuroinflammation 2015; 12:43; PMID:25890041; https://doi.org/https://doi.org/10.1186/s12974-015-0257-0
- Hong JS, Greenlee KJ, Pitchumani R, Lee SH, Song LZ, Shan M, Chang SH, Park PW, Dong C, Werb Z, et al. Dual protective mechanisms of matrix metalloproteinases 2 and 9 in immune defense against Streptococcus pneumoniae. J Immunol 2011; 186:6427-36; PMID:21508260; https://doi.org/https://doi.org/10.4049/jimmunol.1003449
- Vafadari B, Salamian A, Kaczmarek L. MMP-9 in translation: from molecule to brain physiology, pathology, and therapy. J Neurochem 2016; 139 Suppl 2:91-114; PMID:26525923; https://doi.org/https://doi.org/10.1111/jnc.13415
- Bricker AL, Camilli A. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol Lett 1999; 172:131-5; PMID:10188240; https://doi.org/https://doi.org/10.1111/j.1574-6968.1999.tb13460.x
- Yamaguchi M, Hirose Y, Nakata M, Uchiyama S, Yamaguchi Y, Goto K, Sumitomo T, Lewis AL, Kawabata S, Nizet V. Evolutionary inactivation of a sialidase in group B Streptococcus. Sci Rep 2016; 6:28852; PMID:27352769; https://doi.org/https://doi.org/10.1038/srep28852
- Yamaguchi M, Terao Y, Mori Y, Hamada S, Kawabata S. PfbA, a novel plasmin- and fibronectin-binding protein of Streptococcus pneumoniae, contributes to fibronectin-dependent adhesion and antiphagocytosis. J Biol Chem 2008; 283:36272-9; PMID:18974092; https://doi.org/https://doi.org/10.1074/jbc.M807087200
- Nakata M, Kimura KR, Sumitomo T, Wada S, Sugauchi A, Oiki E, Higashino M, Kreikemeyer B, Podbielski A, Okahashi N, et al. Assembly mechanism of FCT region type 1 pili in serotype M6 Streptococcus pyogenes. J Biol Chem 2011; 286:37566-77; PMID:21880740; https://doi.org/https://doi.org/10.1074/jbc.M111.239780
