ABSTRACT
Previous and recent investigations on the innate immune response of Drosophila have identified certain mechanisms that promote pathogen elimination. However, the function of Thioester-containing proteins (TEPs) in the fly still remains elusive. Recently we have shown the contribution of TEP4 in the antibacterial immune defense of Drosophila against non-pathogenic E. coli, and the pathogens Photorhabdus luminescens and P. asymbiotica. Here we studied the function of Tep genes in both humoral and cellular immunity upon E. coli and Photorhabdus infection. We found that while Tep2 is induced after Photorhabdus and E. coli infection; Tep6 is induced by P. asymbiotica only. Moreover, functional ablation of hemocytes results in significantly low transcript levels of Tep2 and Tep6 in response to Photorhabdus. We show that Tep2 and Tep6 loss-of-function mutants have prolonged survival against P. asymbiotica, Tep6 mutants survive better the infection of P. luminescens, and both tep mutants are resistant to E. coli and Photorhabdus. We also find a distinct pattern of immune signaling pathway induction in E. coli or Photorhabdus infected Tep2 and Tep6 mutants. We further show that Tep2 and Tep6 participate in the activation of hemocytes in Drosophila responding to Photorhabdus. Finally, inactivation of Tep2 or Tep6 affects phagocytosis and melanization in flies infected with Photorhabdus. Our results indicate that distinct Tep genes might be involved in different yet crucial functions in the Drosophila antibacterial immune response.
Introduction
Drosophila melanogaster has served as an excellent model system to study innate immune defense mechanisms against microbial infections.Citation1 To detect different types of pathogens, the fly uses specific pattern recognition receptors such as peptidoglycan recognition receptors, Gram-negative binding proteins, scavenger receptors or Thioester-containing proteins (TEPs).Citation2 Most TEPs contain a highly reactive thioester motif that covalently binds to the microbial surfaces and leads to their elimination from the host. Although a vast amount of information is available on various pattern recognition receptors in Drosophila, the specific function of TEPs is still not entirely understood. However, their immune role is widely studied in the mosquitoes Anopheles gambiae and Aedes aegypti, and in vertebrates.Citation3-6 The Anopheles TEP1 is involved in the process of phagocytosis of Escherichia coli and Staphylococcus aureus, as well as in the melanization of Plasmodium parasites.Citation7 Similarly, A. aegypti macroglobulin complement-related (MCR) factor participates in fighting off flavivirus infection.Citation5
Previous studies have shown that Drosophila Tep1, Tep2, Tep3 and Tep4 genes are induced upon certain bacterial, fungal, parasitoid and parasitic challenges.Citation8-10 Moreover, an in-vitro study has shown that phagocytosis of E. coli and S. aureus bacteria is regulated by TEP2, TEP3 and phagocytosis of Candida albicans spores by TEP6.Citation11 Recently we have shown that Tep4 modulates the activation of Toll and IMD immune signaling in Drosophila flies responding to 2 species of the potent pathogen Photorhabdus.Citation12 We further reported that inactivation of Tep4 leads to increased phenoloxidase and melanization activity upon Photorhabdus bacteria, and these effects alter the survival response of the flies to these pathogens.
The Photorhabdus genus contains bacteria that are highly virulent insect or human pathogens, which live in a mutualistic relationship with Heterorhabtidid nematodes.Citation13 The bacteria use distinct defense strategies that allow them to surpass the host immune responses. For example, a toxin secreted by P. luminescens has been shown to target a large number of insect hemolymph proteins encoding molecules that are involved in immune recognition, immune signaling and regulation of the coagulation cascade.Citation14 In addition, Photorhabdus can subvert cellular immune responses by secreting toxins or virulence factors that induce freezing or apoptosis of insect hemocytes.Citation15,16 Other studies have also revealed that Photorhabdus is able to interfere with the insect prophenoloxidase cascade.Citation17-19 Photorhabdus bacteria are closely related to many mammalian pathogens such as Yersenia pestis, E. coli and Salmonella.Citation20 Hence, results from studies on the pathogenicity of Photorhabdus in the context of host immune activity can be extrapolated to other pathogens of agricultural or medical importance.
To further our understanding on the immune role of Drosophila TEPs in the host defense against the pathogen Photorhabdus, here we have investigated the participation of Tep genes in the fly humoral and cellular antibacterial immune response. Using tep mutant flies together with gene expression assays and functional immune tests, we have shown that Tep2 and Tep6 are probably involved and act distinctly in the activation and regulation of immune signaling pathways, phagocytosis and phenoloxidase responses in the fly against the pathogen Photorhabdus.
Results
Tep genes are induced in Drosophila upon Photorhabdus challenge
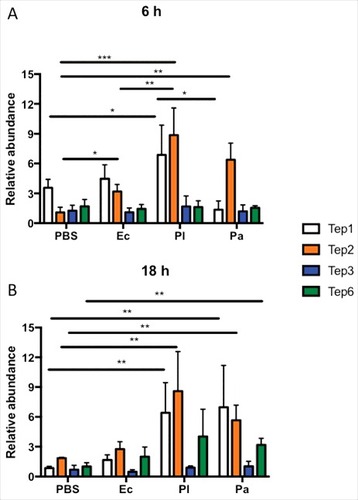
Previously we have shown that Tep4 is transcriptionally activated upon E. coli or Photorhabdus infection. Therefore, we first examined whether other Tep genes (Tep1–3 and Tep6) have altered expression in the background fly strain (w1118) upon infection with these bacteria. We found that certain Tep genes (Tep1, Tep2 and Tep6) were upregulated at 6 and 18 hpi by mostly P. luminescens and P. asymbiotica infection (). Only Tep2 was induced in flies infected with non-pathogenic E. coli bacteria () at 6 hpi. In particular, there was a significant induction of Tep1 and Tep2 genes in P. luminescens infected flies at 6 hpi (), and Tep1, Tep2 and Tep6 genes in P. asymbiotica infected flies at 18 hpi (). While Tep2 was mainly upregulated by P. luminescens and P. asymbiotica at both time points, Tep1 was significantly induced at higher levels by P. luminescens only at the 6 h time-point (). These results show that infection of D. melanogaster with the insect-specific pathogen P. luminescens and the related human pathogen P. asymbiotica as well as E. coli results in significant induction of certain TEP coding genes in the adult fly.
Figure 1. Tep1, Tep2 and Tep6 genes are upregulated in D. melanogaster flies by Photorhabdus infection. Transcript levels of Tep1, Tep2, Tep3 and Tep6 genes are shown in w1118 flies (n = 3–5) after (A) 6 and (B) 18 hpi with 1XPBS (septic injury control), E. coli (Ec), P. luminescens (Pl) and P. asymbiotica (Pa). Gene transcript levels are shown as relative abundance of transcripts normalized to RpL32 and expressed as a ratio compared with untreated flies (negative control). Significant differences are shown with asterisks (#p < 0.05, ##p < 0.01, ###p < 0.001). Bars show the means from 3 independent experiments and error bars represent standard deviation
Drosophila tep mutants have increased survival during the early and mid stages of Photorhabdus infection
To examine the function of the Tep induced genes in the immune response of Drosophila, we first performed survival analysis of the infected mutant flies. We tested the survival response of Tep2 and Tep6 loss-of-function mutants and their background control to infection by the 2 Photorhabdus pathogens and the non-pathogenic E. coli. We excluded tep1 mutants because Tep1 and Tep2 mRNAs were expressed at similar levels () and according to a previous phylogenetic analysis, Tep1 and Tep2 are likely to act redundantly (Bou Aoun et al, 2011). Moreover, we omitted tep3 mutants as we did not observe any changes in the mRNA levels of Tep3 (). To ascertain that we used loss-of-function tep mutants, we estimated the mRNA levels of Tep2 and Tep6 in the tep2 and Tep6 mutant flies injected with PBS, E. coli or Photorhabdus bacteria (Fig. S1 A-B).
Both Tep2 and Tep6 strains died within 36 h after P. luminescens infection and within 48 h after P. asymbiotica infection. We also observed that Tep2 flies died similarly to their background controls when infected with P. luminescens but survived significantly longer when infected with P. asymbiotica (). We found that at 36 hpi with P. asymbiotica, 48% of Tep2 were alive compared with their background controls (∼1%). However, there were 75% of Tep6 mutant flies alive compared with 14% of controls at 24 hpi with P. luminescens whereas 47% of Tep6 mutants were alive at 36 h post P. asymbiotica infection compared with controls (∼2%) (). We further found that injection with non-pathogenic E. coli or sterile PBS did not affect the survival of tep mutant flies and their background controls (Fig. S3A-B). These results indicate that loss-of-function mutations in Tep6 provide a survival advantage to D. melanogaster in response to infection with P. luminescens, while loss-of-function mutations in Tep2 and Tep6 promote the survival of flies against infection with P. asymbiotica.
Figure 2. Survival and bacterial load analysis for tep2 and tep6 mutants after Photorhabdus infection. Survival curves for loss-of-function (A) tep2 mutants and (B) tep6 mutants with w1118 (background control flies) are shown. Flies (n = 20) were injected in the thorax by microinjection with 1XPBS (septic injury control), P. luminescens (Pl) or P. asymbiotica (Pa). Survival was monitored at 6 h intervals for 48 h. The black dotted line represents 50% survival. Colony forming units (CFU) of (C) P. luminescens and (D) P. asymbiotica are shown in tep2, tep6 and control flies (n = 5 per experimental condition) after 6 and 18 hpi. CFU were quantified through quantitative PCR of makes caterpillars floppy (mcf-1) in P. luminescens and the insecticidal toxin complex protein gene (tccC3) in P. asymbiotica. Significant differences are indicated with asterisks (# p < 0.05, ## p <0.01, ### p <0.001). The means from 3 independent experiments are shown and error bars represent standard errors (survival) and standard deviation (bacterial load)
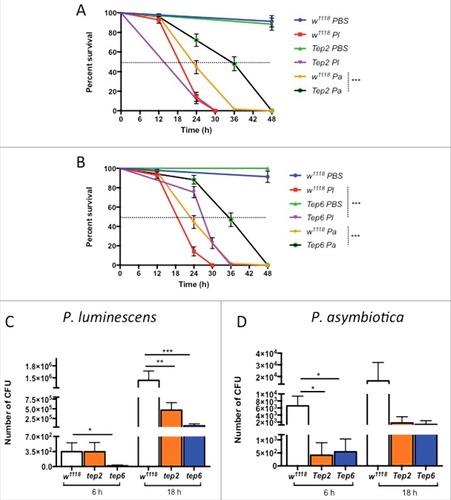
To understand the basis for the increased survival of the tep infected mutant flies, we investigated the bacterial persistence at 2 time-points post infection. To estimate bacterial load, we evaluated the number of colony forming units (CFU) at 6 and 18 hpi. We noticed that although there were no significant differences in survival between Tep2 mutants and their controls, there were 3-times fewer P. luminescens CFU in the mutant flies than in w1118 individuals at 18 hpi (). Similarly, Tep6 mutants had 18-times and 22-times less P. luminescens CFU than the control flies at 6 and 18 hpi, respectively (). In the case of P. asymbiotica infections, there were 3.5-times and 8-times fewer CFU in Tep2 and Tep6 mutants compared with w1118 flies at 6 h only (). Additionally, infections with non-pathogenic E. coli resulted in significantly lower numbers of CFU in Tep2 and Tep6 mutants compared with w1118 controls at both time-points post infection (Fig. S2C). Interestingly, Tep2 mutants contained 20-times more E. coli cells than Tep6 flies (Fig. S2C). These results show that deficiencies in Tep2 and Tep6 genes confer resistance to P. luminescens, P. asymbiotica and E. coli.
Function of Tep2 and Tep6 genes is essential for immune signaling pathway regulation in Drosophila
To explore the increased resistance of Tep2 and Tep6 mutants toward Photorhabdus and E. coli, we examined the transcriptional activation of Toll, Imd, JAK/STAT and JNK immune pathways in loss-of-function Tep2 and Tep6 mutant flies infected with these bacteria. We first tested at the activation of Toll pathway by evaluating the transcript levels of the AMP Defensin, which is a bacterial specific AMP.Citation21 In addition, we have recently found low to moderate transcript levels of Defensin in wild-type flies infected with Photorhabdus.Citation22,23 Here we asked whether flies with inactivated Tep2 or Tep6 have altered Defensin transcript levels upon infection with the pathogens. We found that Defensin was strongly induced in w1118 flies by either Photorhabdus species at 6 hpi but only by P. luminescens at 18 hpi (). We further found that Defensin mRNA levels were significantly higher in Tep6 mutant flies compared with their background controls at 6 hpi with P. luminescens, and at both 6 and 18 hpi upon infection with P. asymbiotica (). We observed significant upregulation in the mRNA levels of Defensin in Tep2 mutants at 6 and 18 hpi with P. luminescens compared with Tep2 mutant flies injected with PBS (). This indicates that Tep6 but not Tep2 gene activity is required in the induction of Toll pathway.
Figure 3. D. melanogaster Tep2 and Tep6 differentially regulate the activation of immune pathways against Photorhabdus. Transcript levels for (A, B) Defensin (Toll pathway), (C, D) Diptericin (IMD pathway), (E, F) Tot-M (JAK/STAT pathway) and (G, H) Puckered (JNK pathway) in loss-of function tep2 and tep6 mutants with their corresponding control strain (w1118) at 6 and 18 hpi with 1XPBS, E. coli (Ec), P. luminescens (Pl) or P. asymbiotica (Pa) (n = 3 individuals per experimental condition). Gene transcript levels are shown as relative abundance of transcripts normalized to RpL32 and expressed as a ratio compared with untreated flies (negative control). Values represent the means from 3 biologic replicates and error bars represent standard deviations. Significant differences are indicated with asterisks; #p <0.05, ##p <0.01, ###p <0.001, ####p <0.0001)
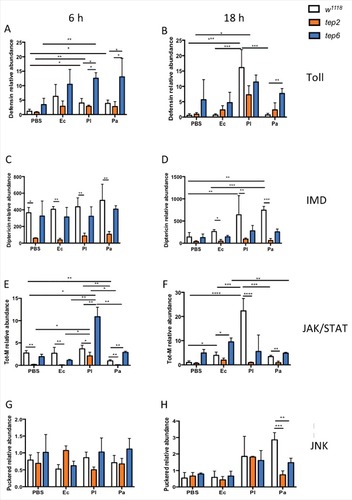
To evaluate Imd pathway activation, we estimated the transcript levels of the AMP-encoding gene Diptericin in infected flies ().Citation24 We observed that Diptericin mRNA levels were significantly induced in the w1118 flies by Photorhabdus and E. coli bacteria at 6 and 18 hpi, as well as in PBS injected flies at 6 hpi (). Moreover, upregulation of Diptericin was significantly higher in w1118 flies infected with P. asymbiotica compared with E. coli infected flies of the same strain at 18 hpi (). Interestingly, we found that transcript levels of Diptericin were consistently lower in Tep2 mutants than in w1118 flies at both time points (). In addition, there were no differences in Diptericin mRNA levels between Tep6 mutants and w1118 background controls (). These results indicate that Tep2 regulates Imd signaling in D. melanogaster adult flies in the context of Photorhabdus infection or response to wounding.
To analyze JAK/STAT and JNK signaling activation in tep mutants and control flies, we assessed the transcript levels of Turandot-M (Tot-M) and Puckered (Puc).Citation22,23,25,26 We first observed that Tot-M was significantly upregulated in w1118 flies at 18 hpi with P. luminescens than flies injected with other bacteria or PBS (). The Tot-M mRNA levels were significantly low in w1118 flies infected with P. asymbiotica than P. luminescens or PBS injected flies at 6hpi (). We found that Tep2 mutants have significantly reduced Tot-M mRNA levels than the w1118 flies injected with any of the bacteria at both time points (). We also observed that in Tep2 mutant flies, Tot-M was slightly upregulated only at 6 hpi with P. luminescens compared with other treatments, but this induction was significantly lower compared with background flies infected by P. luminescens (). We further noticed that Tot-M was significantly upregulated in Tep6 mutants at 6 hpi with Photorhabdus bacteria and at 18 hpi with E. coli in relation to control flies (). We found that Puc mRNA levels were significantly lower in tep mutants compared with the control flies injected with P. asymbiotica at 18 hpi only (). These results indicate that Tep2 is required for full JAK/STAT pathway induction in the presence of certain bacterial infections of adult fruit flies. In addition, Tep2 and Tep6 gene activity is required for JNK signaling in D. melanogaster adult flies upon infection with P. asymbiotica during the late stages of infection.
Functional hemocytes in Drosophila constitute a source of Tep2 and Tep6 transcription
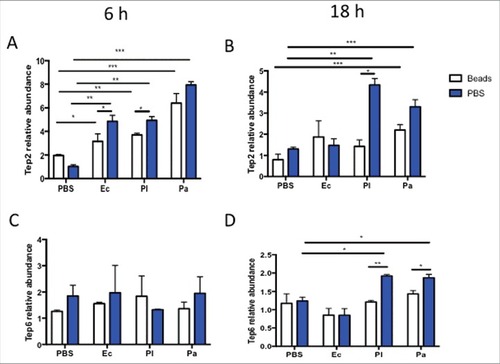
Because TEPs are secreted proteins and they are expressed in larval plasmatocytes,Citation10 we examined whether changes in the function of hemocytes can affect the upregulation of Tep2 and Tep6 in infected adult flies. For this, we pre-injected the w1118 flies with latex beads to ablate the function of hemocytes. A pre-injection with 1X PBS served as control. We found that Tep2 transcript levels were significantly higher in flies pre-injected with beads or PBS followed by any bacterial treatment at 6 hpi, but only by Photorhabdus challenge at 18 hpi (). Moreover, we noticed significant upregulation of Tep2 in flies pre-injected with PBS compared with flies preinjected with beads followed by infection with P. luminescens or E. coli (). There was also significant upregulation of Tep2 in flies pre-injected with PBS compared with those treated with beads, at 18 hpi with P. luminescens (). Furthermore, we found significant upregulation of Tep6 in flies pre-injected with PBS than in those injected with beads at 18 hpi with either Photorhabdus species (). These results indicate that functional hemocytes are one of the sources of Tep2 and Tep6 genes upregulation.
Figure 4. Transcript levels of Tep2 and Tep6 are significantly decreased in control flies (w1118) with functionally ablated hemocytes. Transcript levels of (A) Tep2 and (B) Tep6 at 6 and 18 hpi with 1XPBS, E. coli (Ec), P. luminescens (Pl) or P. asymbiotica (Pa) in w1118 flies (n = 5) pre-injected with beads or 1XPBS. Significant differences are indicated with asterisks (#p < 0.05, ## p < 0.01, ###p < 0.001). Bars show the means from 2 independent experiments and error bars represent standard deviations
Drosophila Tep2 and Tep6 mutants have differential number of hemocytes and fewer dead hemocytes against Photorhabdus infection
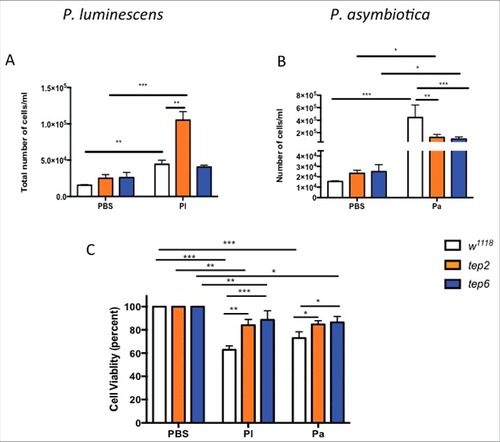
To evaluate whether inactivation of Tep2 and Tep6 genes can affect activation of circulating hemocytes, we then investigated the cellular function of Drosophila against Photorhabdus and E. coli bacteria. We first looked at changes in the total number of hemocytes in infected and uninfected flies. Based on the hemocyte counting protocol, the total number of hemocytes was significantly higher in w1118 as well as Tep2 mutants infected with P. asymbiotica, P. luminescens or E. coli compared with w1118 flies injected with PBS (, Fig. S3A). Interestingly, there were significantly more hemocytes in the Tep2 mutants [(10.52 ± 0.11) X105 or (3.73 ± 0.46) X104] than in w1118 [(0.44 ± 0.05) X105 or (1.17 ± 0.34) X104] flies infected with P. luminescens or E. coli (, Fig. S3A). We did not observe any significant change in the number of hemocytes between Tep6 mutants and control flies infected with P. luminescens or E. coli bacteria (, Fig. S3A). Moreover, both Tep2 [(1.24 ± 0.48) X105] and Tep6 mutants [(0.97 ± 0.35) X105] had significantly fewer hemocytes than the w1118 flies [(4.44 ± 1.98) X105] after P. asymbiotica infection (). However, we observed an increase in hemocyte numbers after P. asymbiotica infection in Tep6 mutants compared with mutants injected with PBS (). We also evaluated cell viability in Tep2, Tep6 mutants and w1118 flies after infection with Photorhabdus and E. coli. We observed reduced cell viability in all the strains after Photorhabdus and E. coli infection (, Fig. S3B). We found that tep mutants contained significantly higher percentage of viable cells compared with w1118 flies infected with Photorhabdus (). These data suggest that Tep2 and Tep6, plays an important role in the activation of hemocytes in Drosophila flies responding to infection with Photorhabdus or E. coli bacteria.
Figure 5. D. melanogaster tep2 and tep6 mutants display variable number of total hemocyte counts and increased hemocyte viability compared with control flies (w1118) after Photorhabdus infection. According to the hemocyte counting protocol, total number of hemocytes (total cells/ml) in tep2 and tep6 mutants with control flies after 18 h of injection with 1XPBS, (A) P. luminescens (Pl) or (B) P. asymbiotica (Pa). The percentage of total viable cells in the control and tep mutant flies at 18 hpi with 1XPBS, (C) P. luminescens and (D) P. asymbiotica. Significant differences are indicated with asterisks (#p < 0.05, ##p < 0.01, ###p < 0.001). Bars show the means from 3 independent experiments and error bars represent standard deviations
Drosophila Tep2 and Tep6 are required for phagocytosis of Photorhabdus or E. coli bacteria
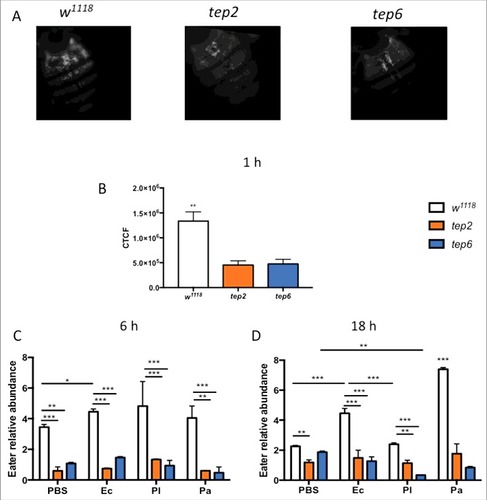
To estimate whether inactivation of Tep2 or Tep6 affects the phagocytosis of bacteria in Drosophila, we injected opsonized inactive E. coli bioparticles in tep mutants and their control flies. We found that the phagocytic activity was significantly reduced (∼3 times lower) in the Tep2 and Tep6 mutants compared with w1118 flies at one hpi with E. coli (). We also looked at the transcript levels of Eater gene, as a marker of phagocytosis,Citation27 in flies injected with PBS, E. coli or Photorhabdus bacteria at 6 and 18 hpi. The mRNA levels of Eater were significantly higher in control flies infected with E. coli at both time points but also with P. asymbiotica at 18 hpi (). We found that Eater was significantly upregulated at 6 hpi with E. coli, P. luminescens or PBS in w1118 flies compared with the tep mutants (). Additionally, we observed that Eater mRNA levels were significantly lower in both tep mutants compared with control flies at 18 hpi following bacterial or buffer injection (). In particular, Eater mRNA levels were significantly lower in Tep6 mutants infected with P. asymbiotica compared with those injected with PBS at 18 hpi (). Our data suggest that inactivation of Tep2 and Tep6 severely prevents the phagocytic activity in flies against certain bacterial infections.
Figure 6. Tep2 and Tep6 are essential for the phagocytosis process in Drosophila. (A) Representative images of phagocytosis in tep2 and tep6 loss-of-function mutants and control flies (w1118) at 1 hpi of lipophilized pHrodo-labeled E. coli particles. Images were taken using fluorescence microscopy at 10X magnification. (B) Corrected total cell fluorescence (CTCF) in tep mutants and w1118 flies (n = 7), 1 h following injection of pHrodo-labeled E. coli. Images were processed in ImageJ and CTCF was estimated. Transcript levels of Eater in tep mutants and w1118 flies (n = 5) at (C) 6 h and (D) 18 hpi of 1XPBS, E. coli (Ec), P. luminescens (Pl) or P. asymbiotica (Pa). Significant differences are indicated with asterisks (#p < 0.05, ##p < 0.01, ###p < 0.001). The means from 2 (Eater transcription) -three (Phagocytosis) independent experiments are shown and error bars represent standard deviation
Tep2 participates in the drosophila melanization and phenoloxidase response against Photorhabdus infection
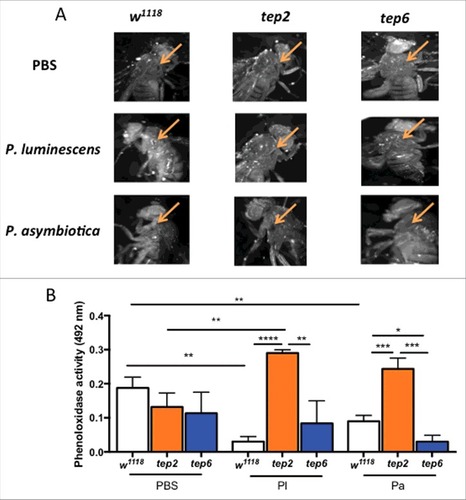
To examine whether inactivation of Tep2 or Tep6 influence the in vivo melanization response in D. melanogaster, we visually inspected the wound site at 3 hpi of mutant and control flies with Photorhabdus, E. coli or PBS. We observed that w1118 flies and Tep2 mutants exhibited strong melanization response against all injection treatments (, Fig. S4A). Melanin spots developed in Tep6 flies following injection with PBS or E. coli only (). We also estimated the phenoloxidase (PO) enzyme activity in the hemolymph plasma of tep mutant and control flies injected with the different bacteria. We noticed that the PO activity was significantly reduced in control flies infected with Photorhabdus bacteria (). We found no significant changes in PO activity between Tep2 or Tep6 mutant flies and w1118 controls injected with PBS or E. coli (, Fig. S4B). Furthermore, Tep2 mutant flies infected with P. luminescens or P. asymbiotica had substantially higher PO activity than Tep6 and w1118 flies infected with the pathogens. Interestingly, we found that upon P. asymbiotica infection, Tep6 mutants displayed significantly lower levels of PO activity than w1118 flies (). These results suggest that the absence of functional TEP2 in D. melanogaster adult flies promotes phenoloxidase activity against infection with pathogenic Photorhabdus.
Figure 7. Melanization response and PO activity are elevated in D. melanogaster tep2 mutants upon Photorhabdus infection. (A) Melanization of the wound site in tep2 and tep6 loss-of-function mutant flies and their background control strains (w1118) is shown at 10X magnification 3 h after injection with PBS, P. luminescens or P. asymbiotica bacteria. Arrows indicate the site of injury. (B) PO activity in the hemolymph plasma of tep2, tep6 mutants and control flies (w1118) at 3 hpi with PBS, P. luminescens (Pl) or P. asymbiotica (Pa) (n = 20 flies) as measured by the optical density at 492 nm after incubation with L-Dopa. Values represent the means from 3 biologic replicates and error bars represent standard deviations. Significant differences are indicated with asterisks (#p < 0.05, ##p < 0.01, ###p < 0.001, ####p < 0.0001). The means from 3 independent experiments are shown and error bars represent standard deviation
Discussion
Despite remarkable advances in the field of innate immunity, our understanding of the role of TEP molecules in the immune defense of Drosophila is mostly unexplored. Recently, we showed the participation of Tep4 in the humoral and phenoloxidase responses of the fruit fly against Photorhabdus infection. Here we investigated the role of other Tep genes in the antibacterial immune response of Drosophila. Previously, we observed induction of Tep4 in flies infected with 2 Photorhabdus species, therefore we hypothesized that presumably other Tep genes might also be activated upon infection with this pathogen.
Previous studies have reported upregulation of Tep1 and Tep2 genes but not Tep6 in adult flies infected with a mixture of E. coli and Micrococcus luteus.Citation10,28 Our results are in accordance to the previous studies, as we observe an early upregulation of Tep2 by E. coli bacteria. Transcriptomic analysis has also shown that Tep1 and Tep2 are induced following P. luminescens, symbiotic Heterorhabditis nematodes (carrying Photorhabdus) or axenic nematodes (lacking Photorhabdus) infection.Citation8,29 In accordance, upregulation of Tep1 and Tep2 in flies after Photorhabdus infection suggests their probable function in the immune response of the fly against the Photorhabdus bacteria. Although there are no reports of Tep3 induction by Photorhabdus or its symbiotic nematode partner, recent work has reported that tep3 loss-of-function mutants are sensitive to Heterorhabditis symbiotic nematode infections.Citation8 No changes in the transcript levels of Tep3 could be due to its specificity only to nematodes. Also, induction of Tep6 in response to P. asymbiotica indicates a specific function of this molecule against this pathogen. However, as previously reported,Citation10 we cannot exclude the possibility that the function of TEPs in the fly immune system might be redundant or that TEP molecules might act in combination with other factors to provide efficient levels of protection to the fly against certain pathogens.
Previously TEP molecules have been shown to be expressed in larval plasmatocytes, adult fat body of the head and digestive tract lining at basal levels.Citation10 Reduced transcript levels of Tep2 and Tep6 in flies containing dysfunctional hemocytes upon Photorhabdus infection indicates that functional plasmatocytes are one of the major sources for Tep gene expression in the adult flies. However, other tissues such as gut or fat body may contribute toward Tep gene upregulation when hemocytes are inactive. This could explain the induction of Tep2 in flies containing non-functional heymocytes in response to E. coli or Photorhabdus infection.
Inactivation of certain genes in D. melanogaster can alter the survival ability of the fly in response to microbial infections.Citation1 A former study has shown that loss-of-function Tep2 mutants are susceptible to Porphyromonas gingivalis infection.Citation30 However, another study failed to identify changes in the survival of single, double or triple tep mutants in response to Gram-positive and Gram-negative bacterial pathogens as well as to fungal infection.Citation10 Hence, prolonged survival of Tep6 mutant flies against P. luminescens as well as of Tep2 and Tep6 mutants in response to P. asymbiotica infection suggests that the survival response is pathogen specific. In addition, the presence of significantly fewer Photorhabdus CFU in Tep2 and Tep6 mutants could explain their increased survival during the initial and intermediate stages of Photorhabdus infection. The finding that Tep2 mutants are resistant to both Photorhabdus and E. coli could probably suggest that Tep2 is evolving in relation to the different pathogen challenges Drosophila flies encounter in the wild. Interestingly, it has been previously proposed that Drosophila Tep2 may have evolved under strong positive selection.Citation31
The modulation of immune pathways by Tep4 Citation12 and the effect of Tep4 gene inactivation on the resistance of mutant flies to bacterial infection formed the basis for testing whether TEP2 and TEP6 can also play a central regulatory role in the D. melanogaster immune system. No effect on the Toll pathway activation and downregulation of IMD pathway in Tep2 mutants suggests different mode of actions of Tep2 and Tep4 genes. Additionally, our results are in agreement with previous findings as JAK/STAT activation was severely impaired in Tep2 mutants.Citation32 Similar to tep4, here we find that Toll signaling activity is upregulated in Tep6 mutants infected with Photorhabdus. Stimulation of JAK/STAT in Tep6 mutants after Photorhabdus infection implies that Tep6 may not participate in the activation of this pathway. Another explanation for the differential induction patterns of the immune signaling pathways in Tep6 mutants could be the absence of a thioester motif in TEP6 that probably affects its function as an effector molecule. Nevertheless, increased activation of certain pathways in tep mutants compared with background controls injected with PBS could suggest that wounding initiates a response in these flies. However, this activation increases in the presence of the non-pathogenic bacteria E. coli, as seen in the case of Tot-M transcript levels in Tep6 mutants. In contrast, Photorhabdus may interfere with the activation of these pathways by either increasing or suppressing them, as seen with the induction of Tot-M in Tep6 mutants injected with P. luminescens or the reduction of Puc in Tep6 mutants infected with P. asymbiotica, respectively. Altogether, the 3 TEP molecules- TEP2, TEP4 and TEP6, may modulate immune signaling pathways in discrete ways.
Complement proteins are known to activate mast and basophils in human lungs and blood as an inflammatory and allergic response.Citation33 Additionally, in Drosophila, dramatic change in the number of circulating hemocytes is observed after pathogenic invasion.Citation34 Nonetheless the function of TEPs in the recruitment and activation of hemocytes in Drosophila after bacterial infection is still undefined. The increase in the number of hemocytes in Tep2 and Tep6 mutants after Photorhabdus or E. coli infection indicates that TEP2 and TEP6 are not directly involved in the induction of hemocytes. However, inactivation of either Tep2 or Tep6 is not entirely insignificant, as we observed larger numbers of hemocytes in the control flies against P. asymbiotica infection. The increase in the number of hemocytes might also be a consequence of other activated TEP molecules, such as TEP4, as we have previously observed a significant decline in the number of hemocytes in the absence of TEP4 following E. coli or Photorhabdus infection (unpublished).
One of the main evasion strategies of Photorhabdus involves targeting and attacking insect hemocytes. Photorhabdus can cause morphological changes to hemocytes by affecting the cytoskeletal components, such as actin, that can in turn disturb their normal functions.Citation35,36 Moreover, Photorhabdus pathogens secrete several toxins that can induce apoptosis in the insect hemocytes.Citation16,37 Increased hemocyte viability in Tep2 and Tep6 mutants indicates that inactivation of these Tep genes is advantageous for the hemocytes to respond against the Photorhabdus insult. This could further support the prolonged survival of the Tep2 and Tep6 mutants during the course of Photorhabdus infection.
An in vitro study has shown that TEP2 and TEP6 in D. melanogaster are involved in the phagocytosis of E. coli and Candida albicans, respectively.Citation11 The decreased phagocytosis of inactive E. coli particles in the Tep2 and Tep6 mutants indicates the significance of TEP2 and TEP6 in the phagocytosis process against E. coli. Moreover, the notably reduced transcript levels of Eater probably suggests a direct role of TEP2 and TEP6 in this process in response to Photorhabdus or E. coli infection. We propose that although Tep2 and Tep6 mutants contain high numbers of hemocytes after bacterial challenge, due to the inactivation of these 2 Tep genes, the phagocytosis function is substantially impaired in the mutants.
We also examined the effect of Tep2 and Tep6 on the melanization response, which forms an essential and rapid cellular immunity process.Citation38,39 The elevated melanization and phenoloxidase activity in Tep2 mutants against Photorhabdus bacteria points out that TEP2 and TEP4 perform similar immune functions in response to Photorhabdus infection in Drosophila. This may also account for the reduced number of Photorhabdus CFU in the Tep2 mutants. It could be possible that the growth of Photorhabdus bacteria is restricted in the Tep2 mutants due to increased PO and melanization during the initial phase of infection. In contrast, inactivation of Tep6 leads to reduced PO and melanization in the flies after Photorhabdus infection. The contrasting findings between Tep2 and Tep6 mutants may be best explained by the structural difference between the 2 proteins. TEP6, which lacks the thioester motif, regulates phenoloxidase activity and melanization in a different manner than TEP2 and TEP4 molecules, which contain the thioester motif.Citation10
In conclusion, we have extended our previous findings that TEPs serve an imperative function in the immune defense of Drosophila. The experiments described herein were focused on critical immune responses of fruit fly in response to Photorhabdus bacteria. We show that inactivation of Tep2 and Tep6 serve a protective and immunomodulatory role against certain insect pathogenic bacteria, such as Photorhabdus. Furthermore, our data suggest that different TEP molecules act in a distinct manner in the Drosophila antibacterial immune system. It has been shown that disruption of C5aR encoding the Complement protein 5a receptor results in increased resistance to acute Gram-negative bacterial infections in mice and this ultimately leads to reduced endotoxic shock.Citation40 Similarly, we propose that the absence of TEP2 or TEP6 leads to lower levels of inflammation in the host following bacterial infection, which successively modulates their survival ability against potent entomopathogenic bacteria. We anticipate that such studies will lead to a better understanding of the complex mechanism of action of TEP molecules in the antibacterial immune reponse of the fruit fly. These findings could also be applied to insects of agricultural or medical importance.
Materials and methods
Fly and bacterial strains
The following D. melanogaster strains were used in the study- w1118 (genetic background strain), Tep2 (f02756, Harvard), and Tep6 (f03851, Harvard). All strains were kept and amplified for experimentation with instant Drosophila media (Carolina Biological Supply) with deionized water. All stocks were maintained at 25°C and a 12:12-hour light:dark photoperiod.
The bacterial strains used were Photorhabdus luminescens subsp. laumondii (strain TT01), P. asymbiotica subsp asymbiotica (strain ATCC 43949) and Escherichia coli (strain K12). Bacteria were cultured in sterile Luria–Bertani (LB) broth for approximately 18–22 h at 30°C on a rotary shaker at 220 rpm. The cultures were then pelleted down, washed and re-suspended in 1x sterile phosphate-buffered saline (PBS, Sigma Aldrich). For infections, bacterial concentrations were brought to an Optical Density (OD, 600 nm) of 0.1 for P. luminescens, 0.25 for P. asymbiotica and 0.015 for E. coli using a spectrophotometer (NanoDropTM 2000c – Thermo Fisher Scientific).
Infection assays and survival experiment
All procedures were performed as described previously.Citation12 In brief, 7–10 d old adult flies were anesthetized with CO2 and then injected in the thorax with 18.4 nl (100–300 CFU) of each bacterial suspension (P. luminescens, P. asymbiotica or E. coli) or sterile 1XPBS (septic injury control) using a Nanoject II apparatus (Drummond Scientific) equipped with glass capillaries prepared with a micropipette puller (Sutter Instruments). Two replicates of 10 flies each were used for each treatment and survival was recorded at 6-hour intervals and up to 48 hours. Each experiment was replicated at least 3 times.
Bacterial load and gene transcription
All procedures were performed as described previously.Citation12 Briefly, 4-five adult flies were injected and subsequently frozen at 6 and 18 hours post infection (hpi). DNA was extracted from the frozen flies using DNeasy Blood and Tissue kit (Qiagen) using the manufacturer's protocol. The DNA samples were adjusted to 500 ng for estimating bacterial load. Samples were run in technical duplicates and Quantitative PCRs were performed in twin-tech. semi-skirted 96 well plates on a Mastercycler® ep realplex.2Standard curves for each bacterium were used to estimate the bacterial load in infected flies.
For gene transcription studies, total RNA was isolated using the PrepEase RNA spin kit (Affymetrix USB), followed by cDNA synthesis and quantitative RT-PCR (qRT-PCR). ΔΔCt method was used to perform analysis. Data are presented as the ratio between injected flies versus uninfected flies (baseline controls). All the experiments were performed at least 3 times. The list of primers used for the PCR assays are listed in .
Table 1. List of primers used in the study
Hemolymph collection, hemocyte counts and viability
Hemolymph was collected from adult female flies (n = 4) at 18 hpi with P. luminescens, P. asymbiotica, E. coli or 1X PBS injection; using a modified version of a previously published protocol.Citation41 Briefly, flies were anesthetized using CO2 and then injected into the thorax with 2–3 uL of incubation solution [60% Grace's Medium (GM) supplemented with 10% of Fetal Bovine Serum (FBS) and 20% of Anticoagulant Buffer (98 mM NaOH, 186 mM NaCI, 1.7 mM EDTA and 41 mM citric acid, pH 4.5)] using a blunt end needle (16 gauge) fitted with a tubing connected to a 20 ml glass syringe. After 20 minutes of incubation on ice, flies were kept on a petri dish and an incision was made between the 2nd and 3rd abdominal segments. Flies were again injected into the thorax with 5 uL of collection solution (90% of GM supplemented with 10% of FBS). Hemolymph was then collected in a 1.5 mL tube and used for further assays. Hemolymph samples (10 uL) were loaded on a hemocytometer and total numbers of cells as well as the different hemocyte types were estimated using 40X magnification of a compound microscope (Olympus CX21). For cell viability, Trypan blue exclusion assay was performed. All experiments were repeated at least 3 times.
Functional ablation of hemocytes
For ablating the function of hemocytes in D. melanogaster, flies were anesthetized with CO2 and then injected with 69 nL of latex beads (0.3 um diameter, Molecular Probes, Invitrogen) into the thorax using a Nanoject II apparatus (Drummond Scientific) equipped with glass capillaries prepared with a micropipette puller (Sutter Instruments). Latex beads were prepared by washing them with sterile 1XPBS and used 4X concentrated in PBS (corresponding to 5–10% solids). 1XPBS served as control for the first round. After 18 h, the flies were injected again with each bacterial suspension (P. luminescens, P. asymbiotica or E. coli) or PBS (septic injury control) and used for further assays.
Phagocytosis assay
All procedures were performed as described previously.Citation22 Briefly, Seven flies from each strain were injected with 50.4 nL of 1 mg/mL pHrodo labeled E. coli (Molecular Probes) and allowed to phagocytose at room temperature for 60 min. The flies were fixed ventrally on a glass slide using clear nail paint. Fluorescent images of the dorsal surface were obtained using Nikon ECLIPSE Ni microscope (10X magnification) fitted with Zyla (ANDOR) 5.5 camera. The images were analyzed using ImageJ software and analyzed. Each experiment was performed 3 times.
Melanization and PO activity
Melanization spots on the site of injury were observed at 3 hpi using a Nikon SMZ18 microscope with Zyla (ANDOR) 5.5 camera. Images were analyzed using Nikon Software Suite at 10X magnification. PO activity was measured as described previously (Shokal and Eleftherianos, 2016). Briefly, at 3 hpi the injected flies (n = 20) were placed on a spin column (Pierce, Thermo fisher) containing 2.5X protease inhibitor (Sigma) and covered with 5 4 mm glass beads (VWR). They were centrifuged at 4°C and 13,000 rpm for 20 min. Protein concentrations were then adjusted using a BCA test. A mixture of 15 μg of protein (diluted in 2.5x protease inhibitor) with 5 mM Cacl2 was added to L-DOPA solution (15 mM in phosphate buffer, pH 6.6) making a final volume of 200 μL. The absorbance (OD 492 nm) for each sample was measured after 36 min incubation at 29°C in the dark against a blank control. Each experiment was performed in biologic duplicates and repeated 3 times.
Statistical analysis
All statistics were performed using the GraphPad Prism7 software. Analysis of survival experiments was conducted using a Log-rank (Mantel-Cox) and Chi-square tests. Unpaired 2-tailed t-test and 2-way analysis of variance (ANOVA) with a Tukey post-hoc test for multiple comparisons were used for analyzing bacterial load, gene expression data, hemocyte cell counts, cell viability and PO activity results. p values below 0.05 were considered statistically significant.
Abbreviations
| Hpi | = | hours post infection |
| PO | = | Phenoloxidase |
| TEPs | = | Thioester-containing proteins |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KVIR_S_1330240.zip
Download Zip (1 MB)Acknowledgments
We thank members of the Department of Biological Sciences at GWU for critical reading of the manuscript. We also thank Dr. Mollie Manier for letting us use the fluorescence microscope and stereomicroscope in her laboratory.
Funding
This research was supported by a start-up fund from the Columbian College of Arts and Sciences at GWU to I.E., the Harlan Summer Fellowships from the Department of Biological Sciences at GWU and a scholarship from the Cosmos Club Foundation (Washington, DC) to US. The I.E. laboratory is funded by the National Institutes of Health, (grant 1R01AI110675–01A1).
References
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Ann Rev Immunol 2007; 25:697-743; PMID:17201680; https://doi.org/https://doi.org/10.1146/annurev.immunol.25.022106.14161510.1146/annurev.immunol.25.022106.141615
- Pal S, Wu LP. Pattern recognition receptors in the fly: lessons we can learn from the Drosophila melanogaster immune system. Fly (Austin) 2009; 3:121-9; PMID:19440043; https://doi.org/https://doi.org/10.4161/fly.882710.4161/fly.8827
- Buresova V, Hajdusek O, Franta Z, Sojka D, Kopacek P. IrAM-An alpha2-macroglobulin from the hard tick Ixodes ricinus: characterization and function in phagocytosis of a potential pathogen Chryseobacterium indologenes. Dev Comp Immunol 2009; 33:489-98; PMID:18948134; https://doi.org/https://doi.org/10.1016/j.dci.2008.09.01110.1016/j.dci.2008.09.011
- Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, Levashina EA. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 2004; 116:661-70; PMID:15006349; https://doi.org/https://doi.org/10.1016/S0092-8674(04)00173-410.1016/S0092-8674(04)00173-4
- Xiao X, Liu Y, Zhang X, Wang J, Li Z, Pang X, Wang P, Cheng G. Complement-related proteins control the flavivirus infection of Aedes aegypti by inducing antimicrobial peptides. PLoS Pathog 2014; 10:e1004027; PMID:24722701; https://doi.org/https://doi.org/10.1371/journal.ppat.100402710.1371/journal.ppat.1004027
- Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol 2010; 11:785-97; PMID:20720586; https://doi.org/https://doi.org/10.1038/ni.192310.1038/ni.1923
- Blandin SA, Marois E, Levashina EA. Antimalarial responses in Anopheles gambiae: from a complement-like protein to a complement-like pathway. Cell Host Microbe 2008; 3:364-74; PMID:18541213; https://doi.org/https://doi.org/10.1016/j.chom.2008.05.00710.1016/j.chom.2008.05.007
- Arefin B, Kucerova L, Dobes P, Markus R, Strnad H, Wang Z, Hyrsl P, Zurovec M, Theopold U. Genome-wide transcriptional analysis of Drosophila larvae infected by entomopathogenic nematodes shows involvement of complement, recognition and extracellular matrix proteins. J Innate Immun 2014; 6:192-204; PMID:23988573; https://doi.org/https://doi.org/10.1159/00035373410.1159/000353734
- Wertheim B, Kraaijeveld AR, Schuster E, Blanc E, Hopkins M, Pletcher SD, Strand MR, Partridge L, Godfray HC. Genome-wide gene expression in response to parasitoid attack in Drosophila. Genome Biol 2005; 6:R94; PMID:16277749; https://doi.org/https://doi.org/10.1186/gb-2005-6-11-r9410.1186/gb-2005-6-11-r94
- Bou Aoun R, Hetru C, Troxler L, Doucet D, Ferrandon D, Matt N. Analysis of thioester-containing proteins during the innate immune response of Drosophila melanogaster. J Innate Immun 2011; 3:52-64; PMID:21063077; https://doi.org/https://doi.org/10.1159/00032155410.1159/000321554
- Stroschein-Stevenson SL, Foley E, O'Farrell PH, Johnson AD. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol 2006; 4:e4; PMID:16336044; https://doi.org/https://doi.org/10.1371/journal.pbio.004000410.1371/journal.pbio.0040004
- Shokal U, Eleftherianos I. Thioester-Containing Protein-4 Regulates the Drosophila Immune Signaling and Function against the Pathogen Photorhabdus. J Innate Immun 2017; 9:83-93; PMID:27771727; https://doi.org/https://doi.org/10.1159/00045061010.1159/000450610
- Joyce SA, Watson RJ, Clarke DJ. The regulation of pathogenicity and mutualism in Photorhabdus. Curr Opin Microbiol 2006; 9:127-32; PMID:16480919; https://doi.org/https://doi.org/10.1016/j.mib.2006.01.00410.1016/j.mib.2006.01.004
- Felfoldi G, Marokhazi J, Kepiro M, Venekei I. Identification of natural target proteins indicates functions of a serralysin-type metalloprotease, PrtA, in anti-immune mechanisms. Appl Environ Microbiol 2009; 75:3120-6; PMID:19304826; https://doi.org/https://doi.org/10.1128/AEM.02271-0810.1128/AEM.02271-08
- Vlisidou I, Dowling AJ, Evans IR, Waterfield N, ffrench-Constant RH, Wood W. Drosophila embryos as model systems for monitoring bacterial infection in real time. PLoS Pathog 2009; 5:e1000518; PMID:19609447; https://doi.org/https://doi.org/10.1371/journal.ppat.100051810.1371/journal.ppat.1000518
- Daborn PJ, Waterfield N, Silva CP, Au CP, Sharma S, Ffrench-Constant RH. A single Photorhabdus gene, makes caterpillars floppy (mcf), allows Escherichia coli to persist within and kill insects. Proc Natl Acad Sci U S A 2002; 99:10742-7; PMID:12136122; https://doi.org/https://doi.org/10.1073/pnas.10206809910.1073/pnas.102068099
- Eleftherianos I, Boundy S, Joyce SA, Aslam S, Marshall JW, Cox RJ, Simpson TJ, Clarke DJ, ffrench-Constant RH, Reynolds SE. An antibiotic produced by an insect-pathogenic bacterium suppresses host defenses through phenoloxidase inhibition. Proc Natl Acad Sci U S A 2007; 104:2419-24; PMID:17284598; https://doi.org/https://doi.org/10.1073/pnas.061052510410.1073/pnas.0610525104
- Eleftherianos I, Waterfield NR, Bone P, Boundy S, ffrench-Constant RH, Reynolds SE. A single locus from the entomopathogenic bacterium Photorhabdus luminescens inhibits activated Manduca sexta phenoloxidase. FEMS Microbiol Lett 2009; 293:170-6; PMID:19243439; https://doi.org/https://doi.org/10.1111/j.1574-6968.2009.01523.x10.1111/j.1574-6968.2009.01523.x
- Ullah I, Khan AL, Ali L, Khan AR, Waqas M, Lee IJ, Shin JH. An insecticidal compound produced by an insect-pathogenic bacterium suppresses host defenses through phenoloxidase inhibition. Molecules 2014; 19:20913-28; PMID:25514230; https://doi.org/https://doi.org/10.3390/molecules19122091310.3390/molecules191220913
- Eleftherianos I, ffrench-Constant RH, Clarke DJ, Dowling AJ, Reynolds SE. Dissecting the immune response to the entomopathogen Photorhabdus. Trends Microbiol 2010; 18:552-60; PMID:21035345; https://doi.org/https://doi.org/10.1016/j.tim.2010.09.00610.1016/j.tim.2010.09.006
- Imler JL, Bulet P. Antimicrobial peptides in Drosophila: structures, activities and gene regulation. Chem Immunol Allergy 2005; 86:1-21; PMID:15976485.
- Shokal U, Yadav S, Atri J, Accetta J, Kenney E, Banks K, Katakam A, Jaenike J, Eleftherianos I. Effects of co-occurring Wolbachia and Spiroplasma endosymbionts on the Drosophila immune response against insect pathogenic and non-pathogenic bacteria. BMC Microbiol 2016; 16:16; PMID:26862076; https://doi.org/https://doi.org/10.1186/s12866-016-0634-610.1186/s12866-016-0634-6
- Castillo JC, Shokal U, Eleftherianos I. Immune gene transcription in Drosophila adult flies infected by entomopathogenic nematodes and their mutualistic bacteria. J Insect Physiol 2013; 59:179-85; PMID:22902989; https://doi.org/https://doi.org/10.1016/j.jinsphys.2012.08.00310.1016/j.jinsphys.2012.08.003
- Wicker C, Reichhart JM, Hoffmann D, Hultmark D, Samakovlis C, Hoffmann JA. Insect immunity. Characterization of a Drosophila cDNA encoding a novel member of the diptericin family of immune peptides. J Biol Chem 1990; 265:22493-8.
- McEwen DG, Peifer M. Puckered, a Drosophila MAPK phosphatase, ensures cell viability by antagonizing JNK-induced apoptosis. Development 2005; 132:3935-46; PMID:16079158; https://doi.org/https://doi.org/10.1242/dev.0194910.1242/dev.01949
- Brun S, Vidal S, Spellman P, Takahashi K, Tricoire H, Lemaitre B. The MAPKKK Mekk1 regulates the expression of Turandot stress genes in response to septic injury in Drosophila. Genes Cells 2006; 11:397-407; PMID:16611243; https://doi.org/https://doi.org/10.1111/j.1365-2443.2006.00953.x10.1111/j.1365-2443.2006.00953.x
- Chung YS, Kocks C. Recognition of pathogenic microbes by the Drosophila phagocytic pattern recognition receptor Eater. J Biol Chem 2011; 286:26524-32; PMID:21613218; https://doi.org/https://doi.org/10.1074/jbc.M110.214007 https://doi.org/10.1074/jbc.A110.214007https://doi.org/10.1074/jbc.A110.214007 https://doi.org/10.1074/jbc.M110.214007
- Irving P, Troxler L, Heuer TS, Belvin M, Kopczynski C, Reichhart JM, Hoffmann JA, Hetru C. A genome-wide analysis of immune responses in Drosophila. Proc Natl Acad Sci U S A 2001; 98:15119-24; PMID:11742098; https://doi.org/https://doi.org/10.1073/pnas.26157399810.1073/pnas.261573998
- Castillo JC, Creasy T, Kumari P, Shetty A, Shokal U, Tallon LJ, Eleftherianos I. Drosophila anti-nematode and antibacterial immune regulators revealed by RNA-Seq. BMC Genomics 2015; 16:519; PMID:26162375; https://doi.org/https://doi.org/10.1186/s12864-015-1690-210.1186/s12864-015-1690-2
- Igboin CO, Tordoff KP, Moeschberger ML, Griffen AL, Leys EJ. Porphyromonas gingivalis-host interactions in a Drosophila melanogaster model. Infect Immun 2011; 79:449-58; PMID:21041486; https://doi.org/https://doi.org/10.1128/IAI.00784-10 https://doi.org/10.1128/IAI.00785-1010.1128/IAI.00784-10 https://doi.org/10.1128/IAI.00785-10
- Jiggins FM, Kim KW. Contrasting evolutionary patterns in Drosophila immune receptors. J Mol Evol 2006; 63:769-80; PMID:17103056; https://doi.org/https://doi.org/10.1007/s00239-006-0005-210.1007/s00239-006-0005-2
- Lagueux M, Perrodou E, Levashina EA, Capovilla M, Hoffmann JA. Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila. Proc Natl Acad Sci U S A 2000; 97:11427-32; PMID:11027343; https://doi.org/https://doi.org/10.1073/pnas.97.21.1142710.1073/pnas.97.21.11427
- Ali H. Regulation of human mast cell and basophil function by anaphylatoxins C3a and C5a. Immunol Lett 2010; 128:36-45; PMID:19895849; https://doi.org/https://doi.org/10.1016/j.imlet.2009.10.00710.1016/j.imlet.2009.10.007
- Vallet-Gely I, Lemaitre B, Boccard F. Bacterial strategies to overcome insect defences. Nat Rev Microbiol 2008; 6:302-13; PMID:18327270; https://doi.org/https://doi.org/10.1038/nrmicro187010.1038/nrmicro1870
- Visschedyk DD, Perieteanu AA, Turgeon ZJ, Fieldhouse RJ, Dawson JF, Merrill AR. Photox, a novel actin-targeting mono-ADP-ribosyltransferase from Photorhabdus luminescens. J Biol Chem 2010; 285:13525-34; PMID:20181945; https://doi.org/https://doi.org/10.1074/jbc.M109.07733910.1074/jbc.M109.077339
- Lang AE, Schmidt G, Schlosser A, Hey TD, Larrinua IM, Sheets JJ, Mannherz HG, Aktories K. Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science 2010; 327:1139-42; PMID:20185726; https://doi.org/https://doi.org/10.1126/science.118455710.1126/science.1184557
- Costa SC, Girard PA, Brehelin M, Zumbihl R. The emerging human pathogen Photorhabdus asymbiotica is a facultative intracellular bacterium and induces apoptosis of macrophage-like cells. Infect Immun 2009; 77:1022-30; PMID:19075024; https://doi.org/https://doi.org/10.1128/IAI.01064-0810.1128/IAI.01064-08
- Tang H. Regulation and function of the melanization reaction in Drosophila. Fly (Austin) 2009; 3:105-11; PMID:19164947; https://doi.org/https://doi.org/10.4161/fly.3.1.774710.4161/fly.3.1.7747
- Eleftherianos I, Revenis C. Role and importance of phenoloxidase in insect hemostasis. J Innate Immun 2011; 3:28-33; PMID:21051882; https://doi.org/https://doi.org/10.1159/00032193110.1159/000321931
- Hollmann TJ, Mueller-Ortiz SL, Braun MC, Wetsel RA. Disruption of the C5a receptor gene increases resistance to acute Gram-negative bacteremia and endotoxic shock: opposing roles of C3a and C5a. Mol Immunol 2008; 45:1907-15; PMID:18063050; https://doi.org/https://doi.org/10.1016/j.molimm.2007.10.03710.1016/j.molimm.2007.10.037
- Castillo JC, Robertson AE, Strand MR. Characterization of hemocytes from the mosquitoes Anopheles gambiae and Aedes aegypti. Insect Biochem Mol Biol 2006; 36:891-903; PMID:17098164; https://doi.org/https://doi.org/10.1016/j.ibmb.2006.08.01010.1016/j.ibmb.2006.08.010
