Klebsiella pneumoniae carbapenamases (KPCs) are class A β-lactamases that efficiently hydrolyze almost all β-lactams including carbapenems, with exception of cephamycins. The blaKPC genes were initially discovered in K. pneumoniae in 1996 and have been frequently detected all over the world in particular among the species belonging to the family Enterobacteriaceae.Citation1 KPC-producing Pseudomonas aeruginosa isolates were initially found in 2006 from Colombia and increasingly being reported in South America, North America, the Caribbean islands, and China,Citation2,3 indicating a potential worldwide spread of KPC-producing P. aeruginosa. The blaKPC genes from P. aeruginosa are located on chromosome or plasmids.Citation4 Up to now, five fully sequenced blaKPC-carrying plasmids from P. aeruginosa are available in public databases: two IncP-6 plasmids pCOL-1 (accession number KC609323) and p10265-KPC (accession number KU578314) from ColombiaCitation5 and China,Citation6 respectively, two IncU plasmids pPA-2 (accession number KC609322) and pD5170990 (accession number KX169264) from ColombiaCitation5 and Brazil, respectively, and pBH6 (accession number CM003767; it could not be assigned into any of known incompatibility groups) from Brazil.Citation7 Each of pCOL-1, p10265-KPC, pPA-2, and pBH6 contains the sole resistance gene blaKPC-2, while pD5170990 carries blaKPC-2, strAB (aminoglycoside resistance), cmx (chloramphicol resistance), qacED1 (quaternary ammonium compound resistance), and sul1 (sulphonamide resistance).
The tet loci responsible for inducible tetracycline resistance commonly contain a tetA gene encoding the transmembrane tetracycline efflux protein, and a tetR gene encoding the tetracycline repressor.Citation8 At least 38 different classes (A, B, etc) of tet efflux have been identified, giving the designations tetA(A)-tetR(A), tetA(B)-tetR(B), etc.Citation8 In P. aeruginosa, the class A tetracycline resistance module tetA(A)-tetR(A) has been detected in only IncP-1α plasmids, including RP1/RP4/R18/R68/RK2 (identical to each other; accession number L27758) from United Kingdom,Citation9 pBS228 (accession number AM261760) from Russia,Citation10 and R1033 (accession number HM804085) from Spain;Citation11 among them RP1/RP4/R18/R68/RK2 and pBS228 were fully sequenced. This study dealt with detailed genomic characterization of two novel plasmids p14057A and p14057B found in a single clinical P. aeruginosa isolate.
On January 1st 2016, a middle-aged male with acute epigastric pain, which occurred upon nocturnal excessive drinking, was hospitalized in a local hospital in Nanjing City, China, and diagnosed to have acute pancreatitis. The symptoms of severe respiratory/renal failure and obnubilation progressed although the patient received the rehydration, acid suppression, and anti-inflammatory therapies. The patient was transferred to Jinling Hospital on January 3rd and received a series of symptomatic treatments, especially including continuous (until January 27th) and subsequent intermittent (until February 7th) mechanical ventilation, and repeated abdominal cavity drainage (until February 18th), and his symptoms associated with pancreatitis gradually improved. The patient started to suffer again from fever and respiratory difficulty since January 13th and was diagnosed to have ventilator-associated pneumonia. P. aeruginosa 14057 was isolated from the sputum specimens after repeated sampling and cultivation from January 13th to 15th. Bacterial species was confirmed by 16S rRNA gene sequencingCitation12 and PCR detection of P. aeruginosa-specific oafA gene.Citation13 According to antimicrobial susceptibility test results (see below), the patient received intravenous administration with amikacin plus ceftriaxone, and his symptoms associated with pneumonia progressively disappeared.
The major plasmid-borne carbapenemase genes were screened for by PCR,Citation14 followed by amplicon sequencing on ABI 3730 Sequencer (LifeTechnologies, CA, USA). Out of all the carbapenemase genes tested, only blaKPC-2 was detected in the 14057 isolate.
Plasmid DNA was isolated from the 14057 isolate using a Qiagen large construct kit, followed by sequencing from a mate-pair library with average insert size of 5,000 bp, using a MiSeq sequencer (Illumina, CA, USA). Sequence assembly and annotation were performed as described previously.Citation15 The contigs were assembled using Newbler 2.6.Citation16 Open reading frames and pseudogenes were predicted using RAST 2.017 combined with BLASTP/BLASTN18 searches against the UniProtKB/Swiss-Prot databaseCitation19 and the RefSeq database.Citation20 Annotation of resistance genes, mobile elements, and other features was carried out using online databases including CARD,Citation21 ResFinder,Citation22 BacMet,Citation23 ISfinder,Citation24 INTEGRALL,Citation25 and the Tn Number Registry.Citation26 Multiple and pairwise sequence comparisons were performed using MUSCLE 3.8.3127 and BLASTN, respectively. Gene organization diagrams were drawn in Inkscape 0.48.1.
It was revealed that the 14057 isolate harbored two circularly closed DNA sequences, designated p14057A and p14057B, which were 51,663 bp and 35,306 bp in length with mean G+C contents of 59.2% and 56.8% and contained 65 and 43 predicted ORFs, respectively (). The modular structure of each of p14057A and p14057B was discriminated as the backbone regions with insertion of a single accessory module, namely the blaKPC-2 region and the tetA(A) region, respectively (). The accessory modules were defined as acquired DNA regions associated with and bordered by mobile elements.
Figure 1. Schematic map of p14057A and p14057B. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and color, respectively. The innermost circle presents GC-skew [(G-C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents GC content. Shown also are backbone and accessory modules.
![Figure 1. Schematic map of p14057A and p14057B. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and color, respectively. The innermost circle presents GC-skew [(G-C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents GC content. Shown also are backbone and accessory modules.](/cms/asset/f09c95b8-75aa-448a-a857-dbb34ab3f6f9/kvir_a_1372082_f0001_oc.gif)
The backbones of p14057A and p14057B, 40.6 kb and 29.7kb in length, respectively, showed very low levels of similarity (≤11% query coverage) to DNA sequences available in public sequence databases. Located in the p14057A backbone were repA and its iterons responsible for plasmid replication initiation, parAB for plasmid partition, higBA encoding the toxin-antitoxin system for post-segregational killing, and a 27-kb conjugal transfer region that harbored tivB genes encoding a type IV secretion system.Citation28 The p14057B backbone contained genes involved in plasmid replication initiation (repA and its iterons) and maintenance (parAB, higBA, xerC, xamIM, and umuCD), but lacked those for conjugal transfer. xerC encoded a site-specific recombinase helping to resolve DNA dimers into monomers after termination of replication.Citation29 xamIM encoded a restriction-modification system involved in post-segregational killing.Citation30 umuCD encoded the SOS stress-inducible DNA polymerase for DNA lesion bypass.Citation31 The deduced RepA proteins of p14057A and p14057B had the highest matches (>97% amino acid similarity) to two different P. aeruginosa RepA proteins (accession numbers ERX63685 and WP_059309776.1), respectively. All the above replication proteins could not be assigned into any of known incompatibility groups.
The blaKPC-2 region () of p14057A was organized in order of a 3.5-kb partial region of the Tn1403 backbone, a 6.5-kb ΔTn6296 region, and an IS6100 element. Tn1403 was a multi-drug resistant transposon with insertion of In28 within the resolution (res) site and that of Tn5393c within orfB.Citation32 Insertion of foreign MDR regions at the sites within res and other regions generated Tn1403 derivatives such as Tn6060, Tn6061, Tn6162 and Tn6249 in P. aeruginosa.Citation33 The ΔTn3:ISKpn27 to ΔrepB region represented a core blaKPC platform, and its insertion into the mcp gene of the cryptic transposon Tn1722 carried in Tn1721,Citation34 leaving mcp to be truncated, generated the prototype Tn6296 as observed in pKP048.Citation35 Various Tn6296 derivatives with different deletions, insertions and rearrangements had been found in KPC-encoding plasmids from China.Citation6,36 In p14057A, ΔTn6296 manifested as a partial fragment of Tn6296 and consisted of res and the core blaKPC platform with further truncation of Tn3 (likely resulted from its connection with downstream IS6100), while the partial Tn1403 backbone region was composed of the 38-bp inverted repeat left (IRL) and the core transposition module tnpA (transposase)-tnpR (resolvase) lacking res. Both Tn1403 and Tn6296 belonged to the Tn21 subgroup of the Tn3 family, and their terminal inverted repeats and core transposition modules shared significant nucleotide sequence homology, which might facilitate homologous recombination between Tn1403- and Tn6296-like elements, generating the blaKPC-2 region of p14057A.
Figure 2. The accessory regions of p14057A and p14057B and comparison to related genetic contents. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). Numbers in brackets indicate the nucleotide positions within the corresponding plasmids. The accession numbers of Tn1721, Tn3, Tn6296, and Tn1403 for reference are X61367, HM749966, FJ628167, and AF313472, respectively.
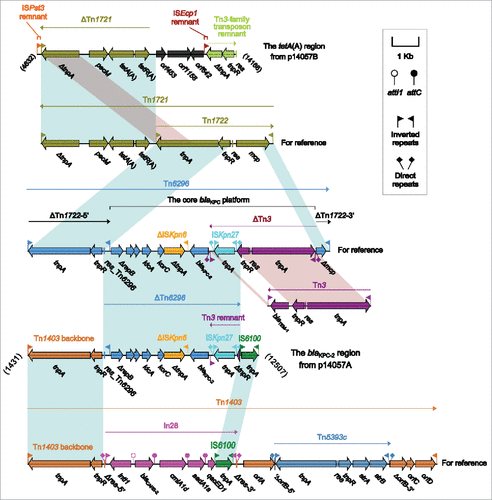
The tetA(A) region () of p14057B was organized sequentially as an 150-bp remnant of ISPst3, a 5.5-kb ΔTn1721 region, orf453, orf1158, orf642, a 201-bp remnant of ISEcp1, and an 1.4-kb remnant of a novel Tn3-family transposon core transposition module. Tn1721 was also a member of the Tn21 subgroup of Tn3 family, and had an unusual structure that included three 38-bp IR elements and a partial duplication of the tnpA gene.Citation34 Tn1722 was a independently transposable transposon with an IRL-tnpAR-res-mcp-IRR structure and located at the 3'-region of Tn1721, while the other end of Tn1721 included the pecM-tetA(A)-tetR(A) region, which represented a core genetic platform of class A tetracycline resistance genes.Citation34 The ΔTn1721 region was a 3'-terminal resistance fragment of Tn1721,Citation34 and carried the class A tetracycline resistance module tetA(A)-tetR(A). This is the first report of detecting tetA(A) and tetR(A) in P. aeruginosa from China.
Plasmids were transferred in attempt from the 14057 isolate into Escherichia coli TOP10 or DH10B and EC600 (highly resistant to rifampicin) through electroporation and conjugal transfer, respectively.Citation15 For selection of electroporant or transconjugant containing tetA(A) or blaKPC, 10 μg/ml tigecycline, 2 μg/ml imipenem, and 1000 μg/ml rifampicin were used in accordance with specific circumstances. Neither p14057A nor p14057B could be transferred into EC600 through conjugal transfer. p14057A was transferred into TOP10 through electroporation, yielding the blaKPC-2-carrying electroporant TOP10(p14057A), but p14057B could not be transferred into DH10B. Activity of Ambler class A/B/D carbapenemases in bacterial cell extracts was determined by a modified CarbaNP test.Citation15 Both 14057 and TOP10(p14057A) had the activity of class A carbapenemase, which was attributable to production KPC-2 enzyme (data not shown). Bacterial antimicrobial susceptibility was tested by the broth dilution method, and interpreted as per CLSI guidelines.Citation37 The 14057 isolate was resistant to ampicillin, ceftazidime, imipenem, aztreonam, azithromycin, minocycline, ciprofloxacin, trimethoprim, and sulfamethoxazole, but susceptible to amikacin, tigecycline, and colistin; TOP10(p14057A) was resistant to ampicillin, ceftazidime, imipenem, and aztreonam, but susceptible to all the other drugs tested ().
Table 1. Antimicrobial drug susceptibility profiles.
p14057A appears to be potentially self-transmissible due to the fact that is harbors a potentially complete set of conjugative transfer genes. p14057B seems unlikely to be self-transmissible because it contains none of putative mobilization or conjugal transfer regions. Each of these two plasmids carries a single accessory module, namely the blaKPC-2 region and the tetA(A) region, respectively, and these two resistance genes serve as the sole resistance determinant of the corresponding plasmid. The core genetic environments of blaKPC-2 and tetA(A)-tetR(A) have been well characterized previously, but they are connected with additional transposon- and insertion sequence-like elements to generate the novel blaKPC-2 and tetA(A) regions inserted in p14057A and p14057B, respectively, which might be resulted from complex recombination events. These two accessory resistance regions cannot be identified as native transposition units because they lack paired terminal inverted repeats at both ends. Co-existence of two different resistance plasmids has been reported in the clinical P. aeruginosa isolates from India,Citation38,39 but none of these plasmids were sequenced. This work presented the first two fully sequenced resistance plasmids coexisting in P. aeruginosa, which is a significant cause of nosocomial infection. Data presented here provides a deeper insight into how P. aeruginosa becomes drug-resistant by diversified mechanisms, and illustrates the way that genomic sequence analysis allows assessment of the genetic basis of plasmid-mediated resistance profile and appears to eventually promote establishment of successful antimicrobial treatment.
Nucleotide sequence accession numbers: The p14057A and p14057B sequences were submitted to GenBank under accession numbers KY296095 and KY296096, respectively.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Special Key Project of Biosafety Technologies (2017YFC1200800) for the National Major Research & Development Program of China.
References
- Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785-96. doi:10.1016/S1473-3099(13)70190-7. PMID:23969216
- Jacome PR, Alves LR, Cabral AB, Lopes AC, Maciel MA. First report of KPC-producing Pseudomonas aeruginosa in Brazil. Antimicrob Agents Chemother. 2012;56:4990. doi:10.1128/AAC.00699-12. PMID:22751532
- Ge C, Wei Z, Jiang Y, Shen P, Yu Y, Li L. Identification of KPC-2-producing Pseudomonas aeruginosa isolates in China. J Antimicrob Chemother. 2011;66:1184-6. doi:10.1093/jac/dkr060. PMID:21393139
- Cuzon G, Naas T, Villegas MV, Correa A, Quinn JP, Nordmann P. Wide dissemination of Pseudomonas aeruginosa producing beta-lactamase blaKPC-2 gene in Colombia. Antimicrob Agents Chemother. 2011;55:5350-3. doi:10.1128/AAC.00297-11. PMID:21844315
- Naas T, Bonnin RA, Cuzon G, Villegas MV, Nordmann P. Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J Antimicrob Chemother. 2013;68:1757-62. doi:10.1093/jac/dkt094. PMID:23569197
- Dai X, Zhou D, Xiong W, Feng J, Luo W, Luo G, Wang H, Sun F, Zhou X. The IncP-6 plasmid p10265-KPC from Pseudomonas aeruginosa carries a novel ΔISEc33-associated blaKPC-2 gene cluster. Front Microbiol. 2016;7:310. doi:10.3389/fmicb.2016.00310. PMID:27014233
- Galetti R, Andrade LN, Chandler M, Varani Ade M, Darini AL. New small plasmid harboring blaKPC-2 in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60:3211-4. doi:10.1128/AAC.00247-16. PMID:26953192
- Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245:195-203. doi:10.1016/j.femsle.2005.02.034. PMID:15837373
- Pansegrau W, Lanka E, Barth PT, Figurski DH, Guiney DG, Haas D, Helinski DR, Schwab H, Stanisich VA, Thomas CM. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J Mol Biol. 1994;239:623-63. doi:10.1006/jmbi.1994.1404. PMID:8014987
- Haines AS, Jones K, Batt SM, Kosheleva IA, Thomas CM. Sequence of plasmid pBS228 and reconstruction of the IncP-1α phylogeny. Plasmid. 2007;58:76-83. doi:10.1016/j.plasmid.2007.01.001. PMID:17320955
- Pinyon JL, Hall RM. Evolution of IncP-1α plasmids by acquisition of antibiotic and mercuric ion resistance transposons. Microb Drug Resist. 2011;17:339-43. doi:10.1089/mdr.2010.0196. PMID:21476866
- Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74:2461-70. doi:10.1128/AEM.02272-07. PMID:18296538
- Choi HJ, Kim MH, Cho MS, Kim BK, Kim JY, Kim C, Park DS. Improved PCR for identification of Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2013;97:3643-51. doi:10.1007/s00253-013-4709-0. PMID:23504075
- Chen Z, Li H, Feng J, Li Y, Chen X, Guo X, Chen W, Wang L, Lin L, Yang H, et al. NDM-1 encoded by a pNDM-BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Front Microbiol. 2015;6:294. doi:10.3389/fmicb.2015.00294. PMID:25926823
- Feng W, Zhou D, Wang Q, Luo W, Zhang D, Sun Q, Tong Y, Chen W, Sun F, Xia P. Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Sci Rep. 2016;6:33419. doi:10.1038/srep33419. PMID:27641711
- Nederbragt AJ. On the middle ground between open source and commercial software – the case of the Newbler program. Genome Biol. 2014;15:113. doi:10.1186/gb4173. PMID:25180324
- Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365. doi:10.1038/srep08365. PMID:25666585
- Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41:W29-33. doi:10.1093/nar/gkt282. PMID:23609542
- Boutet E, Lieberherr D, Tognolli M, Schneider M, Bansal P, Bridge AJ, Poux S, Bougueleret L, Xenarios I. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol Biol. 2016;1374:23-54. doi:10.1007/978-1-4939-3167-5_2. PMID:26519399
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733-45. doi:10.1093/nar/gkv1189. PMID:26553804
- Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2016.
- Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640-4. doi:10.1093/jac/dks261. PMID:22782487
- Pal C, Bengtsson-Palme J, Rensing C, Kristiansson E, Larsson DG. BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014;42:D737-43. doi:10.1093/nar/gkt1252. PMID:24304895
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32-6. doi:10.1093/nar/gkj014. PMID:16381877
- Moura A, Soares M, Pereira C, Leitao N, Henriques I, Correia A. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics. 2009;25:1096-8. doi:10.1093/bioinformatics/btp105. PMID:19228805
- Roberts AP, Chandler M, Courvalin P, Guedon G, Mullany P, Pembroke T, Rood JI, Smith CJ, Summers AO, Tsuda M, et al. Revised nomenclature for transposable genetic elements. Plasmid. 2008;60:167-73. doi:10.1016/j.plasmid.2008.08.001. PMID:18778731
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792-7. doi:10.1093/nar/gkh340. PMID:15034147
- Thomas CM, Thomson NR, Cerdeno-Tarraga AM, Brown CJ, Top EM, Frost LS. Annotation of plasmid genes. Plasmid. 2017;91:61-7. doi:10.1016/j.plasmid.2017.03.006. PMID:28365184
- Hallet B, Arciszewska LK, Sherratt DJ. Reciprocal control of catalysis by the tyrosine recombinases XerC and XerD: an enzymatic switch in site-specific recombination. Mol Cell. 1999;4:949-59. doi:10.1016/S1097-2765(00)80224-5. PMID:10635320
- Mruk I, Kobayashi I. To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res. 2014;42:70-86. doi:10.1093/nar/gkt711. PMID:23945938
- Goldsmith M, Sarov-Blat L, Livneh Z. Plasmid-encoded MucB protein is a DNA polymerase (pol RI) specialized for lesion bypass in the presence of MucA, RecA, and SSB. Proc Natl Acad Sci U S A. 2000;97:11227-31. doi:10.1073/pnas.200361997. PMID:11016960
- Stokes HW, Elbourne LD, Hall RM. Tn1403, a multiple-antibiotic resistance transposon made up of three distinct transposons. Antimicrob Agents Chemother. 2007;51:1827-9. doi:10.1128/AAC.01279-06. PMID:17261631
- Di Pilato V, Pollini S, Rossolini GM. Tn6249, a new Tn6162 transposon derivative carrying a double-integron platform and involved with acquisition of the blaVIM-1 metallo-β-lactamase gene in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2015;59:1583-7. doi:10.1128/AAC.04047-14. PMID:25547348
- Partridge SR. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev. 2011;35:820-55. doi:10.1111/j.1574-6976.2011.00277.x. PMID:21564142
- Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L. Novel genetic environment of the carbapenem-hydrolyzing β-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother. 2009;53:4333-8. doi:10.1128/AAC.00260-09. PMID:19620332
- Wang L, Fang H, Feng J, Yin Z, Xie X, Zhu X, Wang J, Chen W, Yang R, Du H. Complete sequences of KPC-2-encoding plasmid p628-KPC and CTX-M-55-encoding p628-CTXM coexisted in Klebsiella pneumoniae. Front Microbiol. 2015;6:838. doi:10.3389/fmicb.2015.00838. PMID:26347725
- CLSI. Performance standards for antimicrobial susceptibility testing: twenty-fifth informational supplement M100-S25. Wayne, PA, USA: CLSI, 2015.
- Paul D, Dhar Chanda D, Maurya AP, Mishra S, Chakravarty A, Sharma GD, Bhattacharjee A. Co-carriage of blaKPC-2 and blaNDM-1 in clinical isolates of Pseudomonas aeruginosa associated with hospital infections from India. PLoS One. 2015;10:e0145823. doi:10.1371/journal.pone.0145823. PMID:26714034
- Paul D, Dhar D, Maurya AP, Mishra S, Sharma GD, Chakravarty A, et al. Occurrence of co-existing blaVIM-2 and blaNDM-1 in clinical isolates of Pseudomonas aeruginosa from India. Ann Clin Microbiol Antimicrob. 2016;15:31. doi:10.1186/s12941-016-0146-0. PMID:27154587
