ABSTRACT
Legionella pneumophila, the causative agent of Legionnaires’ disease, is widely distributed throughout natural and artificial water systems and can replicate in macrophages and amoebae. Amoebae are the natural hosts of L. pneumophila, whereas macrophages are incidentally infected. The life cycle of L. pneumophila comprises a replicative phase within the Legionella-containing vacuole (LCV) and a transmissive phase during which bacterial cells become motile and are released via killing of the host. Although the host death mechanisms induced by L. pneumophila have been studied, the expression patterns of related L. pneumophila genes have not been reported. The present study compared the expression patterns of host cell death-associated genes in L. pneumophila grown in the human monocytic cell line THP-1 and Acanthamoeba castellanii. Notably, when L. pneumophila was grown in THP-1, expression of the gene flaA, which is involved in the induction of pyroptosis, was downregulated during the course of infection. In contrast, sdhA associated indirectly with host death, was upregulated. Expression of the genes vipD and sidF, which are involved in the induction and suppression of apoptosis, changed by less than 2-fold. Notably, a lower percentage of pyroptotic cells was observed among infected THP-1 cells relative to uninfected cells, and the latter exhibited stronger expression of caspase-1. A different pattern was observed when L. pneumophila was grown in A. castellanii: flaA and vipD were activated, whereas sdhA and sidF were downregulated during the later stage of replication. The percentage of non-viable (annexin-V+ PI+ or annexin-V+PI−) A. castellanii organisms increased with Legionella infection, and the expression of metacaspase-1, which is involved in encystation was up-regulated at late infection time. In summary, L. pneumophila can multiply intracellularly in both amoebae and macrophages to induce cell death and secondary infection, and this characteristic is essential for its survival in water and the lungs. The gene expression profiles observed in this study indicated the increased cytotoxicity of L. pneumophila in A. castellanii, suggesting an increased adaptation of Legionella to this host.
Introduction
Legionella pneumophila is the major causative agent of Legionnaires’ disease, a pneumonia-like illness associated with high mortality among immunocompromised populations.Citation1,2 L. pneumophila is ubiquitously distributed throughout both natural and artificial water systems, including cooling towers, whirlpools, and shower heads which serve as important sources for pathogen transmission,Citation3,4 and it can replicate in a wide range of hosts from protozoa to human macrophages.Citation5,6 Acanthamoeba species are among the major environmental hosts of Legionella,Citation7 and these organisms play key roles in the survival of L. pneumophila in aquatic environments.
The survival of L. pneumophila within different hosts hinges on successful evasion of the hosts’ conserved phagocytic killing pathways.Citation8 The intracellular life cycle of L. pneumophila comprises replicative and transmissive (post-exponential) phases.Citation9 During the replicative phase, L. pneumophila multiplies actively inside Legionella-containing vacuoles (LCV). Here, L. pneumophila effector proteins translocated through the Dot/Icm type IV secretion system (T4SS) can manipulate host cell functions and support intracellular growth in the LCV.Citation10,11 After depleting nutrients inside the host, L. pneumophila enters the transmissive phase, wherein the organism becomes motile and achieves readiness for extracellular release via host cell death.Citation12
In contrast to its standard intracellular replication strategy, L. pneumophila employs distinctive, host type-dependent mechanisms to induce host cell death. In mammalian macrophages, L. pneumophila induces cell death by apoptosis, a non-lytic, caspase-3-induced process, and pyroptosis, a lytic, proinflammatory process triggered by L. pneumophila flagellin and induced by host caspase-1.Citation13,14 However, the mechanism of cell death induction in amoeba hosts is less well-defined. Although protozoan species possess a functional pathway that leads to apoptosis, it remains unclear whether this pathway is triggered by L. pneumophila infection.Citation15,16 However, Legionella-infected amoeba hosts are known to undergo necrosis mediated by pore-forming activity.Citation17 Furthermore, the release of Legionella-containing vesicles from the amoeba host was observed in L. pneumophila-infected hosts before encystation.Citation18 Notably, this encystation process was mediated by metacaspase-1, a caspase-like enzyme.Citation19 A more recent study reported that L. pneumophila infection reduced cell cycle protein production and impaired cell proliferation in the amoeba host.Citation20
Key L. pneumophila virulence factors, including flaA, vipD, sdhA, and sidF, contribute to macrophage cell death.Citation21,22 Of these, flaA encodes L. pneumophila flagellin, a flagellar component produced during the stationary growth phase of L. pneumophila.Citation23 In addition to its role as a structural protein, flagellin exhibits effector functions: for example, the injection of flagellin into the host cytoplasm from the LCV leads to the activation of the inflammasome and caspase-1, and consequently to pyroptosis.Citation24 Several researchers have studied the role of vipD. Gaspar et al. demonstrated that vipD, a phospholipase, could inhibit endosomal fusion with the LCV in COS-1 and CHO cell models, and observed rounding and death in VipD-producing cells.Citation25 Additionally, Zhu et al. described vipD-induced apoptosis in L. pneumophila-infected macrophages, observing that this phospholipase hydrolyzed both phosphatidylethanolamine (PE) and phosphocholine (PC) to destabilize the mitochondrial membrane, and the consequent release of cytochrome c led to caspase-3 activation and apoptosis.Citation26
In contrast to flaA and vipD, which trigger death in L. pneumophila-infected host cells, sdhA and sidF have been shown to inhibit host cell death. The gene sdhA encodes a Dot/Icm-translocated effector protein that prevents pyroptosis by blocking AIM2 inflammasome and caspase-1 activation and is therefore required for L. pneumophila multiplication within macrophages.Citation27 In the absence of sdhA protein, L. pneumophila infection causes nuclear degradation, mitochondrial disruption, and significant cell death in infected macrophages.Citation28 Additionally, a previous infection study reported an association of sdhA-deficient L. pneumophila with increased dendritic cell death.Citation29 The sidF protein inhibits macrophage apoptosis by interacting with the endogenous pro-apoptotic Bcl-2 family proteins to facilitate the intracellular multiplication of L. pneumophila.Citation30
These host cell death-related L. pneumophila genes directly affect the pathogen's intracellular replication cycle. Although studies have evaluated the roles of these genes in host cell death after L. pneumophila infection, no reported study has investigated trends in the expression of these genes in macrophages and amoebae during various stages of L. pneumophila multiplication. This study aimed to compare the expression of flaA, vipD, sdhD, and sidF during intracellular L. pneumophila replication and cell death in Acanthamoeba castellanii and THP-1 monocyte hosts.
Results
Determination of the post-exponential phase of L. pneumophila
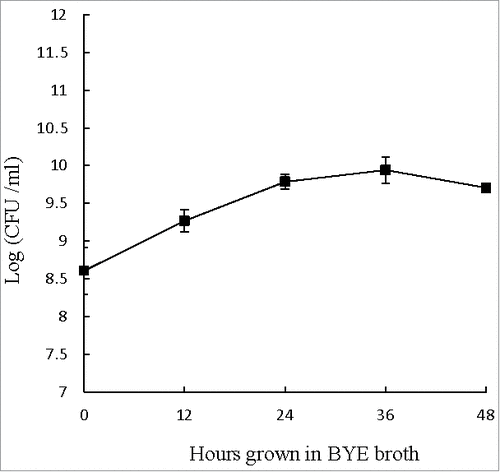
During extracellular growth in BYE broth, the number of L. pneumophila increased from 108 to 1010 CFU ml−1 during the first 24 hours. From 24 to 48 hours, however, the numbers of viable L. pneumophila remained near 1010 CFU ml−1 (), suggesting that the bacteria might have reached the post-exponential phase (reportedly the most virulent phase) and would be ready to infect host cells.Citation31,32 Accordingly, L. pneumophila cultured in BYE broth for 48 hours was used to infect THP-1 cells and A. castellanii in this study.
Figure 1. Extracellular growth curve of L. pneumophila. Number of viable and culturable L. pneumophila grown in BYE broth was enumerated at every 12 hours using plate count method. BCYE agar plates were used for plate count. Wilcoxon-signed rank test was used to compare the bacterial counts at 12 to 48 hours after inoculation with that at 0 hour. A p-value <0.05 was considered significant.
Replication capacities of L. pneumophila in THP-1 cells and A. castellanii
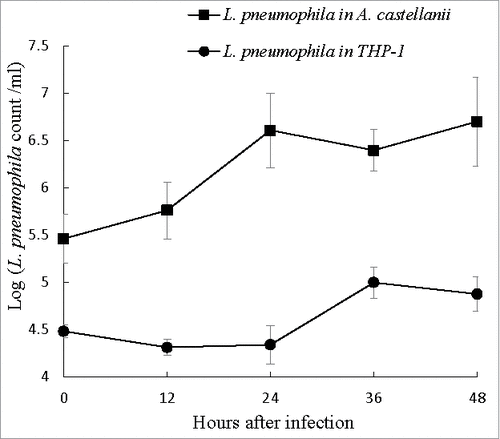
THP-1 cells and A. castellanii were both challenged with L. pneumophila at a multiplicity of infection (MOI) of 10. Following gentamicin treatment, however, the initial intracellular L. pneumophila counts (reported as log CFU ml−1) at 0 hours were 4.48 in THP-1 cells and 5.54 in A. castellanii, which might indicate a difference in the hosts abilities to uptake L. pneumophila. When grown in THP-1 cells, the intracellular Legionella number remained stable during the first 24 hours post-infection, and exhibited a <1-log increase from 24 to 36 hours post-infection followed by another stable period from 36 to 48 hours (). By contrast, in A. castellanii, intracellular L. pneumophila exhibited a 1-log increase in growth during the first 24 hours post-infection, with no further increases up to 48 hours (), thus demonstrating better replication of L. pneumophila in A. castellanii.
Figure 2. Intracellular growth of L. pneumophila. Viable and culturable L. pneumophila grown in THP-1 and A. castellanii were released by lysis of the host cells at 12-hour intervals up to 48 hours. The released L. pneumophila cells were enumerated using plate count method. BCYE agar plates were used for plate count. Wilcoxon-signed rank test was used to compare the bacterial counts at 12 to 48 hours after inoculation with that at 0 hour. A p-value <0.05 was considered significant.
Expression of L. pneumophila virulence genes in different hosts
When grown in THP-1 cells, the expression of the pyroptosis-related genes flaA and sdhA varied more dynamically over time. The expression of flaA decreased from −3.6-fold to −6.7-fold over time, whereas the expression of sdhA was upregulated at all time points, increasing from a 1.5-fold upregulation at 12 hours to 8.1-fold at 48 hours post-infection (). By contrast, the expression of the apoptosis-related genes vipD and sidF was more static, with slight decreases (≤2-fold downregulation) over time ().
Figure 3. L. pneumophila virulence genes expression during intracellular growth in THP-1 and A. castellanii. L. pneumophila grown in THP-1 (upper panel) and A. castellanii (lower panel) at different time points were collected for RNA isolation and used to run quantitative RT-PCR. All virulence genes had been normalized to housekeeping gene gyrB. Data were expressed as the mean fold change (2−ΔΔCT) compared to T0. Error bars show the SEM (Standard error of the mean). Fold changes at T24 to T48 were statistically compared with that at T12 using Wilcoxon signed rank test, asterisks (#) represent significant differences (p < 0.05).
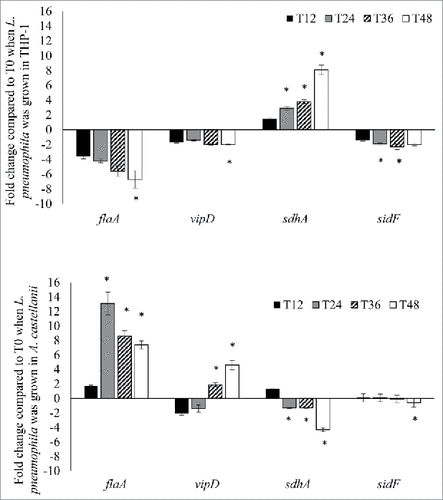
In A. castellanii, the expression of both flaA and vipD was upregulated. During the first infection cycle, flaA expression had not obviously changed (1.7-fold upregulation) at 12 hours post-infection, but peaked (13.1-fold) at 24 hours, followed by decreases during the second infection cycle to 8.6- and 7.4-fold upregulation at 36 hours and 48 hours, respectively. Although vipD exhibited 1.8- and 1.3-fold downregulation at 12 and 24 hours, respectively, the expression levels exhibited an increasing trend in the second infection cycle, with 1.8- and 4.6-fold upregulation at 36 and 48 hours, respectively (). By contrast, sdhA and sidF exhibited changes of <2-fold at most time points, except for a 4.3-fold downregulation of sdhA at 48 hours post-infection (). In summary, these gene expression studies indicate that during growth in THP-1 cells, L. pneumophila genes responsible for pyroptosis activation were downregulated, whereas those involved in suppressing host pyroptosis were activated. By contrast, the L. pneumophila genes responsible for initiating host cell death were upregulated during growth in A. castellanii.
Expression of CASP genes in the host cells
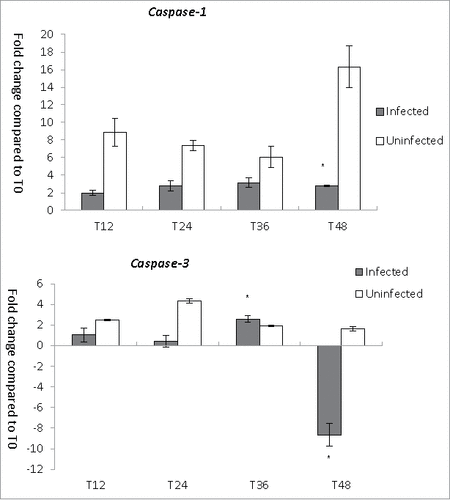
To understand the host cell responses during L. pneumophila infection, we investigated the expression of CASP-1 and CASP-3 in THP-1 cells (). In uninfected THP-1 cells, the expression of CASP-1, which encodes caspase-1, was strongly upregulated, with increases ranging from 6.1- to 16.3-fold over a period of 12–48 hours. Despite the strong upregulation of CASP-1 at 48 hours, however, the viable cell number was not remarkably reduced. By contrast, in L. pneumophila-infected THP-1 cells, CASP-1 was only modestly upregulated, with increases ranging from 2- to 3.2-fold between 12 and 48 hours post-infection. The expression of CASP-3, which encodes the apoptotic protein caspase-3, was lower in L. pneumophila-infected THP-1 cells than in uninfected cells at all time points except for T36, when the trend reversed slightly (). An 8.6-fold downregulation of CASP-3 expression at 48 hours post-infection might have been caused by a decrease in the number of viable THP-1 cells. In summary, our results demonstrate reduced activation of caspase genes in the Legionella-infected cells relative to uninfected cells.
Figure 4. CASP genes expression during L. pneumophila infection in THP-1. Fold changes in the expression of CASP-1 and CASP-3 in THP-1 were detected using quantitative RT-PCR every 12 hours from T0 to T48. Expression levels of the THP-1 genes were normalized to those of its housekeeping gene GAPDH. Data were expressed as the mean fold change. Error bars show the SEM (Standard error of the mean). Fold changes at T24 to T48 were statistically compared with the that at T12 using Wilcoxon signed rank test, asterisks (#) represent significant differences (p < 0.05).
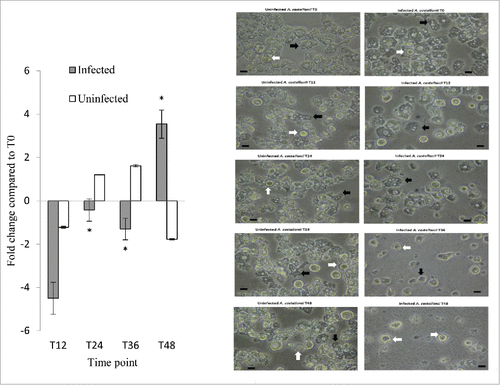
The expression of the A. castellanii metacaspase-1 gene (MCASP-1) was also investigated (, left panel). In uninfected A. castellanii, a <2-fold change in MCASP-1 expression was observed from the 12- to 48-hour time points. In Legionella-infected A. castellanii, however, MCASP-1 expression exhibited an increasing trend from 12 to 48 hours post-infection. At 12 hours, MCASP-1 expression was downregulated by 4.5-fold, downregulated by only 1.5-fold at 24 and 36 hours, and upregulated by 3.5-fold at 48 hours, indicating that MCASP-1 activation occurred at a later time point after L. pneumophila infection. As MCASP-1 promotes amoeba encystment, we microscopically examined both uninfected and infected cells for the presence of amoeba cysts at different time points. At 48 hours, amoeba cysts were present in 1% and 32% of the uninfected and infected A. castellanii, respectively, demonstrating that L. pneumophila led to increased cyst formation (, right panel).
Figure 5. MCASP-1 expression during L. pneumophila grown in A. castellanii that carried change from trophozoites to cysts. Fold changes in the expression of MCASP-1 in A. castellanii were detected using quantitative RT-PCR every 12 hours from T0 to T48 (Left). The expression level of MCASP-1 was normalized to that of A. castellanii 18S rRNA gene. Data were expressed as the mean fold change. Error bars show the SEM (Standard error of the mean). Fold changes at T24 to T48 were statistically compared with the that at T12 using Wilcoxon signed rank test, asterisks (#) represent significant change (p < 0.05). The microscopic images (Right) show the morphologies of the uninfected and infected A. castellanii. Black arrows indicate trophozoites and white arrows indicate cyst forms that have two-layer cell wall (scale bar = 25 μm, magnification 400 ×).
Investigation of host cell death after L. pneumophila infection
We additionally evaluated the occurrence of cell death in the two hosts using annexin-V and propidium iodide (PI) staining. Positive double staining (annexin-V+PI+) served to indicate pyroptosis and late apoptosis, negative staining (annexin-V−PI−) indicated viability, annexin-V+ single positivity denoted early apoptosis, and PI+ single positivity indicated late necrosis or cell debris. In THP-1 cells, the pyroptosis/late apoptosis population (annexin-V+PI+) in the Legionella-infected and uninfected groups increased from 13.9% to 41.5% and from 8.33% to 53.9%, respectively, from 0 to 48 hours (), and significantly lower frequencies of this population were observed in the Legionella-infected group from 12 to 48 hours (p values <0.05). This corroborated the observed reduction in CASP-1 expression in Legionella-infected cells. However, when we compared the percentages of viable cells and the trends of caspase gene expression, we noted that although the percentage of viable cells in the infected group was the lowest at 48 hours, the highest level of CASP-1 expression was not observed at that time point. By contrast, a 16-fold increase in CASP-1 expression was observed in the uninfected cells at 48 hours, even there was no remarkable corresponding decrease in the percentage of viablecells at T48 (). In summary, there was no clear relationship between the annexin-V/PI staining results and the caspase expression patterns.
The increased percentage of dead cells in the infected group at 0 hours might have resulted from experimental manipulation. Notably, both infected and uninfected THP-1 cells exhibited low frequencies of early apoptotic cells at all time points (<3% and 6%, respectively) (), although the infected group had significantly lower percentages at 24 and 48 hours (p values <0.05). The viable population decreased from 82.9% to 53.7% in the infected group and from 89.5% to 39.4% in the uninfected group over time (), and the infected group had a significantly higher percentage of viable cells relative to the uninfected group from 12 to 48 hours (p values <0.05). These results are further summarized in and .
As noted, protozoan species possess an apoptotic pathway, although it is not triggered by L. pneumophila infection.Citation16,17 In our examination of A. castellanii viability, we observed that the annexin-V+PI+, or necrotic, population increased in both Legionella-infected and uninfected organisms from 0 to 36 hours (from 14.5% to 44.7% and from 1.61% to 39.4%, respectively) (). Notably, the percentage of necrotic cells was significantly higher in the L. pneumophila-infected population at all time points (p values <0.05). Furthermore, the percentage of annexin-V+PI+ cells decreased at 48 hours in both L. pneumophila-infected and uninfected A. castellanii, which might indicate the harvesting of the insufficient numbers of cells for flow cytometric analyses at the last time point. In contrast to THP-1 cells, the L. pneumophila-infected A. castellanii had higher percentages of necrotic cells, compared to the uninfected A. castellanii (p values <0.05). This increase in annexin-V+PI+ double-staining suggests the changes in host membrane permeability associated with necrosis.Citation17 The percentages of early apoptotic cells (annexin-V+PI⁻) ranged from 4.1% to 6.5% and from 0.55% to 7.4% in Legionella-infected and uninfected A. castellanii, respectively (), and these percentages were significantly higher in Legionella-infected cells at 0 and 12 hours (p value <0.05). The viable populations (annexin-V−PI−) decreased from 59.1% to 33.8% in the infected group and from 72.4% to 39.1% in the uninfected group (), and the percentages of viable cells were significantly lower among infected A. castellanii relative to uninfected cells at all time points (p value <0.05). The strong reduction in viability among uninfected A. castellanii (72.4% at T0 vs. 39.1% at T48) might be attributable to a higher-than-optimal incubation temperature (37°C vs. the optimal 30°C).Citation33
Discussion
This study compared trends in the expression of L. pneumophila genes involved in the manipulation of host cell death. When L. pneumophila was grown in THP-1 cells, expression of the pyroptotic protein flaA was downregulated over time. By contrast, the expression of sdhA, which stabilizes the LCV and inhibits pyroptosis, increased steadily after L. pneumophila infection. The expression profiles of flaA and sdhA, together with the reduced expression of CASP genes in L. pneumophila-infected THP-1 cells, indicated that cell death pathways were inhibited after infection. Furthermore, the percentage of non-viable cells was higher in uninfected THP-1 cells than in L. pneumophila-infected cells. Although this inhibition of host death favored the intracellular multiplication of L. pneumophila, it might also hinder the egress of L. pneumophila and decrease the overall number of intracellular L. pneumophila in THP-1 cells. Our finding that L. pneumophila had a lower replication level when grown in THP-1 cells supports this possibility.
In a transcriptome study, Faucher et al. demonstrated the upregulation of sidF in Legionella-infected THP-1 cells.Citation34 Legionella sidF is involved in phosphoinositide metabolism and remodeling, and is therefore essential to LCV maintenance.Citation35 In our study, we observed a <2-fold downregulation of sidF expression at all time points. We similarly observed <2-fold changes in vipD expression levels. The involvement of both sidF and vipD in apoptosis suggests that pyroptosis, which involves pore formation and thus facilitates the release of intracellular agents, plays a more major role in THP-1 monocytes during Legionella infection. Despite the small changes in vipD and sidF expression, however, we observed an 8.6-fold downregulation of CASP-3 expression at 48 hours post-infection that may have been caused by a decrease in the total number of viable THP-1 cells. Accordingly, this decrease might also have induced only a slight upregulation in CASP-1 at 48 hours after infection.
In infected THP-1 cells, flaA downregulation occurred gradually, although significant changes in gene expression were observed at the late time point (48 hours) rather than earlier time points. The downregulation of flaA could be due to the low number of intracellular Legionella and slow consumption of nutrients that failed to induce flaA expression. sdhA was upregulated gradually over the time points studied, expression levels were significantly higher at 24 to 48 hours after infection. sdhA has an indirect role in inhibiting cell death by maintaining the integrity of LCV, an increased level of sdhA ensured protection of the Legionella replication niche.
In uninfected THP-1 cells, we observed a 16.3-fold upregulation of CASP-1 expression at 48 hours, with no corresponding remarkable reduction in the viable cell number. Accordingly, we did not identify a clear relationship between the trends in viable cell numbers and CASP-1 expression. Possibly, this was due to a decreased number of viable cells, particularly at 48 hours, as this would affect the RNA yields and quantified gene expression levels.
In this study, we observed a high percentage of non-viable cells among uninfected THP-1 cells. Previous studies have demonstrated crosstalk between pathways related to autophagy and other types of cell death.Citation36,37 Growth in nutrient-deprived conditions causes starving mammalian cells to induce autophagy,Citation38,39 which in turn induces mitochondrial dysfunction and activates apoptosis or inflammatory pyroptosis.Citation37,38 The high rate of cell death among uninfected THP-1 cells might therefore be a result of nutrient deprivation.Citation39 Although the infected THP-1 cells were also grown in a nutrient-poor environment, L. pneumophila might have suppressed host cell death during its replication phase inside the host.
Previous reports have not described the trends of flaA, vipD, sdhA, and sidF expression during a L. pneumophila infection of A. castellanii. In this study, we found that the expression patterns of these four genes differed somewhat from those in THP-1 monocytes. The growth of L. pneumophila in A. castellanii led to the upregulation of flaA and vipD and downregulation of sdhA at later time points. Similar to our results, Bruggemann et al. (Citation2006) demonstrated an upregulation of flaA (lpg1340) from 4 to 11 hours post-infection during intracellular growth in Acanthamoeba.Citation40 The activation of flaA is known to promote cell death. Our findings showed that flaA expression peaked at 24 hours post-infection during the intracellular growth phase of L. pneumophila in A. castellanii. This lined up with the pattern of pyroptosis observed in Acanthamoeba, which exhibited a peak at 24 hours post-L. pneumophila infection. This increase in host cell death after replication facilitates the egress of Legionella and further infection of new host cells.
Although we observed a sharp increase in flaA expression during the first infection cycle, this expression decreased and subsequently rebounded to a lower peak during the second cycle. This decrease in flaA corresponds to the invasion of new hosts and multiplication of L. pneumophila and may be attributable to the increased frequency of encystation. Furthermore, the second infection cycle was slower, leading to a subdued flaA response. Nevertheless, in infected A. castellanii, significant changes in flaA expression were observed at an early time point (24 hours), corresponding to the time point when Legionella had multiplied intracellularly to reach the point of release. Therefore, the Legionella genes responsible for controlling host cell death are better adapted to Legionella replication within A. castellanii, compared with THP-1 cells. Accordingly, Legionella species are better adapted to Acanthamoeba than to macrophages.
Although vipD expression was downregulated during the first infection cycle, the expression levels exhibited an overall increasing trend. Although the slow activation response is not fully understood, vipD might play a less important role relative to flaA in the regulation of Legionella growth within an Acanthamoeba host. In the second infection cycle, no obvious increase in the intracellular L. pneumophila count was observed, despite upregulated levels of vipD. During the second cycle, the proportion of Acanthamoeba cysts increases, and lysosomal proteases might degrade cytosolic proteins and organelles within these cysts.Citation41 Notably, as vipD is an inhibitor of lysosome fusion, its upregulation might have reduced the fusion of lysosomes with LCVs during encystation. Regarding sdhA, the expression levels varied slightly from 12 to 36 hours, followed by a decrease at 48 hours. sdhA is involved in the stabilization of the LCV and protection from cell death. Accordingly, its downregulation would destabilize vacuoles, leading to cell death and the release of L. pneumophila from the host.
In our study, we observed an initial downregulation of MCASP-1 expression in Legionella-infected A. castellanii, followed by a trend toward increased expression during the course of infection. In a previous study, bacteria-infected amoebae grown at lower temperatures exhibited upregulated expression of MCASP-1 at 48 hours, compared to their uninfected counterparts.Citation18 The protozoan metacaspase has been shown to activate encystment.Citation19,42,43 During the course of infection, we observed cysts in both uninfected and L. pneumophila-infected A. castellanii at different time points, but note that more cysts were observed in the latter group at 24 hours and thereafter ().
Our findings regarding the intracellular growth of Legionella species in Acanthamoeba and THP-1 cells were similar to those reported by Weissenmayer et al. (2013)Citation44 and Roland et al. (2013),Citation45 respectively, but differed from those reported by DeyCitation46 and Lebeau.Citation47 We note that Dey reported the exponential replication of Legionella in Acanthamoeba only after 24 hours, while Lebeau observed Legionella duplication for up to 72 hours post-infection. The discrepancies in these findings could be due to differences in the experimental operations, including the MOIs, co-culture media,Citation31 and methods used to lyse host cells.Citation48
We note that our study of Legionella gene expression was subject to several limitations. First, the numbers of viable host cells decreased at later time points. Although we did not observe host cell exhaustion at late time points, a decrease in the number of viable host cells could affect the Legionella replication cycle, which might consequently affect gene expression. Second, growth of A. castellanii in the PYG media at later time points. Although amoebae can grow in PYG medium, our study used an incubation temperature of 37°C, which was not optimal for amoeba multiplication. However, we note that Legionella could not grow in PYG medium, which lacked a growth supplement. Third, the Legionella burdens (i.e., numbers of intracellular bacteria) differed between THP-1 cells and A. castellanii. To overcome this discrepancy, we normalized the expression of the four target genes to that of the reference gene gyrB, thus minimizing the effects of bacterial burden variations on the quantification of gene expression. Forth, accumulation of metabolic wastes in the culture environment could interfere with host viability, to minimize the background interference, uninfected cell controls have been set up in the study.
In summary, we demonstrated that the expression patterns of L. pneumophila genes involved in host cell death differ between THP-1 macrophage and A. castellanii hosts. Notably, in THP-1 cells, genes involved in cell death were downregulated while those involved in cell death suppression were upregulated, and the expression of the cell death-related genes CASP-1 and -3 was lower in L. pneumophila-infected vs. uninfected THP-1 cells. However, the opposite expression pattern was observed in A. castellanii with regard to L. pneumophila genes, and a trend toward increased expression of the A. castellanii MCASP-1 gene was observed in the L. pneumophila-infected group. Both L. pneumophila and A. castellanii are natural biofilm inhabitants, and our results demonstrate that the expression pattern of flaA, the Legionella gene responsible for inducing host death, was better adapted to the Legionella replication cycle within A. castellanii, compared to that within THP1 macrophages. Therefore, Legionella is better adapted to and more readily induces cell death in A. castellanii.
Materials and methods
Bacterial strains, acanthamoeba strain, and cell line used in this study
L. pneumophila Philadelphia strain (ATCC 33152) was used in this study. L. pneumophila was cultured on BCYE agar plates supplemented with αBCYE growth supplement and GVPC selective antibiotics (Oxoid, UK). The cultures were incubated at 37°C with 5% CO2 for 3–5 days. L. pneumophila was also cultured in liquid BYE broth according to previously described procedures.Citation49
A. castellanii (ATCC 30234) was cultured in supplied PYG medium (2% peptone, 0.1% yeast extract, and 1.8% glucoseCitation50) at 25°C for 1–2 weeks.Citation51 THP-1 cells (ATCC TIB-202) were were grown in RPMI 1640 growth medium (Gibco, USA) supplemented with 10% fetal bovine serum (Invitrogen, USA) and 1% penicillin/streptomycin (Gibco, USA) at 37°C with 5% CO2 for 3–5 days. Trypan Blue staining was used to enumerate viable cells. Briefly, 0.5 ml of a suspension of A. castellanii or THP-1 cells was mixed with 0.5 ml of 0.4% Trypan Blue stain, and the mixture was incubated at the ambient temperature for 5 min. Subsequently, the viable (i.e., unstained) cells were counted under a microscope using a hemocytometer.
Extracellular growth of L. pneumophila for determination of the post-exponential phase
A liquid culture was used to determine the time required by L. pneumophila to reach the post-exponential phase. L. pneumophila colonies grown on BCYE plates were suspended in phosphate-buffered saline (PBS), and the absorbance was adjusted to 1.0 at an absorbance of 600 nm (A600). Five hundred microlitres of this bacterial suspension were inoculated into 10 ml of BYE broth, which was then incubated at 37°C for 48 hours with agitation at 250 rpm. One hundred microliters of BYE culture were withdrawn at 12-hour intervals for up to 48 hours to enumerate the bacterial density using the plate count method. All experiments were performed in triplicate. The post-exponential phase was defined as the time point at which there was no further increase in L. pneumophila growth.
Intracellular growth assays
L. pneumophila was co-cultured with THP-1 cells at a MOI of 10.Citation31,52 Briefly, 2 ml of a THP-1 cell suspension (106 cells ml−1) were mixed with 20 μl of a post-exponential L. pneumophila suspension (109 colony-forming units [CFU] ml−1). The mixture was centrifuged in a conical tube at 900 × g for 5 min to enhance bacterial contact with the THP-1 cells, which were then incubated at 37°C with 5% CO2 for 3 hours. After incubation, the pellet was washed once with PBS and incubated with 2 ml of gentamicin (100 μg ml−1) at 37°C for 2 hours to kill extracellular bacteria. Subsequently, the mixture was washed twice with PBS and re-suspended in 2 ml of fresh incomplete RPMI medium (without fetal bovine serum). This point was regarded as “time zero” (0 hours, T0). At T0 and at every 12-hour interval up to 48 hours (12 hours–48 hours, T12 – T48), 106 THP-1 cells were incubated with 2 ml of sterilized distilled water at room temperature for 10 minutes. The THP-1 cells were then lysed by 5–10 forced passages through a 23-gauge syringe needle.Citation48 L. pneumophila released from the lysed co-cultures were enumerated using the plate count method. The experiments were performed in triplicate.
A co-culture of L. pneumophila and A. castellanii was set up as described above for the co-culture of L. pneumophila and THP-1, except that PYG was used as the co-culture medium and Page's amoeba saline (PAS) was used to wash the co-culture contents.Citation50 Two milliliters of A. castellanii suspension (106 cells ml−1) were inoculated into each well of a 6-well plate, followed by the addition of 20 µl of L. pneumophila suspension (109 CFU ml−1) to each well. After centrifugation at 900 × g for 5 min, the plate was incubated at 37°C with 5% CO2 for 3 hours. After a PAS wash, the medium in each well was replaced with 2 mL of gentamicin (100 μg ml−1). After 2-hour incubation, the co-culture mixtures were washed twice with PAS and re-suspended in 2 ml of PYG medium. This time point was regarded as T0. At every time point, 106 A. castellanii were added to 2 ml of sterilized distilled water and incubated at room temperature for 10 minutes, followed by lysis via 5–10 forced passages through a 23-gauge syringe needle. The experiments were performed in triplicate.
RNA isolation and reverse transcription
RNA samples were prepared from lysed co-cultures using the PureLink RNA Mini Kit (Invitrogen, USA) according to the manufacturer's instructions. A NanoDrop spectrophometer (Thermo Fisher Scientific, USA) was used to evaluate the purity and concentration of each isolated total RNA sample. A RNA sample was considered pure if the ratio of absorbance values at 260 nm and 280 nm (A260/280) fell within the range of 1.8–2.1. A consistent quantity of RNA (<800 ng RNA per DNase I reaction) was then digested using a DNaseI kit (Sigma, USA) at 37°C for 30 minutes to remove any DNA contamination. Subsequently, 10 µl of digested RNA were reverse transcribed into cDNA using a RevertAid First Strand cDNA Synthesis kit (Fermentas, USA). To monitor the presence of DNA contamination in the digested RNA sample, a second reverse transcription was without reverse transcriptase (no-reverse transcriptase control) was set up simultaneously.
Quantitative real-time PCR to detect gene expression
The sequences of primers and probes are summarized in . Quantitative real-time PCR was performed in 20-µl PCR mixtures comprising 1 µl of cDNA, 1 × SYBR Green Mastermix (Roche, Germany), and 500 µM of each forward and reverse primer. For TaqMan assays, the 20-µl PCR mix comprised 1 µl of cDNA, 1 × TaqMan Universal PCR Mastermix (ABI, USA), and 1 µl of 20 × TaqMan probes (containing the probe and forward and reverse primers). PCRs were run on an ABI7500 system (Applied Biosystems, Foster City, CA). The thermal cycling conditions comprised an initial denaturation step at 95°C for 10 min, followed by 40 cycles of 94°C for 30 s and 60°C for 1 min. The threshold cycle (CT) values of the target genes were normalized to those of the reference genes. Fold changes in the expression of various target genes and T24 to T48 relative to those at T12 were compared using the Wilcoxon signed rank test. A p value <0.05 was considered significant.
Table 1. Primers and TaqMan probes used in this study. All primers and probes were designed using online Primer-BLAST program (National Centre for Biotechnology Information, US).
Table 2. Flow cytometric analysis of L. pneumophila-infected and uninfected THP-1 cells using annexin-V and PI-staining. THP-1 cells were harvested and stained with annexin-V and PI at 12-hour intervals up to 48 hours. Stained cell samples were analysed on a FACS Aria III flow cytometer and data were analysed using BD FACSDiva software. Paired t-test was used to compared the percentages of cells at various stages between L. pneumophila-infected and uninfected THP-1 cells. Asterisks (#) represent statistical significant differences (p < 0.05).
Table 3. Flow cytometric analysis of L. pneumophila-infected and uninfected A. castellanii cells using annexin-V and PI-staining. A. castellanii cells were harvested and stained with annexin-V and PI at 12-hour intervals up to 48 hours. Stained amoeba samples were analysed on a FACS Aria III flow cytometer and data were analysed using BD FACSDiva software. Paired t-test was used to compared the percentages of cells at various stages between L. pneumophila-infected and uninfected A. castellanii. Asterisks (#) represent statistical significant differences (p < 0.05).
Flow cytometric analysis of cell death using annexin-V and propidium iodide (PI) staining
A total of 106 non-lysed, co-cultured cells (otherwise processed as described for intracellular growth assays) from both the L. pneumophila-inoculated and non-inoculated groups were collected and stained with fluorescein isothiocyanate (FITC)-labeled annexin-V and phycoerythrin (PE)-labeled PI (BD Pharmingen, USA). The labeled cells were then subjected to flow cytometry to determine the cell death frequencies. A. castellanii and THP-1 cells infected with L. pneumophila were collected at different time points (T0, T12, T24, T36 and T48), washed twice with ice-cold PBS, and incubated with 100 μl of 1 × staining buffer containing 5 μl annexin-V and 5 μl propidium iodide (PI). After a 15-minute incubation at room temperature in the dark, the reaction was stopped by adding 400 μl of 1 × annexin-V-stain buffer. The stained cells were analyzed on a FACS Aria™ III flow cytometer (BD, USA) on which the fluorescence compensation had been adjusted to minimize overlap of the FITC and PE signals. A total of 50,000 events were acquired for each samples. The flow cytometry data were analyzed using BD FACSDiva Software (BD, USA). The experiments were performed in triplicate, and data were presented as percentage. L. pneumophila-inoculated and non-inoculated groups were compared using paired t-test, and p value <0.05 was considered significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study was funded by a Central Research Grant of the Hong Kong Polytechnic University.
References
- Adeleke AA, Fields BS, Benson RF, Daneshvar MI, Pruckler JM, Ratcliff RM, Harrison TG, Weyant RS, Birtles RJ, Raoult D, et al. Legionella drozanskii sp nov., Legionella rowbothamii sp nov and Legionella fallonii sp nov.: Three unusual new Legionella species. Int J Syst Evol Micr. 2001;51:1151-1160. http://doi.org/10.1099/00207713-51-3-1151
- Diederen BMW. Legionella spp. and Legionnaires' disease. J Infection. 2008;56:1-12. http://doi.org/10.1016/j.jinf.2007.09.010
- Leoni E, De Luca G, Legnani PP, Sacchetti R, Stampi S, Zanetti F. Legionella waterline colonization: detection of Legionella species in domestic, hotel and hospital hot water systems. J Appl Microbiol. 2005;98:373-379. http://doi.org/10.1111/j.1365-2672.2004.02458.x. PMID:15659192
- Rowbotham TJ. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J Clin Pathol. 1983;36:978-986. http://doi.org/10.1136/jcp.36.9.978. PMID:6350372
- Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980;33:1179-1183. http://doi.org/10.1136/jcp.33.12.1179. PMID:7451664
- Gao LY, Harb OS, Abu Kwaik Y. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect Immun. 1997;65:4738-4746. PMID:9353059
- Stockman LJ, Wright CJ, Visvesvara GS, Fields BS, Beach MJ. Prevalence of Acanthamoeba spp. and other free-living amoebae in household water, Ohio, USA–1990-1992. Parasitol Res. 2011;108:621-627. http://doi.org/10.1007/s00436-010-2120-7. PMID:20978791
- Nash TW, Libby DM, Horwitz MA. Interaction between the Legionnaires-Disease Bacterium (Legionella Pneumophila) and Human Alveolar Macrophages – Influence of Antibody, Lymphokines, and Hydrocortisone. J Clin Invest. 1984;74:771-782. http://doi.org/10.1172/JCI111493. PMID:6470140
- Byrne B, Swanson MS. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66:3029-3034. PMID:9632562
- Coers J, Monahan C, Roy CR. Modulation of phagosome biogenesis by Legionella pneumophila creates an organelle permissive for intracellular growth. Nat Cell Biol. 1999;1:451-453. http://doi.org/10.1038/15687. PMID:10559990
- Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A. 2004;101:841-846. http://doi.org/10.1073/pnas.0304916101. PMID:14715899
- Sturgill-Koszycki S, Swanson MS. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J Exp Med. 2000;192:1261-1272. http://doi.org/10.1084/jem.192.9.1261. PMID:11067875
- Molmeret M, Bitar DM, Han LH, Abu Kwaik Y. Cell biology of the intracellular infection by Legionella pneumophila. Microbes Infect. 2004;6:129-139. http://doi.org/10.1016/j.micinf.2003.11.004. PMID:14738901
- Silveira TN, Zamboni DS. Pore Formation Triggered by Legionella spp. Is an Nlrc4 Inflammasome-Dependent Host Cell Response That Precedes Pyroptosis. Infect Immun. 2010;78:1403-1413. http://doi.org/10.1128/IAI.00905-09. PMID:20048047
- Gao LY, Kwaik YA. The modulation of host cell apoptosis by intracellular bacterial pathogens. Trends Microbiol. 2000;8:306-313. http://doi.org/10.1016/S0966-842X(00)01784-4. PMID:10878765
- Hagele S, Hacker J, Brand BC. Legionella pneumophila kills human phagocytes but not protozoan host cells by inducing apoptotic cell death. FEMS Microbiol Lett. 1998;169:51-58. http://doi.org/10.1111/j.1574-6968.1998.tb13298.x. PMID:9851034
- Gao LY, Abu Kwaik Y. The mechanism of killing and exiting the protozoan host Acanthamoeba polyphaga by Legionella pneumophila. Environ Microbiol. 2000;2:79-90. http://doi.org/10.1046/j.1462-2920.2000.00076.x. PMID:11243265
- Ohno A, Kato N, Sakamoto R, Kimura S, Yamaguchi K. Temperature-dependent parasitic relationship between Legionella pneumophila and a free-living amoeba (Acanthamoeba castellanii). Appl Environ Microb. 2008;74:4585-4588. http://doi.org/10.1128/AEM.00083-08
- Trzyna WC, Legras XD, Cordingley JS. A type-1 metacaspase from Acanthamoeba castellanii. Microbiol Res. 2008;163:414-423. http://doi.org/10.1016/j.micres.2006.06.017. PMID:16891103
- Mengue L, Regnacq M, Aucher W, Portier E, Hechard Y, Samba-Louaka A. Legionella pneumophila prevents proliferation of its natural host Acanthamoeba castellanii. Sci Rep-Uk. 2016;6:36448. http://doi.org/10.1038/srep36448. PMID:27805070
- Lamkanfi M, Dixit VM. Manipulation of host cell death pathways during microbial infections. Cell Host Microbe. 2010;8:44-54. http://doi.org/10.1016/j.chom.2010.06.007. PMID:20638641
- Creasey EA, Isberg RR. The protein SdhA maintains the integrity of the Legionella-containing vacuole. P Natl Acad Sci USA. 2012;109:3481-3486. http://doi.org/10.1073/pnas.1121286109
- Molofsky AB, Shetron-Rama LM, Swanson MS. Components of the Legionella pneumophila flagellar regulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect Immun. 2005;73:5720-5734. http://doi.org/10.1128/IAI.73.9.5720-5734.2005. PMID:16113289
- Silveira TN, Zamboni DS. Pore formation triggered by Legionella spp. is an Nlrc4 inflammasome-dependent host cell response that precedes pyroptosis. Infect Immun. 2010;78:1403-1413. http://doi.org/10.1128/IAI.00905-09. PMID:20048047
- Gaspar AH, Machner MP. VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proc Natl Acad Sci U S A. 2014;111:4560-4565. http://doi.org/10.1073/pnas.1316376111. PMID:24616501
- Zhu WH, Hammad LA, Hsu FS, Mao YX, Luo ZQ. Induction of caspase 3 activation by multiple Legionella pneumophila Dot/Icm substrates. Cell Microbiol. 2013;15:1783-1795. PMID:23773455
- Ge JN, Gong YN, Xu Y, Shao F. Preventing bacterial DNA release and absent in melanoma 2 inflammasome activation by a Legionella effector functioning in membrane trafficking. P Natl Acad Sci USA. 2012;109:6193-6198. http://doi.org/10.1073/pnas.1117490109
- Laguna RK, Creasey EA, Li Z, Valtz N, Isberg RR. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. P Natl Acad Sci USA. 2006;103:18745-18750. http://doi.org/10.1073/pnas.0609012103
- Nogueira CV, Lindsten T, Jamieson AM, Case CL, Shin S, Thompson CB, Roy CR. Rapid pathogen-induced apoptosis: A mechanism used by dendritic cells to limit intracellular replication of Legionella pneumophila. Plos Pathog. 2009;5:e1000478. http://doi.org/10.1371/journal.ppat.1000478. PMID:19521510
- Banga S, Gao P, Shen XH, Fiscus V, Zong WX, Chen LL, Luo ZQ. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. P Natl Acad Sci USA. 2007;104:5121-5126. http://doi.org/10.1073/pnas.0611030104
- Moffat JF, Tompkins LS. A quantitative model of intracellular growth of Legionella-Pneumophila in Acanthamoeba-Castellanii. Infect Immun. 1992;60:296-301. PMID:1729191
- Molofsky AB, Swanson MS. Differentiate to thrive: Lessons from the Legionella pneumophila life cycle. Mol Microbiol. 2004;53:29-40. http://doi.org/10.1111/j.1365-2958.2004.04129.x. PMID:15225301
- Schuster FL. Cultivation of pathogenic and opportunistic free-living amebas. Clin Microbiol Rev. 2002;15:342-354. http://doi.org/10.1128/CMR.15.3.342-354.2002. PMID:12097243
- Faucher SP, Mueller CA, Shuman HA. Legionella pneumophila transcriptome during intracellular multiplication in human macrophages. Front Microbiol. 2011;2:60. http://doi.org/10.3389/fmicb.2011.00060. PMID:21747786
- Hsu FS, Zhu WH, Brennan L, Tao LL, Luo ZQ, Mao YX. Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase. P Natl Acad Sci USA. 2012;109:13567-13572. http://doi.org/10.1073/pnas.1207903109
- Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr Opin Cell Biol. 2004;16:663-669. http://doi.org/10.1016/j.ceb.2004.09.011. PMID:15530778
- Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N. Crosstalk between apoptosis, necrosis and autophagy. Bba-Mol Cell Res. 2013;1833:3448-3459.
- Ryter SW, Mizumura K, Choi AM. The impact of autophagy on cell death modalities. Int J Cell Biol. 2014;2014:502676. http://doi.org/10.1155/2014/502676. PMID:24639873
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Bio. 2007;8:741-752. http://doi.org/10.1038/nrm2239
- Bruggemann H, Hagman A, Jules M, Sismeiro O, Dillies MA, Gouyette C, Kunst F, Steinert M, Heuner K, Coppee JY, et al. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol. 2006;8:1228-1240. http://doi.org/10.1111/j.1462-5822.2006.00703.x. PMID:16882028
- Leitsch D, Kohsler M, Marchetti-Deschmann M, Deutsch A, Allmaier G, Duchene M, Walochnik J Major role for cysteine proteases during the early phase of acanthamoeba castellanii encystment. Eukaryot Cell. 2010;9:611-618. http://doi.org/10.1128/EC.00300-09. PMID:20190073
- Saheb E, Trzyna W, Bush J. Caspase-like proteins: Acanthamoeba castellanii metacaspase and Dictyostelium discoideum paracaspase, what are their functions? J Biosciences. 2014;39:909-916. http://doi.org/10.1007/s12038-014-9486-0
- Saheb E, Trzyna W, Bush J. An Acanthamoeba castellanii metacaspase associates with the contractile vacuole and functions in osmoregulation. Exp Parasitol. 2013;133:314-326. http://doi.org/10.1016/j.exppara.2012.12.001. PMID:23274641
- Weissenmayer BA, Prendergast JG, Lohan AJ, Loftus BJ. Sequencing illustrates the transcriptional response of Legionella pneumophila during infection and identifies seventy novel small non-coding RNAs. PloS one. 2011;6:e17570. http://doi.org/10.1371/journal.pone.0017570. PMID:21408607
- Rolando M, Sanulli S., Rusniok C, Gomez-Valero L, Bertholet C, Sahr T, Margueron R, Buchrieser C. Legionella pneumophila effector roma uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe. 2013;13:395-405. http://doi.org/10.1016/j.chom.2013.03.004 PMID:23601102
- Dey R, Bodennec J, Mameri MO, Pernin P. Free-living freshwater amoebae differ in their susceptibility to the pathogenic bacterium Legionella pneumophila. FEMS Microbiol Lett. 2009;290:10-17. http://doi.org/10.1111/j.1574-6968.2008.01387.x. PMID:19016880
- Lebeau I, Lammertyn E, De Buck E, Maes L, Geukens N, Van Mellaert L, Anne J. Novel transcriptional regulators of Legionella pneumophila that affect replication in Acanthamoeba castellanii. Arch Microbiol. 2004;181:362-370. http://doi.org/10.1007/s00203-004-0664-6. PMID:15034642
- Dietersdorfer E, Cervero-Arago S, Sommer R, Kirschner AK, Walochnik J. Optimized methods for Legionella pneumophila release from its Acanthamoeba hosts. Bmc Microbiol. 2016;16:74. http://doi.org/10.1186/s12866-016-0691-x. PMID:27113731
- Takemura H, Yamamoto H, Kunishima H, Ikejima H, Hara T, Kanemitsu K, Terakubo S, Shoji Y, Kaku M, Shimada J. Evaluation of a human monocytic cell line THP-1 model for assay of the intracellular activities of antimicrobial agents against Legionella pneumophila. J Antimicrob Chemoth. 2000;46:589-594. http://doi.org/10.1093/jac/46.4.589
- Cirillo JD, Falkow S, Tompkins LS. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect Immun. 1994;62:3254-3261. PMID:8039895
- Buse HY, Ashbolt NJ. Differential growth of Legionella pneumophila strains within a range of amoebae at various temperatures associated with in-premise plumbing. Lett Appl Microbiol. 2011;53:217-224. http://doi.org/10.1111/j.1472-765X.2011.03094.x. PMID:21623847
- Tiaden AN, Kessler A, Hilbi H. Analysis of legionella infection by flow cytometry. Methods Mol Biol. 2013;954:233-249. http://doi.org/10.1007/978-1-62703-161-5_14. PMID:23150400
