Staphylococcus aureus colonizes the upper respiratory tract and skin of humans, which constitutes a risk factor for the development of invasive diseases such as pneumonia, meningitis and septicaemia, especially in immune-compromised people.Citation1 The success of S. aureus infections depends on the ability of the pathogen to escape the host's protective mechanisms, allowing invasion and pathogen proliferation. The multitude of evasive mechanisms and the increase in antibiotic resistance have made S. aureus a serious human threat. Moreover, methicillin-resistant S. aureus strains (MRSA) are no longer restricted to the hospital setting and are widespread in the community.Citation2
The innate immune system is the first barrier encountered by the pathogen during host infection. Several studies have reported that internalisation by epithelial cells and phagocytosis by macrophages exposes the microbes to reactive oxygen species (ROS).Citation3 Therefore, the defences against oxidative stress play an important role in pathogen survival. The wax moth larva Galleria mellonella, that only has innate immunity, is a recognized in vivo model for the study of bacterial virulence of several pathogens including S. aureus.Citation4–6 G. mellonella presents advantages over conventional mammalian models, due to the higher temperature required for infection (37°C) and the possibility of the direct injection of a precise inoculum.Citation7,8
Repair of Iron Centers (RIC) proteins are a widespread family of bacterial proteins, which are also present in the genomes of Trichomonas vaginalis and Cryptococcus neoformans eukaryotes.Citation9 The first RIC protein was found in E. coli due to the marked induction of the encoding gene (formerly named ytfE) in cells grown under nitrosative stress conditions.Citation10 Consistent with these results, transcriptomic studies done in several organisms have consistently shown that the expression of ric is induced in stressed cells, and that the ric gene deletion generates strains with lower resistance to nitrosative stress.Citation11,12 However, in vitro studies done in S. aureus indicated that the protein protects from oxidative stress as inactivation of ric decreased the viability of S. aureus when exposed to hydrogen peroxide.Citation9 Furthermore, the S. aureus Δric mutant strain exhibited reduced activity of important iron-sulfur (FeS)-containing enzymes such as aconitase and fumarase, activities that could be recovered to the levels observed in the wild-type strain by addition of the recombinant RIC protein to the cell extracts.Citation9 Following these studies, in this work we have analysed the contribution of RIC to the survival of S. aureus during infection of macrophages, lung epithelial cells, and G. mellonella larvae.
Therefore to investigate the in vivo role of S. aureus RIC, we first tested the behaviour of a strain lacking ric during infection of innate immune cells. For this purpose, macrophages J774A.1 were incubated with S. aureus wild-type and an isogenic Δric mutant, and the survival rate of each strain was determined (). When compared with the parental strain, S. aureus Δric exhibited lower resistance to macrophage killing. Moreover, the susceptibility of the mutant was shown to be dependent on the infection time, i.e., during the first half-hour post-infection no major differences were observed in the number of viable colonies between the two strains, whereas after 6 h a decrease of ∼40% in the survival rate was noted for the Δric mutant strain. Additionally, expression in trans of RIC abolished the increased susceptibility of the mutant strain, that under these conditions exhibited a viability similar to that of the wild-type ().
Figure 1. RIC protects S. aureus from phagocyte producing ROS. Macrophages J774A.1 were infected, at an MOI ∼ 5, with S. aureus JE2 (black bar), JE2 ∆ric (white bar) and JE2 Δric carrying the complementation vector (pMK4-RIC– striped bar) following activation by INF-γ/LPS (A), and in the presence of the mammalian iNOS inhibitor L-NMMA (B) or the NADPH phagocyte oxidase inhibitor apocynin (C). Bacterial counts were determined at 0.5 or 6 h post-infection. In (D) is depicted the quantification of intracellular bacteria after infection of lung epithelial A549 cells with S. aureus JE2 (black bar) and JE2 Δric (white bar), for 2 h. Data represent means of three (A, B and C) and two (D) biological samples analysed in triplicate, with standard error and unpaired Student's t-test (###P < 0.0005; #P < 0.05; ns: not significant).
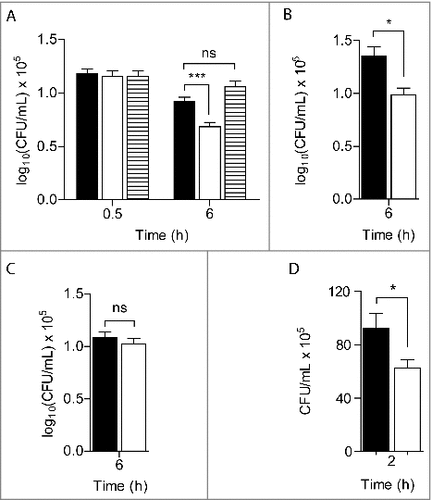
Treatment of macrophages with LPS and γ-IFN induces enzymes such as nitric oxide synthase (iNOS) and the NADPH oxidase (NOX), which produce RNS and ROS that destroy bacteria. To elucidate the role of RIC in the survival of S. aureus against these species, infection experiments were also performed in macrophages treated, separately, with inhibitors of iNOS and NOX, namely L-NMMA and apocynin, respectively.
In macrophages treated with L-NMMA, that did not produce NO as attested by nitrite quantification (Figure S1), the viability of the S. aureus ric mutant was still lower than that of the wild-type strain (). On the contrary, the survival of the ric mutant and of wild-type was similar in macrophages treated with apocynin (). As apocynin is shown to not interfere with production of nitric oxide (Figure S1), these results indicate that RIC protects S. aureus from the oxidative stress imposed by macrophages.
S. aureus is a common inhabitant of the human skin and mucosal surfaces that uses internalization into epithelial cells as an immune evasion mechanism.Citation13 To further understand the role of RIC in S. aureus survival, we carried out invasion/survival assays with human lung epithelial A549 cells, which is the most commonly used epithelial cell model for the study of S. aureus.Citation14 After 30 min of incubation of the A549 cells with a similar number of wild-type and mutant strain bacterial cells (∼107 CFU/mL), no differences in the intracellular survival between the two strains was observed (1.4 × 106 and 1.2 × 106, respectively). However, after 2 h of infection the intracellular viability of the S. aureus ric mutant in the epithelial cells was lower than that of the wild-type strain (). These results show that RIC contributes to the successful infection of non-phagocytic cells, such as the lung epithelial cells, by S. aureus.
Galleria mellonella, that is a model organism for the study of innate immunity,Citation15 was here used to determine the virulence of S. aureus. For this purpose, groups of larvae were injected with S. aureus JE2, incubated at 37°C, and the survival rate was recorded daily for up to 4 days. While inoculation of PBS exhibited no effect (data not shown), the administration of live MRSA (JE2) strain reduced the larval survival in a dose-dependent manner. Inoculation of 103 CFU/larvae reduced larval viability by 30% after 3 days and 108 CFU/larva caused the death of all larvae after 2 days (). The lethal dose of S. aureus showed to cause the death of 50% (LD50) of the G. mellonella population at 48 h was found to be ∼107 CFU/larva (Figure S2).
Figure 2. S. aureus ric mutant has decreased ability to infect Galleria mellonella. A. Survival of G. mellonella after infection with S. aureus JE2 at the following bacterial CFU per larva: 103 (solid line), 105 (dashed line), and 108 (dotted line).B. G. mellonella was infected with S. aureus JE2 (WT; black line), and the isogenic ric mutant strain (Δric; grey line) (###P < 0.0001). For complementation, G. mellonella was also infected by S. aureus JE2 Δric carrying the vector pMK4-RIC (black dashed line), or with the empty vector pMK4 (grey dashed line) (##P < 0.005). In B, approximately 107 bacterial cells were injected per larvae. The survival curves were compared using Mantel-Cox test. Ten larvae were analysed in each condition and larval survival was monitored daily. In all cases, no larval death was observed upon administration of PBS.
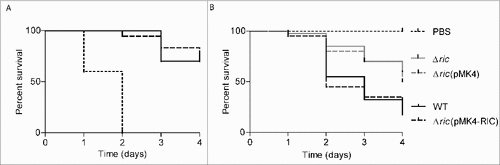
The role of RIC in S. aureus virulence was examined in G. mellonella by inoculating larvae with the S. aureus wild-type and the correspondent isogenic ric mutant. Inoculation of G. mellonella with equivalent doses of S. aureus JE2 and Δric mutant showed that the later had a higher survival rate (). Four days following infection, only ∼20% of the S. aureus JE2 wild-type-infected larvae were still alive, whereas survival of larvae infected with the S. aureus ric mutant was approximately two 2.5-times higher. Complementation in trans with a ric plasmid-borne restored the lethality of the mutant strain to levels similar to those induced by the wild-type strain ().
We also assessed the proliferation of S. aureus within G. mellonella by determination of the bacterial loads in the larvae hemolymph. S. aureus wild-type and Δric mutant were used at concentrations of ∼1 × 107 CFU/larva, and the hemolymph bacterial load was determined at 1 h, 4 h and 8 h post-infection. The viability of the two strains did not decrease significantly after 4 h of infection, but 8 h post-infection the ric mutant exhibited an intracellular viability lower than the wild-type strain (). Moreover, expression of a RIC plasmid-borne in the ric mutant cells led to an increase of the mutant strain viability to levels comparable to the wild-type ().
Figure 3. S. aureus Δric mutant has lower intracellular survival in G. mellonella. A. Bacterial load was analysed in the larvae hemolymph following incubation with S. aureus JE2 (black bar), JE2 Δric (white bar) and JE2 Δric carrying pMK4-RIC (striped bar) for 4 h and 8 h. Data represent means of four independent assays analysed with standard error and unpaired Student's t-test (##P < 0.001; ns: not significant). B. Quantitative RT-PCR analysis of the ric expression done in total RNA extracted from G. mellonella following incubation with S. aureus JE2, for 4 h and 8 h, and relative to the initial expression level (time zero of infection). Data was normalized to the expression of the constitutive 16S rRNA gene. Data represent mean values analysed with standard error (n = 6).
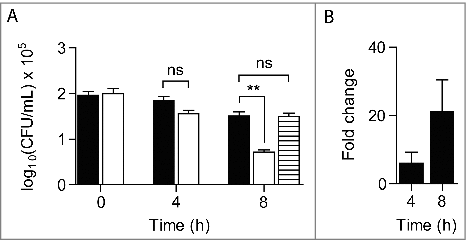
To further investigate the role of ric to the S. aureus survival during infection of G. mellonella, we have determined the expression of the S. aureus ric gene in the larvae by quantitative RT-PCR analysis. For this purpose, total RNA was extracted from G. mellonella after infection with S. aureus wild-type up to 8 h. The expression of the S. aureus ric gene within the larvae increased along the course of infection, been the highest after 8 h of infection (). These results are consistent with the observed contribution of RIC to the survival of S. aureus within the larvae ().
Previous studies done in E. coli and H. influenzae have implicated RIC in nitrosative stress resistance on the basis of marked increase of the ric gene expression and enhanced sensitivity of the Δric mutants to nitrosative stress.Citation10,16 However, in NO-treated S. aureus cells, the ric mRNA levels were only slightly increased and the growth of the S. aureus Δric mutant was not significantly compromised.Citation9,12 Although, it is not possible to exclude that the presence in S. aureus of other RNS defences, particularly flavohaemoglobinCitation12,17,18, may mask the role of RIC in NO stress resistance of this microorganism, several results indicate the involvement of RIC in oxidative stress resistance. RIC is herein shown to defend S. aureus from the effects caused by oxidative stress imposed by macrophages and consistent with these results, we previously reported that S. aureus ric mutant is more sensitive to oxidative stress.Citation9 Also, the mRNA ric levels increase upon exposure of S. aureus to hydrogen peroxideCitation11, and a marked up-regulation of the ric gene was detected in Salmonella enterica and Yersinia pestis upon infection of macrophages and rats, respectively.Citation19,20 In addition, inactivation of ric decreased the survival of H. influenzae within macrophages.Citation16
S. aureus uptake by epithelial cells is a rapid process that is dose-dependent.Citation21 We observed that the deletion of ric reduced the rate of S. aureus internalization and survival into human epithelial cells. Interestingly, the S. aureus ric gene is upstream of the lytSR gene cluster that encodes a two-component system regulating murein hydrolase activity and cell death.Citation22 Moreover, Brunskill and co-workers previously reported that the absence of ric in S. aureus causes morphological defectsCitation23, while other authors reported that the ric transcription increases upon internalization of S. aureus by epithelial A549 cells, along with other genes involved in iron metabolism and oxidative stress resistance.Citation13
Galleria mellonella wax moth larval infection model has been used to study bacterial pathogenesis, including infections by S. aureus.Citation4–6,24–26 In particular, G. mellonella allowed revealing several S. aureus virulence factors, such as the accessory gene regulator (Agr), a global regulator of the staphylococcal virulon that includes secreted virulence factors and surface proteins, RelA, a guanosine pentaphosphate synthase that regulates amino acid biosynthesis under nutrient-limited conditions, and two glyceraldehyde-3-phosphate dehydrogenase homologs called GapAB, involved in glycolysis and gluconeogenesis, respectively.Citation4–6,27,28 Our data show that RIC also contributes to the virulence of S. aureus.
The inactivation of RIC in S. aureus was previously reported to decrease the activity of oxidatively damaged Fe-S containing proteins such as aconitase and fumaraseCitation9, enzymes that are required for a functional TCA and fully respiratory activity of S. aureus. The contribution of RIC to the survival in phagocytes and epithelial cells and to the virulence towards G. mellonella shows the in vivo importance of this protein, which is most probably related with protection that it confers against the oxidative stress imposed by the host cells.
Experimentally the study included the S. aureus strains listed in Table S1, namely the methicillin-resistant S. aureus (MRSA) strain JE2 (wild-type), derived from community-associated methicillin-resistant S. aureus strain USA300 LACCitation29, and the JE2 Δric strain, which were provided by the Nebraska Transposon Mutant Library.Citation30 The media used for growth of bacteria was purchased from BD Difco and antibiotics from Sigma. S. aureus was cultured overnight in Tryptic Soy Broth (TSB), at 37°C, and used to inoculate fresh Luria-Bertani (LB) medium and grown to the indicated optical density at 600 nm (OD600). Bacterial viability was determined by CFUs counting by performing serial dilutions of S. aureus in phosphate-buffered saline (PBS), plating on Tryptic Soy Agar (TSA) and incubation overnight at 37°C.
The infecting dose was optimized by injecting G. mellonella with increasing inocula of S. aureus JE2 cell suspensions (∼103–108 cells/larva). For LD50 determination, eight bacterial concentrations were tested and the number of dead larvae was evaluated after 48 h, 72 h and 96 h. GraphPad Prism program was used to plot a non-linear fitting curve and obtain the LD50 value.
S. aureus JE2 and the isogenic Δric mutant derivative were grown overnight and diluted in PBS (to final 107–108 CFU/mL).
A DNA fragment of ∼1500 bp containing the complete coding region of the S. aureus ric gene plus a ∼1000 bp upstream region, that contains the ric own promoter and lacks any other complete open reading frame, was PCR-amplified from S. aureus NCTC8325 genomic DNA using the oligonucleotides SA_RICcomp_fw and SA_RICcomp_rev (Table S2) and ligated into pMK4 digested with EcoRI and SalI. After confirming the integrity of the cloned gene, the resulting vector (pMK4-RIC) was electroporated (∼100 ng) into S. aureus RN4220 and transformants were selected on TSA medium containing 5 μg/mL chloramphenicol. The recombinant plasmid extracted from S. aureus RN4220 cells was electroporated (∼20 µg) into S. aureus JE2 Δric, and its integration confirmed by colony PCR and digestion with appropriated enzymes. Restriction enzymes were obtained from New England Biolabs, DNA polymerase from Roche.
For the eukaryotic cell culture assays, murine macrophages J774.1 and human lung epithelial A549 cells (ATCC CCL 185) were routinely cultivated for 2 days in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (DMEMi), 50 U/mL penicillin and 50 μg/mL streptomycin (DMEMc), at 37 °C in a 5% CO2-air atmosphere. S. aureus JE2 and JE2 Δric mutant cells grown to OD600 of ∼0.4 were collected, washed with PBS and diluted in DMEMi to obtain a culture at OD600 ∼0.03. Prior to infection, macrophages (∼5 × 105 cells/mL) were seeded in 24-well plates and cultivated under 5% CO2-air atmosphere at 37°C, for 48 h, and activated by incubation with 1 μg/mL gamma interferon (IFN-γ) and 5 μg/mL E. coli lipopolysaccharides (LPS), for 5 h; when indicated, 800 μM NG-monomethyl-l-arginine acetate salt (L-NMMA) or 400 μM apocynin were added to inhibit inducible nitric oxide synthase or NADPH phagocyte oxidase, respectively. Bacterial suspensions at ∼107 cells/mL were used to infect macrophages at a multiplicity of infection (MOI) of 5. After a 30 min incubation period, non-phagocytosed bacterial cells were removed by incubation with 50 µg/mL gentamicin for 10 min. Macrophages were washed with PBS, lysed (2% saponin, Sigma), and the intracellular bacterial CFUs determined after 30 min and 6 h of infection. Nitric oxide levels were inferred by the oxidative product nitrite using the Griess assay.Citation31
For the internalization assays in lung epithelial cells, 24 h prior to infection, human lung epithelial cells A549 (ATCC CCL 185) (∼2 × 105 cells/mL) were seeded in 24-well plates and cultured in 5% CO2-air atmosphere at 37 °C. S. aureus wild-type and ric mutant strains grown in LB to OD600 of ∼0.5–0.6 were diluted in DMEMi to OD600 ∼0.05 (∼107 cells/mL) and incubated for 30 min and 2 h in A549 cells (MOI of 20). Extracellular bacteria were removed by incubation with 4 μg/mL lysostaphin (Sigma) at 37 °C for 20 min. Lung epithelial cells were PBS-washed, trypsinised and lysed with cold Triton X-100 (0.1%), and viable intracellular bacteria were counted.
Larval infection assays were performed with G. mellonella larvae reared in an iBB insectarium and maintained on a beeswax and pollen grains diet at 25 °C in darkness, and used in killing assays at the final instar larval stage. The infecting dose was optimized by injecting G. mellonella with increasing inocula of S. aureus JE2 cell suspensions (∼103–108 cells/larva). S. aureus JE2 and the isogenic Δric mutant were grown overnight and diluted in PBS (to final 107–108 CFU/mL). For each bacterial dilution, 3.5 μL aliquots were injected into the hindmost left proleg of each larva using a microsyringe adapted to a micrometer that controls the volume of injection. Control larvae injected with equal volumes of PBS were also monitored. Larvae were placed in Petri dishes and stored in the dark at 37 °C up to 4 days. Larval survival was monitored daily by inspection for dead organisms which were identified by development of a black colour resultant from larval melanisation, and immobility.
Bacterial load in the G. mellonella hemolymph was evaluated in three living larvae that were punctured in the abdomen with a sterile needle and after 1 h (time zero of infection), 4 h and 8 h of infection the plasma was collected to a sterile microtube containing a few crystals of phenylthiourea. The hemolymph was serially diluted in PBS, plated on TSA plates and CFU were determined after incubation at 37 °C, for ∼14 h.
Statistical analysis was carried out using GraphPad Prism version 5.01 for Windows. Survival curves were plotted using the Kaplan-Meier method, and differences in survival were calculated using the Mantel-Cox test for curve comparisons. Ten larvae were examined for each condition, and each experiment was repeated at least three times, over two different weeks and for up to four weeks. Differences between mean values were tested for significance by performing unpaired two-tailed Student's t-test with P<0.05.
The S. aureus ric expression during G. mellonella infection was analysed by incubating the larvae with S. aureus (∼107 CFU/larva). For each time point (0 h, 4 h and 8 h after infection), three living larvae were cryopreserved, sliced and homogenized in 1 mL TRIzol (ThermoFisher Scientific). Total animal/bacteria RNA was extracted according the manufacturer's protocol, treated with RNase-Free DNase (Quiagen), and its concentration and purity was evaluated in a Nanodrop ND-1000 UV–visible spectrophotometer (Thermo Fisher Scientific) and agarose gel. Total RNA (900 ng) was reverse transcribed with the Transcriptor High Fidelity cDNA Synthesis Kit (Roche) using the Anchored-oligo (dT)18 and Random Hexamer primers. Quantitative real-time RT-PCR assays were done in a LightCycler® 480 (Roche) using the oligonucleotides listed in Table S2 and the LightCycler® 480 SYBR Green I Master kit (Roche). Relative quantification of ric gene is shown in relation to the 16S rRNA reference gene, whose expression does not vary under the tested conditions, and using the comparative CT method.
KVIR_S_1389829.docx
Download MS Word (57.1 KB)References
- Liu GY. Molecular pathogenesis of Staphylococcus aureus infection. Pediatr. Res. 2009;65:71R–77R. doi: 10.1203/PDR.0b013e31819dc44d
- Chambers HF, DeLeo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–41. doi: 10.1038/nrmicro2200
- Alva-Murillo N, López-Meza JE, Ochoa-Zarzosa A. Nonprofessional phagocytic cell receptors involved in Staphylococcus aureus internalization. Biomed Res. Int. 2014;2014:538546. doi: 10.1155/2014/538546. PMID: 24826382
- Peleg AY, Monga D, Pillai S, Mylonakis E, Moellering, RC Jr., Eliopoulos GM. Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J Infect Dis. 2009;199:532–6. doi: 10.1086/596511
- Desbois AP, Coote PJ. Wax moth larva (Galleria mellonella): An in vivo model for assessing the efficacy of antistaphylococcal agents. J Antimicrob Chemother. 2011;66:1785–90. doi: 10.1093/jac/dkr198
- Gibreel TM, Upton M. Synthetic epidermicin NI01 can protect Galleria mellonella larvae from infection with Staphylococcus aureus. J Antimicrob Chemother. 2013;68:2269–73. doi: 10.1093/jac/dkt195
- Conti F, Abnave P, Ghigo E. Unconventional animal models: a booster for new advances in host-pathogen interactions. Front Cell Infect Microbiol. 2014;4:1–4. doi:10.3389/fcimb.2014.00142. PMID: 24592357
- Nathan S. New to Galleria mellonella. Virulence. 2014;5:371–4. doi:10.4161/viru.28338
- Overton TW, Justino MC, Li Y, Baptista JM, Melo AMP, Cole J a, Saraiva LM. Widespread distribution in pathogenic bacteria of di-iron proteins that repair oxidative and nitrosative damage to iron-sulfur centers. J Bacteriol. 2008;190:2004–13. doi:10.1128/JB.01733-07
- Justino MC, Vicente JB, Teixeira M, Saraiva LM. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J Biol Chem. 2005;280:2636–43. doi: 10.1074/jbc.M411070200
- Chang W, Small DA, Toghrol F, Bentley WE. Global transcriptorne analysis of Staphylococcus aureus response to hydrogen peroxide. J Bacteriol. 2006;188:1648–59. doi: 10.1128/JB.188.4.1648-1659.2006
- Richardson AR, Dunman PM, Fang FC. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol Microbiol. 2006;61:927–39. doi: 10.1111/j.1365-2958.2006.05290.x
- Garzoni C, Francois P, Huyghe A, Couzinet S, Tapparel C, Charbonnier Y, Renzoni A, Lucchini S, Lew DP, Vaudaux P, et al. A global view of Staphylococcus aureus whole genome expression upon internalization in human epithelial cells. BMC Genomics. 2007;8:171. doi: 10.1186/1471-2164-8-171. PMID: 17570841
- Maya S, Indulekha S, Sukhithasri V, Smitha KT, Nair S V, Jayakumar R, Biswas R. Efficacy of tetracycline encapsulated O-carboxymethyl chitosan nanoparticles against intracellular infections of Staphylococcus aureus. Int J Biol Macromol. 2012;51:392–9. doi: 10.1016/j.ijbiomac.2012.06.009
- Kavanagh K, Reeves EP. Insect and Mammalian innate immune responses are much alike. Microbe. 2007;2:596–9. doi: 10.1128/microbe.2.596.1
- Harrington JC, Wong SMS, Rosadini C V., Garifulin O, Boyartchuk V, Akerley BJ. Resistance of Haemophilus influenzae to reactive nitrogen donors and gamma interferon-stimulated macrophages requires the formate-dependent nitrite reductase regulator-activated yfE gene. Infect Immun. 2009;77:1945–58. doi:10.1128/IAI.01365-08
- Gonçalves VL, Nobre LS, Vicente JB, Teixeira M, Saraiva LM. Flavohemoglobin requires microaerophilic conditions for nitrosative protection of Staphylococcus aureus. FEBS Lett. 2006;580:1817–21. doi: 10.1016/j.febslet.2006.02.039
- Nobre LS, Gonçalves VL, Saraiva LM. Flavohemoglobin of Staphylococcus aureus. Methods Enzymol. 2008;436:203–16. doi: 10.1016/S0076-6879(08)36011-X
- Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JCD. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–18. doi: 10.1046/j.1365-2958.2003.03313.x
- Sebbane F, Lemaître N, Sturdevant DE, Rebeil R, Virtaneva K, Porcella SF, Hinnebusch BJ. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc Natl Acad Sci. 2006;103:11766–71. doi:10.1073/pnas.0601182103
- Kahl BC, Goulian M, van Wamel W, Herrmann M, Simon SM, Kaplan G, Peters G, Cheung a L. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line. Infect Immun. 2000;68:5385–92. doi:10.1128/IAI.68.9.5385-5392.2000
- Patton TG, Yang SJ, Bayles KW. The role of proton motive force in expression of the Staphylococcus aureus cid and lrg operons. Mol Microbiol. 2006;59:1395–404. doi:10.1111/j.1365-2958.2006.05034.x
- Brunskill EW, De Jonge BLM, Bayles KW. The Staphylococcus aureus scdA gene: A novel locus that affects cell division and morphogenesis. Microbiology. 1997;143:2877–82. doi: 10.1099/00221287-143-9-2877
- Mil-Homens D, Fialho AM, Loutet S, Vinion-Dubiel A, Datta A. A BCAM0223 mutant of Burkholderia cenocepacia is deficient in hemagglutination, serum resistance, adhesion to epithelial cells and virulence. PLoS One. 2012;7:e41747. doi:10.1371/journal.pone.0041747. PMID: 22848588
- Alghoribi MF, Gibreel TM, Dodgson AR, Beatson SA, Upton M. Galleria mellonella infection model demonstrates high lethality of ST69 and ST127 uropathogenic E. coli. PLoS One. 2014;9:e101547. doi: 10.1371/journal.pone.0101547. PMID: 25061819
- Giannouli M, Palatucci AT, Rubino V, Ruggiero G, Romano M, Triassi M, Ricci V, Zarrilli R. Use of larvae of the wax moth Galleria mellonella as an in vivo model to study the virulence of Helicobacter pylori. BMC Microbiol. 2014;14:228. doi:10.1186/s12866-014-0228-0. PMID: 25170542
- Gao W, Chua K, Davies JK, Newton HJ, Seemann T, Harrison PF, Holmes NE, Rhee HW, Hong JI, Hartland EL, et al. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 2010;6:e1000944. doi:10.1371/journal.ppat.1000944
- Purves J, Cockayne A, Moody PCE, Morrissey JA. Comparison of the regulation, metabolic functions, and roles in virulence of the glyceraldehyde-3-phosphate dehydrogenase homologues gapA and gapB in Staphylococcus aureus. Infect Immun. 2010;78:5223–32. doi: 10.1128/IAI.00762-10
- Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Mathema B, Mediavilla JR, Byrne KA, Parkins LD, Tenover FC, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: Recent clonal expansion and diversification. Proc Natl Acad Sci. 2008;105:1327–32. doi:10.1073/pnas.0710217105
- Bose JL, Fey PD, Bayles KW. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol. 2013;79:2218–24. doi:10.1128/AEM.00136-13
- Tarpey MM, Wink DA, Grisham MB. Methods for detection of reactive metabolites of oxygen and nitrogen: in vitro and in vivo considerations. Am J Physiol – Regul Integr Comp Physiol. 2004;286:R431–44. doi: 10.1152/ajpregu.00361.2003
