ABSTRACT
Neisseria meningitidis is the causative agent of cerebrospinal meningitis and that of a rapidly progressing fatal septic shock known as purpura fulminans. Meningococcemia is characterized by bacterial adhesion to human endothelial cells of the microvessels. Host specificity has hampered studies on the role of blood vessels colonization in N. meningitidis associated pathogenesis. In this work, using a humanized model of SCID mice allowing the study of bacterial adhesion to human cells in an in vivo context we demonstrate that meningococcal colonization of human blood vessels is a prerequisite to the establishment of sepsis and lethality. To identify the molecular pathways involved in bacterial virulence, we performed transposon insertion site sequencing (Tn-seq) in vivo. Our results demonstrate that 36% of the genes that are important for growth in the blood of mice are dispensable when bacteria colonize human blood vessels, suggesting that human endothelial cells lining the blood vessels are feeding niches for N. meningitidis in vivo. Altogether, our work proposes a new paradigm for meningococcal virulence in which colonization of blood vessels is associated with metabolic adaptation and sustained bacteremia responsible for sepsis and subsequent lethality.
Introduction
Neisseria meningitidis, referred to as the meningococcus, is the causative agent of cerebrospinal meningitis after crossing of the blood brain barrier and/or septicemia, which can progress toward a rapidly progressing fatal septic shock known as purpura fulminans. In Europe, the average mortality rate of invasive meningococcal diseases (IMD) is around 8%, with a higher rate for purpura fulminans.Citation1,2 A specific step in meningococcal pathogenesis is the ability of N. meningitidis to interact with microvessels throughout the body. In the brain, this interaction is believed to be responsible for invasion of the meninges.Citation3-5 Interactions of N. meningitidis with peripheral microvessels are associated with fibrin deposition, platelet accumulation, and perivascular inflammation foci.Citation6,7 The consequences of this peripheral interaction remain unknown, even though in patients, a high bacterial load in the blood is associated with the more severe forms of IMD (i.e. purpura fulminans).Citation8
Previous works performed using various in vitro and in vivo models have identified several meningococcal virulence factors. These valuable studies gave important insights into the role of factors required for survival in the extra cellular fluids such as the polysaccharide capsule, lipooligosaccharides (LOS), iron uptake and aromatic amino acid metabolismCitation2,9,10 and the Factor H binding protein FhBP.Citation11 However, these models do not allow assessing the consequences of bacterial interactions with endothelial cells of the peripheral vasculature during meningococcal infections. Such close interactions are restricted to human cells and are mediated by type IV pili (TFP), the only means by which virulent capsulated meningococci can interact with endothelial cells.Citation12,13 This species specificity has hampered the in vivo study of the role of meningococcal colonization of blood microvessels. Recently, a model using SCID mice grafted with human skin was engineered to study meningococcal interactions with human endothelial cells during blood-borne infection.Citation12,13 In this model, vessels of the graft are of human origin thus allowing meningococci to interact with human endothelial cells. Previous studies using this model have shown that piliated capsulated meningococci strongly adhere to the human microvessels of the graft in a TFP-dependent manner, whereas no bacteria/endothelial cell interaction was observed in mouse tissues. In addition, histological examination of biopsies of the graft revealed elementary lesions similar to those observed in human disease, i.e. vascular leakage associated with thrombosis and foci of vasculitis.Citation6,12,13
By using the humanized SCID mice model we demonstrate that bacterial interaction with the microvessels leads to a sustained colonization of human vessels and sustained bacteremia that are correlated with mice mortality. Furthermore, we explored the meningococcal factors that are involved in colonization by combining the use of a human skin grafted mice model with Tn-seq analysis (Transposon sequencing).Citation14,15 Unexpectedly, the comparison of Tn-seq data obtained from skin grafts and that of the blood of non-grafted mice revealed that many genes that are important for growth in the blood of mice are dispensable when bacteria colonize human vessels. These genes are mainly related to metabolism and nutrient uptake suggesting that endothelium of the blood vessels is a nutritional niche for N. meningitidis in vivo. Altogether, our work proposes a new paradigm of meningococcal virulence in which N. meningitidis colonization of blood vessels is associated with metabolic adaptation leading to a sustained bacteremia responsible for sepsis and subsequent lethality.
Results
Bacterial colonization of human microvessels is a prerequisite to mice mortality
As mentioned above, the humanized SCID mice model of meningococcal infection allows the study of bacterial adhesion in an in vivo context. Indeed, previously published results have shown that meningococci adhere in a TFP-dependent manner to the endothelial cells of the graft.Citation12,13,16 In this work we initially focused at determining the consequences of this interaction for the outcome of infection. To achieve this goal, grafted and non-grafted SCID mice were injected intravenously (IV) with 5.106 colony-forming units (CFU) of 2C4.3, a virulent adhesive wild type (wt) piliated and capsulated N. meningitidis strain.Citation17 Results reported in clearly show that N. meningitidis is particularly virulent in grafted SCID mice as 87.5% of the grafted mice did not survive by day 2, whereas only 12.5% of the non-grafted mice died (p-value = 0,003; Log-rank Mantel-Cox survival analysis). To confirm that this increase of virulence observed in grafted mice was a consequence of the bacterial adhesion to the human skin capillaries, we infected grafted SCID mice with 5.106 CFU of a non-adhesive piliated PilC1 defective mutant of strain 2C4.3.Citation18 As previously described this mutant is indeed unable to adhere in vitro to endothelial cellsCitation18 and cannot colonize the graft capillaries ().Citation19 As shown in , all mice infected with the PilC1 defective strain survived, thus demonstrating the role of TFP-mediated adhesion to the graft vessels in the outcome of infection.
Figure 1. Neisseria meningitidis colonization of the human skin graft leads to mice lethality. (A) Kaplan-Meier plot showing the survival of grafted (n = 8; dot) and non-grafted (n = 8; square) mice after intravenous challenge with 5.106 CFU of N. meningitidis wt 2C4.3 strain (p-value = 0,003; Log-rank Mantel-Cox survival analysis) or grafted mice infected with 5.106 CFU of N. meningitidis non-adhesive PilC1 defective mutant (n = 6; triangle). (B) Transversal human skin graft sections after infection by N. meningitidis wt 2C4.3 strain and the PilC1 non-adhesive defective derivative (bacteria, red) associated with human vessels (collagen IV-basal lamina, green) in the human grafted skin 4 hrs after infection. (C) Blood and graft bacterial loads of grafted and non-grafted SCID mice infected intravenously with the wt N. meningitidis 2C4.3 strain. Bacterial counts are expressed in CFU/ml for blood and in CFU/g for human skin grafts (non-grafted mice, n = 7; grafted mice, n = 5). Horizontal lines represent the mean bacterial load. Statistical significance was determined using the Wilcoxon-Mann-Whitney U-test (p = 0.0025). (D) Total bacterial load of grafted and non-grafted SCID mice shown in panel B (see material and methods section). Bacterial counts are expressed in CFU/mice for the inoculum and 18 hrs after infection (non-grafted mice, n = 7; grafted mice, n = 5). Horizontal lines represent the mean bacterial load. Statistical significance was determined using One-Way ANOVA (##: p<0.01). (E) Blood bacterial loads of grafted SCID mice infected intravenously with the PilC1 defective N. meningitidis 2C4.3 strain (ΔPilC1). Bacterial counts are expressed in CFU/ml of blood. Horizontal lines represent the mean bacterial load. P-value was determined using the Wilcoxon-Mann-Whitney U-test
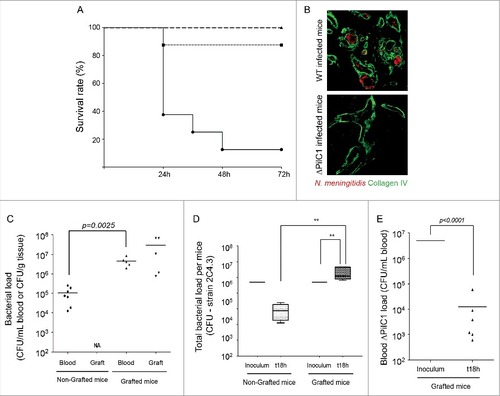
We subsequently determined the amount of bacteria in both the graft and the bloodstream. Mice were infected IV with 5.106 CFU of wt piliated and capsulated 2C4.3 strain and sacrificed 18 hrs after infection, before death of the animals. Results showed that the bacteremia in the blood of infected animals was higher in grafted animals by 2 orders of magnitude than in non-grafted animals. In addition, bacteria heavily colonized the grafted human skin (). We then estimated the total bacterial load after 18 hrs of infection in grafted and non-grafted animals by summing the blood bacterial load and the graft bacterial load (see material and methods section). As shown in , the total bacterial load per animal was significantly increased when compared to that of the number of bacteria that were injected (p-value<0.01; One-Way ANOVA), whereas it dropped to approximately 1.105 CFU in non-grafted animals. To confirm that non-adhesive meningococci were cleared from the blood of mice we followed the bacteremia of a PilC1 defective strain in human skin grafted animals. Indeed, bacteremia of this mutant dramatically dropped to approximately 104 bacteria/ml 18 hrs after infection ().
Altogether these data demonstrate that in the absence of meningococcal interaction with endothelial cells bacteria are cleared from the bloodstream, and that adhesion to endothelial cells in the human graft leads to a sustained colonization correlated with the observed increase of virulence.
Tn-seq identification of meningococcal genes required for blood vessels colonization
To identify the meningococcal components required for bacterial colonization of human microvessels we assessed the fitness of insertion mutants of a previously described saturated random transposon insertion library of strain Z5463Citation15 in SCID mice grafted with human skin. As shown in figure S1A, we first assessed the total meningococcal load in grafted and non-grafted mice infected with the Tn-seq library 18 hrs after injection. The total bacterial load in grafted mice was significantly higher than in non-grafted mice (p-value<0.01; Wilcoxon-Mann-Whitney U test), thus following the same trend of the 2C4.3 strain shown in . Non-grafted and grafted SCID mice were infected intravenously with 1.107 CFU of the input library. Bacteria were recovered from the grafts of the human-skin grafted SCID mice, and from the blood of the non-grafted SCID mice at 4 hrs or 18 hrs after infection for further deep sequencing analysis (output libraries were labeled 4h Graft; 18h Graft; 4h Blood NG mice; 18h Blood NG mice) ( and S1B). Bacteria recovered 4 and 18 hrs after infection were allowed to grow in broth to amplify the DNA content of each sample for only 2 hrs to limit the amplification bias. Deep sequencing of the libraries was performed as described in the experimental procedures section thus allowing determination of the fitness of each gene-disrupted mutant in each condition.
Figure 2. Tn-seq analysis. (A) Distribution of the CDS containing random Tn insertions on the genome map of N. meningitidis Z2491. Outer circles: Green bars indicate the N. meningitidis Z2491 CDS. Inner circles: Red and black bars indicate the essential/growth-defective and non-growth defective gene-disrupted mutants, respectively, in broth; turquoise and purple bars indicate the gene-disrupted mutants with a significant fitness change in the blood of non-grafted mice 4 hrs and 18 hrs after infection; dark blue and pink bars indicate the gene-disrupted mutants with a significant fitness change in the human skin grafts of grafted mice 4 hrs and 18 hrs after infection. (B) Venn diagram representing the absolute number of gene-disrupted mutants with a significant differential fitness decrease (log2FC< −2 with a p-value<0.05) in any of the four tested conditions: blood from non-grafted mice 4 hrs and 18 hrs after infection, human skin grafts 4 hrs and 18 hrs after infection
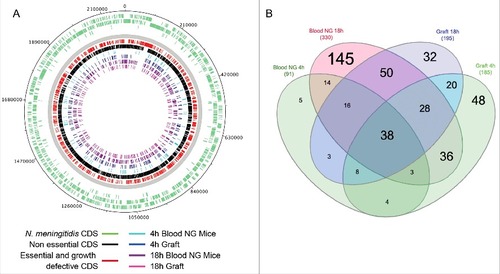
First, we compared the expected and measured number of transposon insertion sites (TIS) in the input libraries, as previously described,Citation15 in order to identify the essential and growth-defective genes in the iron deprived growth conditions used for in vivo experiments (Table S1-B, Figure S2A). Out of the 2065 genes, 798 were found to be important for growth and were not included in our analysis. These data were in agreement with those of our previous studyCitation15 (Pearson's correlation r = 0.95, p-value < 0.0001) (Figure S2B), further validating our approach. Importantly, the genes that were defined as essential or important for growth in our conditions are evenly distributed around the chromosome (). Secondly, genes were assigned a log2 Fold Change (log2FC) and a statistical score, based on the comparison of the number of reads between the input and the output libraries, using the ESSENTIAL tool kit as described previouslyCitation15,20 (Table S1-C). Thus, a gene whose disruption results in a loss of fitness in vivo would be underrepresented in the output library compared to that of the input library and would have a log2FC<0. On the contrary, a gain of fitness is associated to a positive log2FC value. A total of 501 transposon-disrupted mutant genes were found to have a significant differential fitness in at least one of the 4 tested conditions (log2FC> 2 or log2FC< -2 with a p-value<0.05) (Figure S2C).
Comparison with previous studies on meningococcal genes required for blood vessel colonization
Previous studies on various models in which bacteria were unable to adhere to endothelial cells identified a total of 85 virulence genes important for survival/growth in the extra cellular fluidsCitation9,21-26 (Table S2A). To validate our in vivo Tn-seq approach, we first focused on the fitness of these 85 gene-disrupted mutants in the blood of non-grafted SCID mice obtained at 4 and 18 hrs after infection ( and Tables S1 and S2A). Tn insertions in 12 out of these 85 virulence genes were among the 798 mentioned above growth defective gene-disrupted mutants that were discarded. Based on their log2FC, 41 out of the 85 previously reported genes were identified here as necessary for growth in the blood of non-grafted SCID mice obtained 4 and/or 18 hrs after infection. These mutants have insertions in genes encoding classical virulence factors such as those involved in the synthesis of the capsular polysaccharide, LOS and the iron uptake machinery (Table S2A). Conversely, the fitness of mutants of 32 out of these 85 virulence genes previously described was not significantly changed in our screen. No key factors were present in these 32 virulence genes, mainly involved in metabolism and transport of ion and metabolites. Several explanations could account for this apparent discrepancy. Firstly, the humanized SCID mice model is different from the models used in the previous studies since it allows adhesion of bacteria to the vascular wall. Secondly, a possible heterogeneity may exist within the N. meningitidis strains in term of metabolic requirement. Thirdly, the consequence of a mutation is likely to be compensated by the other bacterial cells of the population.
We further identified 307 genes that have not been described before whose disruption turned to be required for blood survival. These mutants have insertions in genes related to classical virulence factors such as the synthesis and export of the capsular polysaccharide, LOS, the iron and amino acid uptake machinery. Transposons are also inserted in genes necessary for cofactors synthesis such as chorismate synthesis. We confirmed that sulfur acquisition is crucial for the growth of meningococci in the blood in vivo as suggested earlier by Rusniok et al.Citation17 and Joseph et al.Citation27 (Table S2). Overall 13% of these 307 genes were involved in cellular processes, 14% in information storage and processing, and 38% involved in metabolism (Table S2B). This list of 307 gene-disrupted mutants involved in blood survival is not conflicting with previous results as Tn-seq approach offers a deeper analysis that allows the identification of mutants with decreased fitness.
To further validate the results of the Tn-seq analysis, we assessed the fitness of 3 defective mutants among those with a mild decrease of their fitness in the blood (i.e. −4.5< log2FC<−3.5). The in vivo fitness of N. meningitidis defective for cysG, putP and NMA0486, known to be involved in biosynthesis of secondary metabolites, solute transport, and biogenesis of outer-membrane vesicles (OMVs)Citation28 respectively, was tested in competition assays by simultaneously injecting IV the wt parental strain and the single defective mutants in BalbC mice. As control we tested the fitness of a pilC2 defective mutant that showed no defect in any of the 4 conditions (Table S1C). Importantly these four mutants were neither growth-defective in iron deprived growth conditions (Table S1-B) nor in GC broth (Figure S3). A significant decrease of the competition indexes (CI) of the ΔputP mutant 18 hrs after infection was shown, while the ΔNMA0486 and ΔcysG mutants already showed a significantly decreased of their CI 4 hrs after infection (). No modification of the CI of the ΔpilC2 mutant was observed. These data are in agreement with the Tn-seq results. Altogether, results obtained in the blood of BalbC mice validated our approach and allowed us to further analyze the data obtained in this work.
Figure 3. In vivo competition assay of defective mutants in blood of non-grafted mice 18 hrs after infection. A mixture of the N. meningitidis wt Z5463 strain and one of the 3 different mutant (ΔputP, ΔcysG, ΔNMA0486) displaying a mild fitness decrease for survival in the blood of non-grafted mice 18 hrs infection in our Tn-seq screening were tested for competition in vivo in Balb/C mice. A mutant derivative strain not having a fitness change in vivo (ΔpilC2) was used as a control. Competition indexes confirmed the Tn-seq fitness predictions. The 3 defective mutants were out-competed by the wt parental strain 18 hrs after infection, which was not the case of the neutral mutant ΔpilC2. Statistical significance was determined for each mutant using One-Way ANOVA (#: p < 0.05; ##: p < 0.01; ####: p < 0.0001)

Genes required for the initial colonization
Meningococcal colonization of the microvessels requires an initial step of adhesion to endothelial cells followed by replication of aggregative bacteria in the lumen of the microvessels. Therefore, we first concentrated on identifying genes required for graft colonization 4 hrs after infection. A total of 68 meningococcal genes were required for early colonization of the human graft and not for bloodstream growth and/or survival (, Tables 1A, S1-C). Twenty of those genes were also required for colonization of grafts harvested 18 hrs after infection. Among these 68 genes, 14 encode type IV pilus related genes including the major pilin pilE (pilC1, -E, -F, -G, -I, -J, -K, -M, -N, -P, -Q, -S5, -V, -W). These results were not unexpected considering that type IV pili have long been identified as essential for initial adhesion of capsulated meningococci to host cells. Interestingly, disruption of pilT, which abolishes pilus retraction and promotes the formation of large aggregates, was positively selected. Moreover, we observed that pilC2 is not important for adhesion, which is consistent with earlier work.Citation18 These results confirmed the critical role of the type IV pili machinery in allowing the initial step of adhesion onto human vessels.Citation13,16,19,29 Besides the type IV piliation machinery, 54 gene-disrupted mutants were found to have a fitness decrease upon early colonization. Among those, 10 genes were involved in information storage and processing, including 3 genes involved in translation processes, such as rsuA, responsible forribosome 16S pseudouridylation, while 5 genes coded for putative transposases. Furthermore, 9 genes were involved in metabolic processes and 25 encoded hypothetical proteins.
Most of the meningococcal genes required for colonization of human vessels are also needed for blood-borne dissemination in non-grafted mice
As we demonstrated that colonization of the human skin graft is crucial for bacterial virulence in vivo, we focused on the 195 genes for which Tn-insertion induced a significantly decreased fitness for survival and growth in the human grafts 18 hrs after infection (, Tables 1B and S1C). Hundred and forty-three of these genes were also important for survival and growth in the bloodstream of non-grafted mice. This indicates that most of the genes required for graft colonization are also important for survival in the blood of grafted mice. Among them we first found genes described above as classical virulence factors, involved in the synthesis of the capsular polysaccharide, LOS, iron and amino acid uptake machinery and chorismate synthesis. We also identified ORFs of the nuo and nqr operon, encoding the respiratory complex I and the sodium pumping NADH:quinine oxidoreductase respectively; the entire pur operon, important for nucleotide metabolism, was also important for survival under such condition. A total of 71 genes involved in metabolic pathways were necessary for sustaining graft colonization, including 18 genes related to amino acid metabolism, and 3 genes involved in coenzyme metabolism (hemD; lipA2; panB). Uptake of specific nutrients was also important since 6 genes encoding transporters were required for graft colonization. We further identified 23 genes involved in information, storage and processing, among which 13 were involved in DNA replication or repair functions (hupA; nth; rdgC; topA), 5 genes were involved in transcription and 4 in translation (pnp; rluC; rph; rpsR). We also identified 46 hypothetical uncharacterized proteins as necessary for colonization of human blood vessels in vivo.
Next, we concentrated on the 52 genes that were required for colonization of the graft after 18 hrs of infection, 20 of which were also necessary for early graft colonization 4 hours after infection (, Tables 1C, S1-C). The above 52 genes included open reading frames coding for the type IV pilus machinery (pilD, -E, -S2, -S5, -S7, -W); hpuA, the hemoglobin-haptoglobin utilization lipoprotein A,Citation30 and 27 proteins for which no clear function is described. Four genes involved in cellular processes were critical for long-term graft colonization (rfbC1, NMA0710, NMA1833, NMA2145), as well as 4 genes involved in amino acid and carbohydrate metabolism, and 5 genes involved in replication or DNA repair (NMA0471, NMA0531, NMA1946) or translation (NMA0861, pnp).
The wall of microvessels seems to be a more favorable environment for meningococci than the bloodstream
Analysis of unexpectedly showed that 145 out of the 330 genes required for growth in whole blood of non-grafted SCID mice 18 hrs after infection were not required for colonization of the human graft (Table S1). This suggests that colonization of the graft may offer a favorable environment. The graft may provide metabolites and factors required by mutants of related uptake systems or biosynthetic pathways. Out of these 145 genes, we focused on the 119 whose log2FC was at least 3 times higher in the graft than in the blood of non-grafted animals after 18 hrs of infection ( and Table S1-C). Seventeen of these genes were involved in cellular processes, such as cell envelope biogenesis (galU; lgtF; mtgA; mtrE; NMA0436) or two-component systems (TCSs), developed by bacteria to sense and respond to changes in different environmental conditions (NMA0160, NMA1805 and NMA1698). Furthermore, 19 genes were involved in DNA processes of which 8 are involved in DNA repair (such as mutL, uvrA, gcr), and 39 in metabolism. Remarkably, for gene-disrupted mutants involved in sulfur acquisition (cysD, -G, -H, -I, -J, -N, -T; metZ; NMA1243) a fitness decrease was only observed in the blood, which indicates that bacteria require less sulfate and thiosulfate while colonizing the graft. Similarly, mutants of biotin and thiamine synthesis genes can survive while colonizing the graft (bioD and bioH’ or thiC, respectively). Moreover, 15 genes coding for transporters and 43 genes coding for hypothetical proteins displayed this same pattern. Altogether, these data suggest that many metabolites are dispensable for growth when colonizing human vessels and that this environment provides N. meningitidis with niche-specific nutrients.
Table 1. N. meningitidis genes with a decreased fitness for growth/survival in mice blood or for human skin graft colonization
Table 2. N. meningitidis genes necessary for early and late growth/survival in blood of mice that are not involved during graft colonization (119 genes).Footnote#
Discussion
The most severe forms of IMD in human (i.e. purpura fulminans and meningococcal meningitis) are associated with colonization of the peripheral blood vessels. In this study, we demonstrated that meningococcal colonization of the endothelium is important for sustained bacteremia in vivo and that bacterial load in mice is directly correlated with mortality. This demonstrates that bacterial sustained colonization of human vessels in vivo is crucial for meningococcal virulence and suggests that colonization of the endothelium may provide new nutrients and a better environment for survival and growth.
In this work we characterized the meningococcal genes involved in early adhesion and colonization of human vessels in vivo by combining the use of the human skin grafted mice model with Tn-seq analysis. We performed a genome-wide analysis to screen for the genes required to colonize the human vessels at an early and late stage of infection. In parallel, we used non-grafted mice as a control, which allowed us to study the genes responsible for the survival of the meningococcus in the blood of mice.
As expected, genes encoding the type IV pilus machinery are essential for early colonization of the microvessels. Few other genes, mostly of unknown function, were also found to be required for early adhesion and/or colonization. Interestingly, some of these genes involved exclusively in early colonization have only a slight fitness defect in the graft 18 hrs after infection, not reaching statistical significance. This may be explained by the concomitant colonization of the graft by other mutants of the library, which can complement adhesion-defective mutants in the graft via aggregative type IV pili. In addition, the local formation of thrombosis could possibly trap non-adhesive mutants in the graft.
We analyzed genes important for sustained colonization of human vessels 18 hrs after infection. Unexpectedly, we discovered that 36% of the genes that were involved in the growth in blood of mice were no longer required to colonize the human graft. The majority of these genes were involved in metabolic processes, which is in accordance with the concept of nutritional virulence, where bacteria would take advantage of the nutrients available on the host to proliferate. For instance, gene-disrupted mutants compensated by the colonization of the graft were involved in sulfur acquisition, biotin and thiamine synthesis, as well as some inorganic ion transporters. Further studies will be needed to understand how and why these nutrients and factors are available in this niche. Type IV pili interaction with endothelial cells may be directly involved since meningococcal adhesion to endothelial cells promotes very specific cell signaling events. On the other hand, colonization of the human vessels in a biofilm-like manner may induce several modifications in the metabolism of N. meningitidis that may compensate for several mutations or it may also favor the production of meningococcal secondary messengers that could induce specific signaling pathway in endothelial cells, which will eventually increase the synthesis of nutrients and factors required by the bacteria for growth.
Interestingly, the study of the late colonization of human grafts revealed that most of the genes involved in graft colonization are also important for survival in the blood. Adhesion to microvessels may be considered as a strategy to evade the innate immune system especially considering that N. meningitidis inhibits monocytes and neutrophils docking to endothelial cells.Citation31 Here, we show that adhesion to human vessels is responsible for mice lethality and probably provides N. meningitidis with important nutrients and/or metabolites. Thus, we propose a new paradigm for meningococcal virulence in which meningococcal colonization of blood vessels is associated with metabolic adaptation and sustained bacteremia responsible for sepsis and subsequent lethality.
Taken altogether, interaction of meningococcus with the vasculature seems to be the support of growth and bacterial virulence in vivo. Our findings open a door to better identify the host metabolic pathways exploited by meningococci so that new strategies to tackle this strict human pathogen can be undertaken.
Materials and methods
Bacterial strains and growth conditions
N. meningitidis NEM 8013 strain 2C4.3, a piliated and capsulated serogroup C strain and its isogenic non-adhesive PilC1 defective mutant were used for in vivo virulence experiments.Citation17,32 N. meningitidis Z5463, a naturally highly transformable piliated and capsulated serogroup A strainCitation33,34 was used instead of 2C4.3 to generate saturating Tn insertion mutant librariesCitation15 and for in vivo competition experiments. Bacterial strains were stored frozen at −80°C and routinely grown at 37°C under 5% CO2 on GC agar plates (Difco) containing Kellogg's supplements. Kanamycin was added when required at a concentration of 100 and 200 µg/ml for 2C4.3 and Z5463 strains, respectively.
Animal experiments: bacteria recovered either from GC agar plates or Tn-seq libraries aliquots were cultured in RPMI – 1% BSA medium with 10 µM deferoxamine under agitation to reach the exponential phase of growth (also referred to as iron deprived growth conditions).
The input pool and the bacteria recovered from the blood or the graft were cultured two hours in GC broth in order to amplify the bacteria content.
Transposon (Tn) mutant library
The Tn-seq library used in this work is a merge of the 3 Tn-seq libraries described previously.Citation15 Briefly, an aliquot (750 μl) of each library was mixed in a single culture with 42.75 ml GC liquid medium (Difco) under shaking conditions containing Kellogg's supplementsCitation35 and kanamycin. The resultant Tn mutant library was split up into multiple starter cultures and stored at −80°C in 10% glycerol GC medium.
Construction of mutant strains
Deletion mutants of the selected candidate genes (cysG, putPNMA0486 and pilC2) were obtained from an available library of signature-tagged mutants of N. meningitidis.Citation21 The disrupted gene was PCR amplified from genomic DNA and the purified product was introduced by transformation into N. meningitidis strain Z5463. The transformants were selected in the presence of kanamycin. Genomic DNA from the resulting mutants was extracted and used to transform a second time our template strain Z5463. The resulting mutants were then stored at −20°C in 10% glycerol GC medium. Primers used to generate these mutants are listed in supplemental materials.
Animals and ethics
Six week old CB17/Icr-Prkdcscid (Severe Combined Immunodeficiency: SCID) and Balb/C female mice were obtained from Charles River Laboratories (Saint Germain sur l'Arbresle, France) and Janvier Labs (Saint-Berthevin, France). The experimental procedures described in this paper were conformed to the European ethical legislation (Directive 2010/63/EU). The experimental protocol was approved by the Comité d'Expérimentation Animale de l'Université Paris Descartes (project number CEEA 12–030). Human skin grafts were obtained from surgical waste from patient undergoing plastic surgery at Groupe Hospitalier Paris Saint-Joseph (Paris, France). According to the French legislation, the patients were informed of the research finality and their non-opposition was orally received.
In vivo humanized mice infection model
CB17 SCID mice were grafted with normal human skin as described by Join-Lambert et al.Citation13 The experiments were performed in independent occasions, with two different skin donors for survival experiments and with two different skin donors for bacterial load at 18 hours after infection.
N. meningitidis strains grown overnight at 37°C on GC agar plates without iron (without Kellogg's supplement II) and supplemented with deferoxamine (Desferal, Novartis) at a final concentration of 8 µM were cultured in iron deprived growth conditions as explained above and resuspended in physiological saline at the required OD and mice were infected intravenously. Inocula of 1.107 and 5.106 colony-forming units were used for the strains Z5463 and 2C4.3 respectively. These inocula correspond to the minimum inoculum necessary to obtain sustained bacteremia in grafted mice 18 hrs after infection. Ten mg of human holotransferrin (R&D Systems) was administered intraperitoneally just before infection. Bacteremia was assessed on blood samples obtained by tail vein puncture at different time-points of infection while crushing and homogenizing a sample of the human skin determined the skin graft bacterial load. Bacterial counts were determined by plating serial dilution of the samples onto GC agar plates. The bacterial aggregates were disrupted before plating. The total bacterial load, referring to the overall quantity of bacteria present in a mouse at a time-point of infection was expressed as CFU per mice. Considering that N. meningitidis is unable to colonize mouse organs in this humanized model,Citation13 it was estimated by adding up the blood bacterial load and the graft bacterial load (for grafted mice). For this calculation, we considered a mean blood volume of 1.4 ml for CB17 SCID mice, and a mean graft mass of 600 mg. For instance a bacteremia of 5.106 CFU/ml associated with a graft bacterial load of 1.107 CFU/g corresponds to a total bacterial load of 1.3.107 bacteria (5.106 × 1,4 + 1.107 × 0,6 = 1,3.107).
Survival assay
For survival experiments, CB17 SCID mice were infected IV with an inoculum of 5.106 CFU of Neisseria meningitidis 2C4.3 strain as described above. Mice survival was assessed twice daily during 3 days. Bacteremia, expressed in CFU/ml, were measured 1 hour after infection and then daily on blood samples obtained by tail vein puncture. Mice were sacrificed when presenting signs of lethal infection.
In vivo screening of Tn library for colonization
A starter culture of the N. meningitidis Tn library was thawed and grown for 2–3 h in RPMI – 1% BSA medium at 37°C under shaking conditions to reach the exponential growth phase. The starter culture was then grown under iron-deprived conditions for 2–3 hrs. Mice were infected with an inoculum of 1.107 CFU. DNA was extracted from the rest of the inoculum culture for subsequent deep sequencing analysis (input pool). Bacteremia was quantified at 1, 4 and 18 hrs post-infection and the graft bacterial load was assessed at the time of sacrifice. A total of five independent infections were performed to recover bacteria 4 hrs post-infection, and six independent infections were also performed to recover bacteria 18 hrs post-infection. The bacteria recovered at 4 hrs and 18 hrs post infection were allowed to grow in GC broth for 2 hrs and were then harvested by centrifugation. This additional growth step was performed in order to reach a sufficient amount of bacterial DNA to perform the Tn-seq experiment. We expected that the 2 hrs amplification step did not cause any bias since the input pool was also amplified in GC broth.Citation15 The number of mice used to obtain the different output libraries is indicated on sheet A from Table S1. Bacterial pellets from blood samples were resuspended in erythrocytes lysis solution called ACK (150 mM ammonium chloride, 10 mM potassium bicarbonate, 0.1 mM EDTA). Chromosomal DNA extraction was performed using the Wizard® Genomic DNA Purification Kit (Promega) and the DNeasy Blood and Tissue Kit (Qiagen) on the input and output mutant pools, respectively.
Bioinformatic analysis of Tn libraries
Transposon insertion sites (TIS) were identified using a strategy of capture by hybridization combined to next generation sequencing (Illumina technology) as described before,Citation15 with minor modifications. Libraries enriched in Tn containing bacterial fragments were sequenced on an Illumina HiSeq (Paired-End sequencing 250+250 bases). The Illumina sequencing reads were filtered on phred30 quality and 150 bp length using the fastx toolkit.Citation36 Reads that contained the Tn inverted terminal repeat (ITR) sequence were identified in the raw fasta files of the Tn-seq results and trimmed of the Tn sequence via the cutadapt tool.Citation37 Reads shorter than 15 bp were removed. Tn containing reads were processed using the ESSENTIALS software with default settings as described before.Citation15,20 Processed sequence reads of Tn-seq libraries, typically 50 bp in length, were mapped on the genome of N. meningitidis Z2491.Citation33 Transposon containing reads in the control and target samples were tested for significant differences (adjusted p-value < 0.05) by the Cox-Reid paired test implemented in EdgeRCitation38 assuming moderated tagwise dispersion of replicates. The prior n value to determine the amount of smoothing of tagwise dispersions was set at 5.
Gene essentiality in the input libraries was determined by taking the log2 of the measured number of transposon containing reads per gene divided by the expected number of transposon containing reads per gene (based on the number of possible TIS per gene, the mutant library size, and the sequencing depth) as determined by the TMM normalization (see Table S1-B). Determination of the log2FC for identification of genes with a fitness change in any of the four tested conditions (4h Graft; 18h Graft; 4h Blood NG mice; 18h Blood NG mice) was calculated as the binary logarithm of the number of reads of the target sample (output libraries harvested after selection in vivo) divided by the number of reads of the gene within the control sample (input libraries grown before selection) (See Table S1-C). Reads have been submitted to Sequence Read Archive (SRA) under accession numberPRJEB20828 (http://www.ebi.ac.uk/ena/data/view/PRJEB20828).
In vitro and in vivo competition assays
To confirm the results obtained by Tn-seq, we tested four gene interruption mutants in competition with the parental Z5463 strain for survival in the blood of Balb/C mice. We first verified in vitro that insertion mutants had no growth defect compared to the parental strain. Each mutant and wild type parental strains were grown as above. For both in vitro and in vivo experiments, mutants and wild type strains were mixed in a 1:10 ratio. For in vitro assays, bacterial counts were performed at the set of competition and 6 hrs after a mixed culture in GC liquid medium containing Kellogg's supplements. For in vivo assays, Balb/C mice were infected IV with a mix of 1.107 CFU of the wt Z5463 strain and 1.106 CFU of the mutant strain. Bacterial counts were performed at the set of competition and at 4 hrs and 18 hrs post-infection. Bacterial counts were determined by plating in parallel serial dilutions of the samples onto GC agar plates with or without 200 µg/ml of kanamycin for both in vitro and in vivo experiments. Results were expressed in CFU/ml. The competition index (CI) was calculated as the mutant/wt ratio in the output divided by the initial mutant/wt ratio. The data from at least five independent experiments were examined for 1% significance using a two-tailed Student's t-test.
Immunofluorescence microscopy
After infection, human skin grafts were removed and fixed in paraformaldehyde-lysine-periodate for 24 hrs at 4°C, dehydrated in successive sucrose gradient solutions (10%, 20%, 30% prepared in phosphate buffer 0.1 M) for 2 hrs at 4°C each, and embedded in OCT before being snap frozen on liquid nitrogen. Seven μm sections of the dermis were immobilized on Superfrost plus microscope slides and analyzed via immunofluorescence. Sections were incubated with the following primary antibodies for 1 h in PBS/BSA 0.1%: monoclonal anti-human collagen IV (1/100) and a rabbit polyclonal serum against the N. memingitidis 2C4.3 strain (1/1000). Alexa Fluor–conjugated secondary antibodies (1/300) for 2 hrs. After additional washing, coverslips were mounted in mowiol (Sigma) and processed for image acquisition with an epifluorescence Axio Observer.Z1microscope (Zeiss, Germany). Images were processed using the ImageJ software.
Web tools used for analysis
Metabolic pathways and subsystems for N. meningitidis strain Z2491 were obtained from the Kyoto Encyclopedia of Genes and Genomes orthology (http://www.genome.jp/kegg/). Amino acid sequences of hypothetical proteins were scanned against the Conserved Domain Database (cdd v3.15, E-Value cut-off: 0.01) using the NCBI's CD-Search tool.Citation39
Disclosure of interest
AZ has received funding from Janssen Pharmaceuticals to pursue studies on meningococcal vaccine development, unrelated to this research. The other authors declare no conflict of interest.
KVIR_S_1391446.zip
Download Zip (1.6 MB)Acknowledgments
We are grateful to Jean-Philippe Jaïs from Hospital Necker-Enfants Malades Biostatistics Department for advice on biostatistics and to Marc García-Garcerà from the Microbial Evolutionary Genomics Unit at the Institut Pasteur for advice on bioinformatics support. We thank N. Goudin of the Necker Institute imaging facility for their technical support and the Imagine foundation for the Leica SP5 microscope funding.
This work was supported by an ANR grant ANR-14-IFEC-0006-01 call ERANET INFECT-ERA 2014, a postdoctoral fellowship supported by the DIM Malinf from the Conseil Régional de l'Ile-De-France. The laboratory of XN is supported by INSERM, CNRS, Université Paris Descartes and the Fondation pour la Recherche Médicale.
References
- Rouphael NG, Stephens DS. Neisseria meningitidis: biology, microbiology, and epidemiology. Methods Mol Biol. 2012;799:1-20. doi:https://doi.org/10.1007/978-1-61779-346-2_1. PMID:21993636
- Read RC. Neisseria meningitidis; clones, carriage, and disease. ClinMicrobiol Infect. 2014;20:391-5. doi:https://doi.org/10.1111/1469-0691.12647.
- Pron B, Taha MK, Rambaud C, Fournet JC, Pattey N, Monnet JP, Musilek M, Beretti JL, Nassif X. Interaction of Neisseria maningitidis with the components of the blood-brain barrier correlates with an increased expression of PilC. J Infect Dis. 1997;176:1285-92. doi:https://doi.org/10.1086/514124. PMID:9359730
- Dupin N, Lecuyer H, Carlotti A, Poyart C, Coureuil M, Chanal J, Schmitt A, Vacher-Lavenu MC, Taha MK, Nassif X, et al. Chronic meningococcemia cutaneous lesions involve meningococcal perivascular invasion through the remodeling of endothelial barriers. Clin Infect Dis. 2012;54:1162-5. doi:https://doi.org/10.1093/cid/cis120. PMID:22412064
- Coureuil M, Lecuyer H, Bourdoulous S, Nassif X. A journey into the brain: insight into how bacterial pathogens cross blood-brain barriers. Nat Rev Microbiol. 2017;15:149-59. doi:https://doi.org/10.1038/nrmicro.2016.178. PMID:28090076
- Pathan N, Faust SN, Levin M. Pathophysiology of meningococcal meningitis and septicaemia. Arch Dis Child. 2003;88:601-7. doi:https://doi.org/10.1136/adc.88.7.601. PMID:12818907
- Lecuyer H, Borgel D, Nassif X, Coureuil M. Pathogenesis of meningococcal purpura fulminans. Pathog Dis. 2017;75(3):ftx027. doi:https://doi.org/10.1093/femspd/ftx027. PMID:28334263
- Ovstebo R, Brandtzaeg P, Brusletto B, Haug KB, Lande K, Hoiby EA, Kierulf P. Use of robotized DNA isolation and real-time PCR to quantify and identify close correlation between levels of Neisseria meningitidis DNA and lipopolysaccharides in plasma and cerebrospinal fluid from patients with systemic meningococcal disease. J Clin Microbiol. 2004;42:2980-7. doi:https://doi.org/10.1128/JCM.42.7.2980-2987.2004. PMID:15243048
- Sun YH, Bakshi S, Chalmers R, Tang CM. Functional genomics of Neisseria meningitidis pathogenesis. Nat Med. 2000;6:1269-73. doi:https://doi.org/10.1038/81380. PMID:11062540
- Deghmane AE, Parent du Chatelet I, Szatanik M, Hong E, Ruckly C, Giorgini D, Lévy-Bruhl D, Alonso JM, Taha MK. Emergence of new virulent Neisseria meningitidis serogroup C sequence type 11 isolates in France. J Infect Dis. 2010;202:247-50. doi:https://doi.org/10.1086/653583. PMID:20515410
- Madico G, Welsch JA, Lewis LA, McNaughton A, Perlman DH, Costello CE, Ngampasutadol J, Vogel U, Granoff DM, Ram S. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J Immunol. 2006;177:501-10. doi:https://doi.org/10.4049/jimmunol.177.1.501. PMID:16785547
- Melican K, MicheaVeloso P, Martin T, Bruneval P, Dumenil G. Adhesion of Neisseria meningitidis to dermal vessels leads to local vascular damage and purpura in a humanized mouse model. PLoS Pathog. 2013;9:e1003139. doi:https://doi.org/10.1371/journal.ppat.1003139. PMID:23359320
- Join-Lambert O, Lecuyer H, Miller F, Lelievre L, Jamet A, Furio L, Schmitt A, Pelissier P, Fraitag S, Coureuil M, et al. Meningococcal interaction to microvasculature triggers the tissular lesions of purpura fulminans. J Infect Dis. 2013;208:1590-7. doi:https://doi.org/10.1093/infdis/jit301. PMID:23840047
- Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A. 2009;106:16422-7. doi:https://doi.org/10.1073/pnas.0906627106. PMID:19805314
- Capel E, Zomer AL, Nussbaumer T, Bole C, Izac B, Frapy E, Meyer J, Bouzinba-Ségard H, Bille E, Jamet A, et al. Comprehensive Identification of Meningococcal Genes and Small Noncoding RNAs Required for Host Cell Colonization. mBio. 2016;7(4):e01173-16. doi:https://doi.org/10.1128/mBio.01173-16. PMID:27486197
- Bernard SC, Simpson N, Join-Lambert O, Federici C, Laran-Chich MP, Maissa N, Bouzinba-Ségard H, Morand PC, Chretien F, Taouji S, et al. Pathogenic Neisseria meningitidis utilizes CD147 for vascular colonization. Nat Med. 2014;20:725-731. doi:https://doi.org/10.1038/nm.3563. PMID:24880614.
- Rusniok C, Vallenet D, Floquet S, Ewles H, Mouze-Soulama C, Brown D, Lajus A, Buchrieser C, Médigue C, Glaser P, et al. NeMeSys: a biological resource for narrowing the gap between sequence and function in the human pathogen Neisseria meningitidis. Genome Biol. 2009;10:R110. doi:https://doi.org/10.1186/gb-2009-10-10-r110. PMID:19818133
- Nassif X, Beretti JL, Lowy J, Stenberg P, O'Gaora P, Pfeifer J, Normark S, So M. Roles of pilin and PilC in adhesion of Neisseria meningitidis to human epithelial and endothelial cells. Proc Natl Acad Sci U S A. 1994;91:3769-73. doi:https://doi.org/10.1073/pnas.91.9.3769. PMID:7909606
- Melican K, Dumenil G. A humanized model of microvascular infection.Future Microbiol. 2013;8:567-9.
- Zomer A, Burghout P, Bootsma HJ, Hermans PW, van Hijum SA. ESSENTIALS: software for rapid analysis of high throughput transposon insertion sequencing data. PloS one. 2012;7:e43012. doi:https://doi.org/10.1371/journal.pone.0043012. PMID:22900082
- Geoffroy MC, Floquet S, Metais A, Nassif X, Pelicic V. Large-scale analysis of the meningococcus genome by gene disruption: resistance to complement-mediated lysis. Genome Res. 2003;13:391-8. doi:https://doi.org/10.1101/gr.664303. PMID:12618369
- Echenique-Rivera H, Muzzi A, Del Tordello E, Seib KL, Francois P, Rappuoli R, Pizza M, Serruto D. Transcriptome analysis of Neisseria meningitidis in human whole blood and mutagenesis studies identify virulence factors involved in blood survival. PLoS Pathogens. 2011;7:e1002027. doi:https://doi.org/10.1371/journal.ppat.1002027. PMID:21589640
- Exley RM, Goodwin L, Mowe E, Shaw J, Smith H, Read RC, Tang CM. Neisseria meningitidis lactate permease is required for nasopharyngeal colonization. Infect Immun. 2005;73:5762-6. doi:https://doi.org/10.1128/IAI.73.9.5762-5766.2005. PMID:16113293
- Lo H, Tang CM, Exley RM. Mechanisms of avoidance of host immunity by Neisseria meningitidis and its effect on vaccine development. Lancet Infect Dis. 2009;9:418-27. doi:https://doi.org/10.1016/S1473-3099(09)70132-X. PMID:19555901
- Mendum TA, Newcombe J, Mannan AA, Kierzek AM, McFadden J. Interrogation of global mutagenesis data with a genome scale model of Neisseria meningitidis to assess gene fitness in vitro and in sera. Genome Biol. 2011;12:R127. doi:https://doi.org/10.1186/gb-2011-12-12-r127. PMID:22208880
- Schoen C, Kischkies L, Elias J, Ampattu BJ. Metabolism and virulence in Neisseria meningitidis. Front Cell Infect Microbiol. 2014;4:114. doi:https://doi.org/10.3389/fcimb.2014.00114. PMID:25191646
- Joseph B, Schneiker-Bekel S, Schramm-Gluck A, Blom J, Claus H, Linke B, Schwarz RF, Becker A, Goesmann A, Frosch M, et al. Comparative genome biology of a serogroup B carriage and disease strain supports a polygenic nature of meningococcal virulence. J Bacteriol. 2010;192:5363-77. doi:https://doi.org/10.1128/JB.00883-10. PMID:20709895
- Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, Klug L, Gadermaier B, Weinzerl K, Prassl R, et al. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun. 2016;7:10515. doi:https://doi.org/10.1038/ncomms10515. PMID:26806181
- Virji M, Kayhty H, Ferguson DJ, Alexandrescu C, Heckels JE, Moxon ER. The role of pili in the interactions of pathogenic Neisseria with cultured human endothelial cells. Mol Microbiol. 1991;5:1831-41. doi:https://doi.org/10.1111/j.1365-2958.1991.tb00807.x. PMID:1722554
- Rohde KH, Gillaspy AF, Hatfield MD, Lewis LA, Dyer DW. Interactions of haemoglobin with the Neisseria meningitidis receptor HpuAB: the role of TonB and an intact proton motive force. Mol Microbiol. 2002;43:335-54. doi:https://doi.org/10.1046/j.1365-2958.2002.02745.x. PMID:11985713
- Doulet N, Donnadieu E, Laran-Chich MP, Niedergang F, Nassif X, Couraud PO, et al. Neisseria meningitidis infection of human endothelial cells interferes with leukocyte transmigration by preventing the formation of endothelial docking structures. J Cell Biol. 2006;173:627-37. doi:https://doi.org/10.1083/jcb.200507128. PMID:16717131
- Morand PC, Tattevin P, Eugene E, Beretti JL, Nassif X. The adhesive property of the type IV pilus-associated component PilC1 of pathogenic Neisseria is supported by the conformational structure of the N-terminal part of the molecule. Mol Microbiol. 2001;40:846-56. doi:https://doi.org/10.1046/j.1365-2958.2001.02452.x. PMID:11401692
- Parkhill J, Achtman M, James KD, Bentley SD, Churcher C, Klee SR, Morelli G, Basham D, Brown D, Chillingworth T, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502-6. doi:https://doi.org/10.1038/35006655. PMID:10761919
- Omer H, Rose G, Jolley KA, Frapy E, Zahar JR, Maiden MC, Bentley SD, Tinsley CR, Nassif X, Bille E. Genotypic and phenotypic modifications of Neisseria meningitidis after an accidental human passage. PloS One. 2011;6:e17145. doi:https://doi.org/10.1371/journal.pone.0017145. PMID:21386889
- Kellogg DS, Jr., Cohen IR, Norins LC, Schroeter AL, Reising G. Neisseria gonorrhoeae. II. Colonial variation and pathogenicity during 35 months in vitro. J Bacteriol. 1968;96:596-605. PMID:4979098
- Pearson WR, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24-36. doi:https://doi.org/10.1006/geno.1997.4995. PMID:9403055
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:200.
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-40. doi:https://doi.org/10.1093/bioinformatics/btp616. PMID:19910308
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43:D222-6. doi:https://doi.org/10.1093/nar/gku1221. PMID:25414356
