ABSTRACT
Background: Aspergillus fumigatus is the most prevalent airborne fungal pathogen, causing invasive fungal infections mainly in immunosuppressed individuals. Death rates from invasive aspergillosis remain high because of limited treatment options and increasing antifungal resistance. The aim of this study was to identify key fungal-specific genes participating in vitamin B biosynthesis in A. fumigatus. Because these genes are absent in humans they can serve as possible novel targets for antifungal drug development. Methods: By sequence homology we identified, deleted and analysed four key A. fumigatus genes (riboB, panA, pyroA, thiB) involved respectively in the biosynthesis of riboflavin (vitamin B2), pantothenic acid (vitamin B5), pyridoxine (vitamin B6) and thiamine (vitamin B1). Results: Deletion of riboB, panA, pyroA or thiB resulted in respective vitamin auxotrophy. Lack of riboflavin and pantothenic acid biosynthesis perturbed many cellular processes including iron homeostasis. Virulence in murine pulmonary and systemic models of infection was severely attenuated following deletion of riboB and panA, strongly reduced after pyroA deletion and weakly attenuated after thiB deletion. Conclusions: This study reveals the biosynthetic pathways of the vitamins riboflavin and pantothenic acid as attractive targets for novel antifungal therapy. Moreover, the virulence studies with auxotrophic mutants serve to identify the availability of nutrients to pathogens in host niches.
Abbreviations: BPS: bathophenanthrolinedisulfonate; BSA: bovine serum albumin; CFU: colony forming unit; -Fe: iron starvation; +Fe: iron sufficiency; hFe: high iron; NRPSs: nonribosomal peptide synthetases; PKSs: polyketide synthaseses; wt: wild type
Introduction
Invasive fungal infections have increased dramatically in number over the last 40 years, largely because of the increase in the global number of immunocompromised patients. It is estimated that over 1.5 million people/year are infected worldwide by the major invasive fungal pathogens Candida albicans, Aspergillus fumigatus and Cryptococcus neoformans [Citation1,Citation2]. Despite modern antifungal treatments, mortality rates remain between 20 to 90%. Treatment efficacy has plateaued and fungal resistance is developing [Citation3].
Development of new drugs is hampered by the fact that fungi are eukaryotes and share with humans many of the same metabolic pathways. Therefore, most existing antifungals are not sufficiently selective and have various side effects.
Research has therefore focused on identifying essential genes and pathways not shared with the infected host [Citation4–Citation7]. Using a combination of comparative genomics, transcriptomics and metabolic flux analysis, we recently identified 64 fungal-specific targets including metabolic enzymes participating in amino acid, lipid and vitamin biosynthesis [Citation8]. Of these, 18 targets have already been validated in the literature, including enzymes participating in the biosynthesis of aromatic amino acids [Citation9], lysine [Citation10,Citation11], histidine [Citation12] and cysteine/methionine [Citation13] and in the biosynthesis of lipids, including phospholipids [Citation14], fatty acids [Citation6] and oxylipins [Citation15]. However, the assessment of fungal-specific targets in the vitamin biosynthesis pathways and their importance during infection has not been well established.
In this study, we analysed for the first time the vitamin B biosynthetic pathways for thiamine (B1), riboflavin (B2), pantothenic acid (B5) and pyridoxine (B6) in the important human pathogenic mold A. fumigatus. These fungal pathways, including the encoding genes are shown in Supplementary Figure 1. They are not found in humans, who need to obtain the required vitamin intake from their food. We deleted key fungal-specific genes in each pathway and phenotypically evaluated the resulting auxotrophic mutants in vitro and in vivo during infection. We show that riboflavin and pantothenic acid biosynthesis are important for iron homeostasis in vitro and for establishing a lethal infection in mice, suggesting strategies for their development as antifungal targets.
Materials and methods
Strains and growth conditions
Fungal strains were generally cultured on/in Aspergillus minimal medium (AMM) containing 1% glucose as carbon source and 20 mM glutamine as nitrogen source or on complete medium containing 2 g/L peptone and 1 g/L yeast extract [Citation16]. Alternatively, YAG medium consisting of 0.5% yeast extract, 1% glucose, 10 mM MgCl2, trace elements and vitamins, served as nutrient source. Supplements are indicated in the respective experiments. Iron was omitted for iron-depleted conditions (-Fe) and 30 µM FeSO4 was added into the media for iron-replete (+Fe) conditions. Vitamin stock solution includes 2.5 µM pantothenic acid, 10 µM pyridoxine and 10 µM niacin. Blood agar medium contained 1.8% agar, 0.5% sodium chloride and 10% or 25% blood. A hypoxic chamber (C-Chamber and Pro-Ox, Pro-CO2 controller; Biosherics) with the settings 1% O2, 5% CO2 and 94% N2 was used for hypoxic conditions. For growth assays, 104 conidia were point-inoculated on minimal medium agar plates and incubated for 48 h at 37°C. For inoculation of 100 ml liquid media, 106/ml conidia were used. The akuA-deficient derivative of ATCC46645 AfS77, termed wild-type (wt) here, served as the reference recipient [Citation17]. Primers used in this study are listed in . Generation of the A. fumigatus mutant strains and their verification by PCR and Southern analysis is outlined in detail in Supplementary Figure 2A-M. Strains used in this study are listed in Supplementary Table 2.
Table 1. Flavoprotein-dependent pathways in A. fumigatus. The number of proteins are given within brackets. The complete list of identified flavoproteins (238 proteins) is found in supplementary table 7.
Analysis of extra-and intracellular siderophores and biomass production
For quantification of extra- or intracellular siderophores, culture supernatants or lyophilized mycelia were saturated with FeSO4 and extracted as described previously [Citation18]. To analyse biomass production, mycelia from liquid cultures were freeze-dried and weighed.
For the vitamin-shift experiment, fungal strains were inoculated in liquid minimal medium supplemented with vitamins (riboflavin 2 μM, pantothenic acid 2 μM, pyridoxine 0.1 μM and thiamine 0.1 μM) for 12 h (supplementation phase). Germlings were extensively washed with water before shifting to vitamin-starved minimal medium for 36 h (starvation phase). Finally, the cultures were either vitamin supplemented (resupplementation phase) or not supplemented (continued starvation phase) for another 12 h. Dry weight was determined from freeze-dried mycelia.
Identification of A. fumigatus flavoproteins from the proteome
For screening flavin-dependent A. fumigatus proteins, the A. fumigatus Af293 proteome data set (AspGD; http://www.aspergillusgenome.org/download/domains/A_fumigatus_Af293/) from the Aspergillus database [Citation19] was searched with the terms ”flavo”, ”flavin”, ”FMN” and ”FAD”. The resulting list was compared with A. fumigatus proteins identified in AspGD (http://www.aspgd.org) with terms ”flavo”, ”flavin”, ”FMN” and ”FAD” [Citation20].
Pulmonary and systemic mouse infection
Three immunocompromised murine models for invasive aspergillosis were used: (i) non-neutropenic model; six-week-old female ICR mice were immunocompromised by subcutaneous injection with cortisone acetate (300 mg/kg) 3 days prior to infection, on the day of infection, and 3, 7, and 11 days postinfection. Fungal strains were grown for 3 days at 37°C on MM agar with appropriate vitamin supplementation. Spores were collected in PBS with 0.2% Tween 20 and counted by a hemocytometer. For the ΔriboB strain, conidial viability and germination were not affected on MM supplemented with 2.5 µM riboflavin (Supplementary Figure 3). The mice were infected intranasally with 5 × 105 dormant spores, suspended in 20 μl of PBS plus 0.2% Tween 20 (10 μl in each nostril). Neutropenic (ii) pulmonary and (iii) disseminated models; six-week-old female ICR mice were immunocompromised with cyclophosphamide (150 mg/kg in PBS) injected intraperitoneally at 3 days prior to and at 2 days post-conidial infection. Cortisone acetate (150 mg/kg PBS with 0.1% Tween 80) was injected subcutaneously at 3 days prior to conidial infection. For pulmonary infection, mice were infected intranasally as described above. For disseminated infection, 5 × 105 dormant spores were injected through the lateral tail vein. Survival was monitored for up to 21 days. For histopathology, mice were sacrificed two days after infection, their lungs were removed and stained with Grocott’s methenamine silver stain (GMS; fungal staining) and hematoxylin and eosin (H&E; tissue and nuclear staining). For fungal burden, infected mice were sacrificed on the second day post- infection, their lungs were removed and homogenized, and the homogenates were plated on YAG medium supplemented with the appropriate vitamin for colony forming unit (CFU) enumeration. Experiments were ethically approved by the Ministry of Health (MOH) Animal Welfare Committee, Israel.
Results
Generation of vitamin B auxotrophic mutant strains in A. fumigatus
To analyse the role of defined vitamin B biosynthetic pathways of A. fumigatus, we first identified by bioinformatics analysis, genes encoding key enzymes of the respective pathways for the synthesis of riboflavin (riboB/Afu1g13300, encoding GTP cyclohydrolase II), pantothenic acid (panA/Afu5g11040, encoding pantoate-beta-alanine ligase), pyridoxine (pyroA/Afu5g08090, encoding pyridoxal 5-phosphate synthase) and thiamine (thiB/Afu2g08970, encoding thiamine-phosphate diphosphorylase and hydroxyethylthiazole kinase) (Supplementary Figure 1 and Supplementary Tables 3–6). These genes were deleted in the A. fumigatus akuA::loxP recipient strain derived from ATCC46645 (AfS77, termed wt here), largely lacking non-homologous recombination [Citation21,Citation22], by homologous recombination and hygromycin selection as described in the Materials and Methods (Supplementary Figure 2). To confirm gene deletion-specific phenotypes, mutant strains were complemented (C strains) by re-integration of functional gene copies. Notably, we also attempted to generate a riboflavin auxotroph by deletion of Afu2g16360, the ortholog of Histoplasma capsulatum rib2, which is essential for riboflavin biosynthesis in this dimorphic fungal species [Citation23]. Surprisingly, deletion of Afu2g16360/rib2 did not result in riboflavin auxotrophy in A. fumigatus (data not shown), most likely because of the presence of paralogous genes (Supplementary Figure 1A).
Deletion of riboB, panA, pyroA and thiB leads to auxotrophy
The growth of the mutant strains was compared to the wt and complemented strains by spotting 104 conidia on solid minimal medium, complete medium or blood agar, containing different concentrations of vitamin as shown in Figure 1. No visible growth was seen in the mutant strains in the absence of their respective vitamin supplementation, confirming that the gene deletions resulted in auxotrophy. Radial growth of the ΔriboB and ΔpanA mutants was fully restored by addition of 2.5 µM riboflavin or pantothenic acid (), respectively, whereas growth of the ΔpyroA and ΔthiB strains necessitated addition of only 0.1 µM of their respective vitamin (). Complemented strains showed wild-type phenotypes in all assays performed, confirming the specificity of gene deletion phenotypes (). Complete medium allowed full growth and sporulation of the ΔpanA, ΔpyroA and ΔthiB strains, whereas in ΔriboB conidiation was blocked (reflected by the white instead of greenish colonies), most likely because the amount of riboflavin in the medium, which derives from the yeast extract component in this medium, is not sufficient to support wt-like growth for ∆riboB.
Figure 1. Deletion of riboB, panA, pyroA and thiB leads to respective auxotrophy.
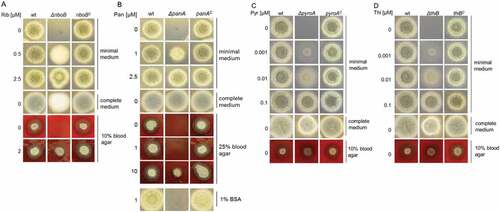
In contrast to the ΔpyroA and ΔthiB strains, both ΔriboB and ΔpanA did not grow on blood agar without vitamin supplementation, indicating that blood riboflavin and pantothenic acid levels are too low to support their growth (). Interestingly, for growth of ΔpanA on blood agar, increased pantothenic acid supplementation (10 µM) was required. This effect is most likely because pantothenic acid is adsorbed by serum albumin in the blood. In agreement, 1% bovine serum albumin (BSA) blocked growth of ΔpanA in the presence of 1 µM pantothenic acid in minimal medium, which supports growth without BSA ()).
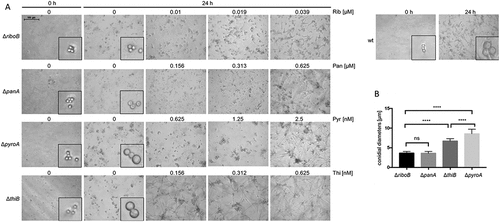
In liquid minimal medium without vitamin, after 24 h at 37°C, ΔriboB and ΔpanA conidia remained compact, whereas ΔpyroA and especially ΔthiB conidia underwent significant swelling, indicating partial early germination (Figure 2(a), insets, )). Interestingly, in liquid minimal medium, vitamin supplementation restored mutant growth at concentrations far lower than those needed on minimal medium agar plates ().
Figure 2. Microscopic analysis of germination and growth of the deletion strains.
Riboflavin and its derivatives FAD and FMN play a crucial role in NO detoxification and sporulation
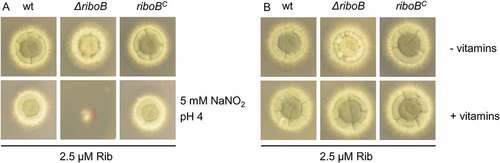
Riboflavin is the precursor of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) and as such used by approximately 238 flavoproteins in A. fumigatus (Supplementary Table 7). Flavoproteins have central metabolic and regulatory roles; displays 22 pathways comprising 91 of the 238 A. fumigatus flavoproteins. FAD is a crucial coenzyme of flavohemoglobins, which provide protection against NO and related reactive nitrogen species [Citation24]. We therefore tested the growth of ΔriboB during NO stress, induced by addition of 5 mM NaNO2 at a low pH of 4.0 (acidic conditions stimulate generation of nitric oxide from nitrate [Citation25]), and limiting 2.5 µM riboflavin for growth. Growth of the mutant was inhibited compared to the wt and complemented strains, indicating an important role for riboflavin in nitric oxide (NO) detoxification (Figure 3(a)). FAD is also a crucial coenzyme for the biosynthesis of the B vitamins niacin, pantothenic acid and pyridoxine. We therefore tested whether we can correct the impaired sporulation of ΔriboB on minimal medium (reflected by a decrease in greenish pigmentation of the colony) in the presence of 2.5 µM riboflavin, by adding these vitamins on minimal medium. We found that supplementation of ΔriboB with 2.5 µM pantothenic acid, 10 µM pyridoxine and 10 µM niacin restored conidiation, suggesting a role of riboflavin in biosynthesis of these vitamins (), ).
Figure 3. Riboflavin and its derivatives FAD and FMN play a crucial role in NO detoxification and sporulation.
The vitamins riboflavin, pantothenic acid, pyridoxine and thiamine have potent anti- oxidative activity. We therefore tested the sensitivity of the ΔriboB, ΔpanA, ΔpyroA and ΔthiB strains to survive oxidative stress induced by H2O2 or menadione under partial supplementation with their respective vitamin. However, we found no differences in the sensitivity of the mutants compared to the wt (data not shown).
Decreased riboflavin availability reduces production of siderophores in A. fumigatus
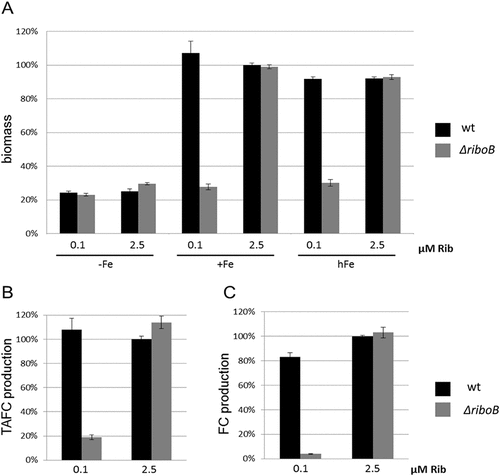
To survive in the human host under conditions of iron starvation, A. fumigatus employs two high-affinity iron-uptake systems, reductive iron assimilation and the extracellular siderophores fusarinine C (FsC) and triacetylfusarinine C (TAFC), as well as the intracellular siderophore ferricrocin (FC) for iron storage and distribution [Citation26,Citation27]. To further characterize the role of riboflavin biosynthesis in iron utilization and siderophore production, wt and ΔriboB were analysed after growth in liquid minimal medium during iron starvation (-Fe), iron sufficiency (+Fe, 30 µM FeSO4) and iron excess (hFe, 5 mM FeSO4) supplemented with either a low (0.1 µM) or a high (2.5 µM) riboflavin concentration (Figure 4). Biomass production was significantly decreased in ΔriboB compared to wt under low riboflavin and iron sufficiency (74% less, P < 0.0001 vs. wt) or iron excess (67% less, P < 0.0001 vs. wt) ()). Siderophore biosynthesis was not observed in the wt or ΔriboB under +Fe or hFe (data not shown). During iron starvation, biomass production of the mutant displayed wt-like levels even under the low riboflavin concentration ()), most likely due to the approximately fourfold decrease in biomass compared to iron sufficiency or excess that results in decreased vitamin requirement for growth. Interestingly, at low (0.1 µM) riboflavin concentration under iron starvation, siderophore production in ΔriboB decreased significantly: 82% less TAFC (P < 0.0001 vs. wt) and 96% less FC (P < 0.0001 vs. wt) compared to the wt, despite the fact that under these conditions both strains achieve the same biomass (). These findings indicate a crucial role for riboflavin in siderophore biosynthesis, most likely as a cofactor for ornithine-N5-monooxygenase SidA (), which catalyzes the initial step in siderophore biosynthesis [Citation28].
Figure 4. Decreased riboflavin availability reduces production of siderophores in A. fumigatus.
Iron supplementation reduces pantothenic acid requirement ofΔpanA
Siderophore biosynthesis involves the enzymatic activity of nonribosomal peptide synthetases (NRPS), which contain 4´-phosphopantetheine, derived from pantothenic acid, as an essential prosthetic group. In A. fumigatus one such enzyme is encoded by npgA/pptA. Previous studies confirmed that NpgA function is essential for biosynthesis of TAFC and FC [Citation29] and consequently for iron homeostasis. To characterize the effects of PanA deficiency on iron homeostasis, we analyzed the biomass production of the pantothenic acid auxotrophic mutant strain ΔpanA relative to the wt strain under iron depleted (-Fe), iron replete (+Fe, 30 µM FeSO4) and iron excess (hFe, 5 mM FeSO4) conditions, with either low (0.5 µM) or high (2.5 µM) concentrations of pantothenic acid. Under -Fe and +Fe conditions, ΔpanA exhibited significantly decreased biomass production of 80% during -Fe (P < 0.001 vs. wt) and 90% during +Fe (P < 0.0001 vs. wt) when supplemented with the low pantothenic acid concentration (while biomass production under high iron conditions was only mildly reduced by 28% compared to the wt). However, wt-like growth of the mutant strain was achieved by the addition of 2.5 µM pantothenic acid into the medium (Figure 5(a)). The beneficial effect of iron excess on biomass production of the pantothenic acid auxotroph was also demonstrated on solid medium ()). In contrast, iron excess did not enhance growth of the riboB, pyroA and thiB null strains under limiting vitamin supplementation, further indicating the specificity of this effect in the ΔpanA mutant (data not shown). These results suggest a role for pantothenic acid in iron metabolism. In agreement, pantothenic acid is essential for activation of nonribosomal peptide synthetases, including siderophore biosynthetic SidC and SidD [Citation30,Citation31].
Figure 5. Iron supplementation reduces pantothenic acid requirement of ΔpanA.
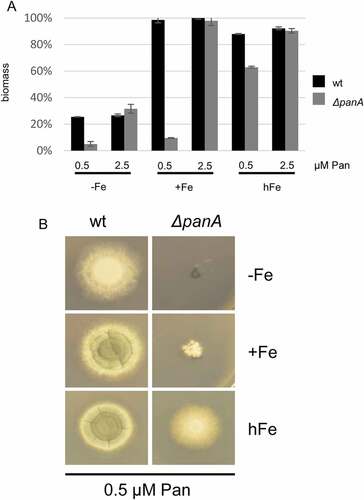
Hypoxia decreases pantothenic acid requirement ofΔpanA
To survive in the hypoxic necrotic tissue during infection, A. fumigatus is able to adapt to extremely low oxygen and low iron surroundings. On minimal medium agar plates, the growth defect of ΔpanA supplemented with limiting 1 µM pantothenic acid was less pronounced under hypoxic (1% oxygen) conditions compared to normoxic (21% oxygen) conditions during iron starvation (-Fe), iron sufficiency (+Fe) and iron excess (hFe, 5 mM FeSO4) (Figure 6). The most remarkable phenotype was observed during iron starvation and iron sufficiency, where the ΔpanA strain under hypoxic conditions reached radial wt-like growth but completely lacked sporulation. The role of pantothenic acid during hypoxic conditions could be explained by reduced pantothenic acid requirement or by increased pantothenic uptake under these conditions [Citation32]. In contrast, the riboB, pyroA and thiB null strains under limiting vitamin supplementation and normoxic or hypoxic iron-sufficient conditions grew like the wt (data not shown).
RiboB, panA, pyroA and thiB deletion mutants are not killed by vitamin starvation
To evaluate the suitability of RiboB, PanA, PyroA and ThiB as possible drug targets, we carried out vitamin-shift experiments to measure whether growth in the absence of vitamin causes growth arrest (i.e. inhibiting the pathway is fungistatic) or cell death (i.e. inhibiting the pathway is fungicidal). Auxotrophic strains were germinated in liquid minimal medium for 12 h in the presence of the respective vitamin (corresponding to early wt growth in the lungs during infection), and then washed repeatedly and grown for 36 h in the absence of the vitamin (corresponding to inhibition of the pathway). Finally, to determine viability, the vitamin was added to the medium for 12 h and the dry weight of the lyophilized mycelium was measured relative to non-supplemented strains. For comparison, the wt strain was starved for nitrogen (instead of vitamins as in the case of the auxotrophic mutants) and re-supplemented with nitrogen for 12h, assuming that wt strains are capable of coping with nitrogen starvation conditions. We found that the vitamin–starved mycelia of the ΔriboB, ΔpanA, ΔpyroA and ΔthiB strains grew rapidly when re-supplemented with their respective vitamin, comparable to the nitrogen-starved wt (Figure 7, black bars), while without supplementation, mycelial growth remained minimal (, grey bars). This indicates that the mycelium remained alive during the 36 h of vitamin-starvation and suggests that inhibition of these pathways will have a fungistatic (and not a fungicidal) effect.
Figure 6. Hypoxia decreases pantothenic acid requirement of ΔpanA.
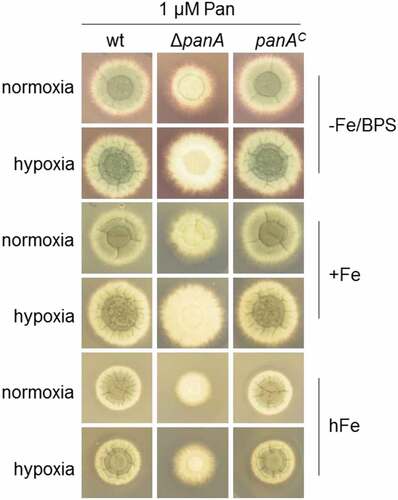
Figure 7. riboB, panA, pyroA and thiB deletion mutants are not killed by starvation of the lacking vitamin.
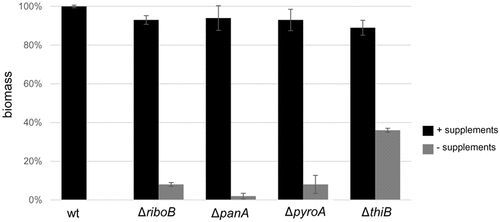
Riboflavin and pantothenic acid biosynthesis are required for A. fumigatus virulence in murine infection models
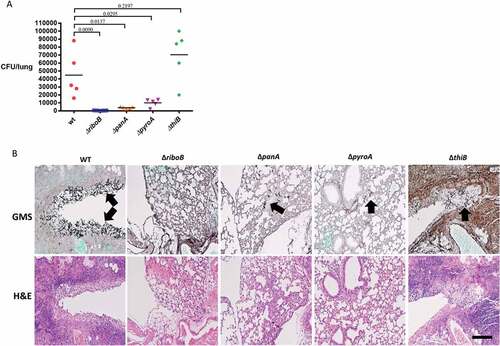
The ΔriboB, ΔpanA, ΔpyroA, ΔthiB strains were assessed for virulence in three immunocompromised murine models of infection: (i) a non-neutropenic pulmonary infection model in which mice were immunosuppressed with cortisone acetate (CA), (ii) a neutropenic pulmonary infection model in which mice were immunosuppressed with cyclophosphamide (CY), (iii) a neutropenic disseminated infection model in which mice were immunosuppressed with cyclophosphamide (CY) and infected via the lateral tail vein. The ΔriboB mutant was avirulent (100% survival) and ΔpanA mutant was strongly attenuated (90% survival) in the non-neutropenic pulmonary model of infection, both strains were avirulent (100% survival) in the disseminated neutropenic model of infection (P < 0.0001 vs. wt), and strongly attenuated in virulence (90% survival ΔpanA, 60% survival ΔriboB) in the neutropenic pulmonary model of infection (P < 0.0005 vs. wt) (Figure 8(a–c)). The ΔpyroA and ΔthiB mutants showed attenuated virulence in both pulmonary models of infection (60 to 70% survival ΔpyroA, 40% survival ΔthiB, P < 0.005 vs. wt) and in disseminated infection (P < 0.05 vs. wt) although in this last model 100% mortality of ΔpyroA and 90% mortality of ΔthiB was observed, possibly suggesting that there are lower levels of available pyridoxine and thiamine in the lungs compared to blood (–)). It should be noted that in the neutropenic models, profound neutropenia resolves after ~ 7 days and that, had an additional round of immunosuppression been administered at day + 6, the virulence data may have been slightly less compelling for some of the strains. Fungal load and histopathology of infected lungs (CA, non-neutropenic host model) further reflected these results. No CFUs or visible fungal growth were found in the lungs of mice infected for 48 h with the ΔriboB strain. Low lung CFU counts were found in ΔpanA (~ 2% of wt), increasing in ΔpyroA (~ 20% of wt), and reaching wt levels in ΔthiB (Figure 9(a)). Histopathology followed by GMS (stains fungal elements black) and H&E (stains lung tissue) further corroborated these findings, showing extensive hyphal growth (arrows) and granulocyte infiltration (purple granulation-lower panel) in the airways of mice infected with the wt strain, which was slightly attenuated following ΔthiB infection, greatly reduced following ΔpyroA and ΔpanA infection and totally absent in the ΔriboB–infected mice ()).
Figure 8. Riboflavin and pantothenic acid biosynthesis are essential for virulence of A. fumigatus in murine infection models.
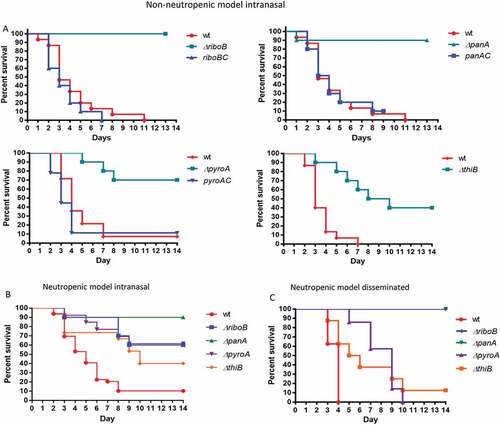
Figure 9. Deficiency in riboflavin, pantothenic acid and pyridoxine biosynthesis significantly reduce A. fumigatus lung burden in cortisone-acetate immunocompromised mice.
Discussion
A central problem in the development of antifungals is the lack of suitable drug targets not found in humans. In this report, using the pathogenic mold A. fumigatus as our model, we analysed four fungal-specific pathways involved in the biosynthesis of riboflavin (vitamin B2), pantothenic acid (vitamin B5), pyridoxine (vitamin B6) and thiamine (vitamin B1) as potential drug targets.
We deleted genes encoding key enzymes in each of these pathways (riboB, panA, pyroA and thiB), which resulted in auxotrophy. Our underlying hypothesis was that the vitamins produced by these pathways might not be available for fungal growth in the host, resulting in dependence on endogenous biosynthesis. Past work in A. fumigatus has shown this to be the case for folate (Vitamin B9) biosynthesis. Deletion of pabA, encoding PABA synthase, catalysing an early step in folate biosynthesis, abrogated virulence in infected mice, designating this pathway as an excellent candidate for the development of novel antifungals [Citation33]. Antibacterial sulfa drugs that inhibit dihydropteroate synthase in the folic acid biosynthetic pathway already exist and show weak antifungal activity [Citation34]. Recent efforts have focused on developing antifungals to target other enzymes in this pathway [Citation35].
Our main finding was that deletion of riboB and panA abolished or strongly attenuated virulence (depending on the infection model), pyroA deletion attenuated virulence and loss of thiB weakly reduced virulence. Virulence was tested in three models of murine infection that recapitulate the most common types of invasive aspergillosis infections [Citation36]. We modelled invasive pulmonary aspergillosis in the neutropenic host using cyclophosphamide and in the non-neutropenic compromised host using cortisone acetate. Disseminated aspergillosis, which frequently occurs in the neutropenic host, was performed by intravenous infection. Our results suggest that there are insufficient levels of riboflavin and pantothenic acid in the infected host to support growth of the ΔriboB and ΔpanA mutants. In contrast, the ΔpyroA and ΔthiB mutants need only very low levels (0.1–2.5 nM) of external pyridoxine and thiamine for growth, which are available in the host at far higher concentrations [Citation37,Citation38], most likely explaining their milder influence on virulence. Notably, the virulence behaviour of the four mutants matched their growth pattern on blood agar plates: in contrast to the ΔpyroA and ΔthiB strains, ΔriboB and ΔpanA mutant strains were unable to grow on blood agar without vitamin supplementation. Virulence studies with auxotrophic mutants also serve to describe nutrient availability in different host niches. The strongly attenuated virulence of ΔriboB and ΔpanA mutants in all tested virulence models demonstrates that both riboflavin and pantothenic acid biosynthesis are not only essential for invasion (intranasal infection model) but also during dissemination (intravenous infection model). In contrast, lysine biosynthesis was previously found to be essential exclusively for invasion but not during dissemination [Citation10,Citation11]. Virulence analysis of auxotrophs was previously tested in the closely related Aspergillus nidulans during the 1970s. Interestingly, they gave slightly different results. As found here for A. fumigatus, thiamine auxotrophy did not reduce virulence. However, in A. nidulans, riboflavin auxotrophy attenuated but did not abolish virulence whereas pyridoxine auxotrophy abolished virulence [Citation39]. However, these studies were performed with mutagenized strains in which the target genes were not deleted and they lacked the complemented strains as controls.
Riboflavin is the precursor of flavin mononucleotide (FMN) and flavin adenine nucleotide (FAD) and as such is an essential cofactor for many flavoprotein-catalyzed physiological processes. Remarkably, A. fumigatus encodes approximately 238 flavoproteins (Supplementary Table 7) involved in central metabolic pathways and regulatory circuits () including for example NO-detoxification (flavohemoglobins FhpA and FhpB), siderophore biosynthesis (SidA), reductive iron assimilation (metalloreductase FreB), amino acid biosynthesis, ergosterol biosynthesis, vitamin biosynthesis, and secondary metabolism. In contrast, S. cerevisiae possesses only 68 flavoproteins [Citation40]. The greatly increased number of flavoproteins in A. fumigatus compared to S. cerevisiae might indicate a higher demand for riboflavin. We confirmed here the importance of riboflavin for NO-detoxification and siderophore biosynthesis in A. fumigatus. In contrast to NO-detoxification, which appears to be dispensable for virulence of A. fumigatus [Citation41], siderophore biosynthesis has been shown to be essential for virulence of several animal and plant pathogenic fungi [Citation42]. Similarly, pantothenic acid is of central importance for primary metabolism as it is essential for coenzyme A biosynthesis and consequently for fatty acid metabolism, carbohydrate metabolism and tricarboxylic acid cycle [Citation43]. Moreover, it is an essential prosthetic group for polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs) including the siderophore biosynthetic NRPSs. The impact of PKSs and NRPSs is reflected by the avirulence of the A. fumigatus mutant lacking the pantothenic acid dependent enzyme phosphopantetheinyl transferase PptA/NpgA [Citation30]. The requirement of PptA/NpgA and consequently pantothenic acid for activation of NRPSs including the siderophore biosynthetic NRPSs SidD and SidC is consistent with the beneficial effect of iron supplementation on biomass production of ΔpanA during limited pantothenic acid supplementation ().
How suitable are the riboflavin and pantothenic acid biosynthetic pathways for development as drug targets? The riboflavin biosynthesis pathway contains many genes absent in humans, including rib3, rib4, rib5, rib7 and rib1/riboB described here (see supplementary Table 3). We show that deletion of riboB in A. fumigatus leads to avirulence in cortisone-compromised mice and in neutropenic mice with disseminated infection. Lung CFU enumeration and histology show complete clearance of the fungus from the lungs 48 h after infection. Riboflavin auxotrophy also leads to avirulence in Candida albicans [Citation6], Histoplasma capsulatum [Citation23] and in many species of gram-negative bacteria including Mycobacteria that lack a riboflavin uptake system and are therefore totally dependent on endogenous biosynthesis [Citation44]. Therefore, this pathway is particularly suitable for the development of antimicrobials. Based on detailed enzymatic and structural data, substrate inhibitors have been developed for the last two steps of riboflavin biosynthesis, catalysed by lumazine synthase and riboflavin synthase, respectively. To the best of our knowledge, in vivo use of these compounds has not been described, suggesting there were pharmacological problems in their development [Citation45,Citation46].
The pantothenic acid biosynthesis pathway is also rich in possible antimicrobial targets including pan2, pan5, and pantothenate synthase pan6/panA (see supplementary Table 4) [Citation47]. A major effort has been made to develop pantothenate synthase inhibitors active against Mycobacterium tuberculosis, as this enzyme is essential for virulence [Citation48]. Although several showed in vitro activity, none was described as acting in vivo [Citation49]. Subsequent CoA synthesis from pantothenate is controlled by pantothenate kinase, encoded by the essential gene cab1 (S. cerevisiae)/panK (A. nidulans) [Citation50]. Several pantothenic acid analogues that inhibit this enzyme have shown antibacterial activity in vitro and in vivo [Citation47] but none were tested for fungal infections.
Two drawbacks should be noted regarding the suitability of the enzymes RiboB and PanA as antifungal targets- first, our in vitro vitamin-shift analysis suggests that inhibiting these enzymes will have a fungistatic rather than fungicidal action. Second, inhibition of a single cellular target often results in mutations leading to resistance. Nevertheless, these problems can be circumvented by developing inhibitors that (i) react with the target enzyme to generate toxic products or (ii) block additional essential fungal targets, and (iii) by using drug combinations [Citation3].
Future work needs to test existing inhibitors of riboflavin and pantothenic acid biosynthesis for activity in pathogenic fungi both in vitro and in vivo. Crystallographic data of key fungal enzymes (RiboB, PanA etc.) needs to be generated and used to plan more specific and potent antifungal inhibitors. Compound library screens can make use of the fact that inhibitors specifically acting on these pathways are neutralized by the addition of exogenous riboflavin and pantothenic acid, respectively [Citation51].
Supplemental Material
Download Zip (1.1 MB)Acknowledgments
We acknowledge funding by the Israel Ministry of Health Infect-ERA (grant 3-0000-11080 to N.O.) and the Austrian Science Fund/Infect-ERA program (FWF grant I1616/Infect-ERA project AspMetNet to H.H.). A.-M.D. is an associate student of the HOROS program (W1253).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Brown GD, Denning DW, Levitz SM. Tackling human fungal infections. Science. 2012;336(6082):647. PubMed PMID: 22582229.
- Bongomin F, Gago S, Oladele RO, et al. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi (Basel). 2017;3(4). PubMed PMID: 29371573; PubMed Central PMCID: PMCPMC5753159. DOI:10.3390/jof3040057
- Perfect JR. The antifungal pipeline: a reality check. Nat Rev Drug Discov. 2017;16(9):603–616. PubMed PMID: 28496146.
- Lu Y, Deng J, Rhodes JC, et al. Predicting essential genes for identifying potential drug targets in Aspergillus fumigatus. Comput Biol Chem. 2014;50:29–40. PubMed PMID: 24569026.
- Thykaer J, Andersen MR, Baker SE. Essential pathway identification: from in silico analysis to potential antifungal targets in Aspergillus fumigatus. Med Mycol. 2009;47 Suppl 1:S80–S87. PubMed PMID: 19253142.
- Becker JM, Kauffman SJ, Hauser M, et al. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc Natl Acad Sci U S A. 2010;107(51):22044–22049. PubMed PMID: 21135205; PubMed Central PMCID: PMCPMC3009777.
- Ianiri G, Idnurm A. Essential gene discovery in the basidiomycete Cryptococcus neoformans for antifungal drug target prioritization. MBio. 2015;6(2). PubMed PMID: 25827419; PubMed Central PMCID: PMCPMC4453551. DOI:10.1128/mBio.02334-14
- Kaltdorf M, Srivastava M, Gupta SK, et al. Systematic identification of anti-fungal drug targets by a metabolic network approach. Front Mol Biosci. 2016;3:22. PubMed PMID: 27379244; PubMed Central PMCID: PMCPMC4911368.
- Sasse A, Hamer SN, Amich J, et al. Mutant characterization and in vivo conditional repression identify aromatic amino acid biosynthesis to be essential for Aspergillus fumigatus virulence. Virulence. 2016;7(1):56–62. PubMed PMID: 26605426; PubMed Central PMCID: PMCPMC4871646.
- Liebmann B, Muhleisen TW, Muller M, et al. Deletion of the Aspergillus fumigatus lysine biosynthesis gene lysF encoding homoaconitase leads to attenuated virulence in a low-dose mouse infection model of invasive aspergillosis. Arch Microbiol. 2004;181(5):378–383. PubMed PMID: 15052376.
- Schobel F, Jacobsen ID, Brock M. Evaluation of lysine biosynthesis as an antifungal drug target: biochemical characterization of Aspergillus fumigatus homocitrate synthase and virulence studies. Eukaryot Cell. 2010;9(6):878–893. PubMed PMID: 20363898; PubMed Central PMCID: PMCPMC2901645.
- Dietl AM, Amich J, Leal S, et al. Histidine biosynthesis plays a crucial role in metal homeostasis and virulence of Aspergillus fumigatus. Virulence. 2016;7(4):465–476. PubMed PMID: 26854126; PubMed Central PMCID: PMCPMC4871644.
- Amich J, Dumig M, O’Keeffe G, et al. Exploration of sulfur assimilation of Aspergillus fumigatus reveals biosynthesis of sulfur-containing amino acids as a virulence determinant. Infect Immun. 2016;84(4):917–929. PubMed PMID: 26787716; PubMed Central PMCID: PMCPMC4807484.
- Tao L, Gao N, Chen S, et al. The choC gene encoding a putative phospholipid methyltransferase is essential for growth and development in Aspergillus nidulans. Curr Genet. 2010;56(3):283–296. PubMed PMID: 20379720.
- Fischer GJ, Keller NP. Production of cross-kingdom oxylipins by pathogenic fungi: an update on their role in development and pathogenicity. J Microbiol. 2016;54(3): 254–264. PubMed PMID: 26920885; PubMed Central PMCID: PMCPMC5107414.
- Pontecorvo G, Roper JA, Hemmons LM, et al. The genetics of Aspergillus nidulans. Adv Genet. 1953;5: 141–238. Epub 1953/01/01.PubMed PMID: 13040135.
- Hartmann T, Dumig M, Jaber BM, et al. Validation of a self-excising marker in the human pathogen Aspergillus fumigatus by employing the beta-rec/six site-specific recombination system. Appl Environ Microbiol. 2010;76(18):6313–6317. PubMed PMID: 20656854; PubMed Central PMCID: PMCPMC2937505.
- Schrettl M, Beckmann N, Varga J, et al. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 2010;6(9):e1001124. Epub 2010/10/14.
- Nierman WC, Pain A, Anderson MJ, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438(7071):1151–1156. PubMed PMID: 16372009.
- Cerqueira GC, Arnaud MB, Inglis DO, et al. The Aspergillus genome database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014;42(Database issue):D705–10. PubMed PMID: 24194595; PubMed Central PMCID: PMCPMC3965050.
- Krappmann S, Sasse C, Braus GH. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end- joining-deficient genetic background. Eukaryot Cell. 2006;5(1):212–215. Epub 2006/01/10.
- Hartmann T, Dumig M, Jaber BM, et al. Validation of a self-excising marker in the human pathogen Aspergillus fumigatus by employing the beta-rec/six site-specific recombination system. Appl Environ Microbiol. 2010;76(18):6313–6317. Epub 2010/07/27.
- Garfoot AL, Zemska O, Rappleye CA. Histoplasma capsulatum depends on de novo vitamin biosynthesis for intraphagosomal proliferation. Infect Immun. 2014;82(1):393–404. PubMed PMID: 24191299; PubMed Central PMCID: PMCPMC3911860.
- Bonamore A, Boffi A. Flavohemoglobin: structure and reactivity. IUBMB Life. 2008;60(1):19–28. Epub 2008/ 04/02.
- Schinko T, Berger H, Lee W, et al. Transcriptome analysis of nitrate assimilation in Aspergillus nidulans reveals connections to nitric oxide metabolism. Mol Microbiol. 2010;78(3):720–738. PubMed PMID: 20969648; PubMed Central PMCID: PMCPMC3020322.
- Haas H. Iron - a key nexus in the virulence of Aspergillus fumigatus. Front Microbiol. Epub 2012/02/22. 2012;3:28. PubMed PMID: 22347220; PubMed Central PMCID: PMC3272694.
- Schrettl M, Bignell E, Kragl C, et al. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 2007;3(9):1195–1207. PubMed PMID: ISI:000249768300003.
- Eisendle M, Oberegger H, Zadra I, et al. The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding L-ornithine N-5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC). Mol Microbiol. 2003;49(2):359–375. PubMed PMID: ISI:000184224700007.
- Oberegger H, Eisendle M, Schrettl M, et al. 4 ‘-phosphopantetheinyl transferase-encoding npgA is essential for siderophore biosynthesis in Aspergillus nidulans. Curr Genet. 2003;44(4):211–215. PubMed PMID: ISI:000186823000005.
- Johns A, Scharf DH, Gsaller F, et al. A nonredundant phosphopantetheinyl transferase, PptA, is a novel antifungal target that directs secondary metabolite, siderophore, and lysine biosynthesis in Aspergillus fumigatus and is critical for pathogenicity. MBio. 2017;8(4). PubMed PMID: 28720735; PubMed Central PMCID: PMCPMC5516258. DOI:10.1128/mBio.01504-16
- Oberegger H, Eisendle M, Schrettl M, et al. 4ʹ-phosphopantetheinyl transferase-encoding npgA is essential for siderophore biosynthesis in Aspergillus nidulans. Curr Genet. 2003;44(4):211–215. PubMed PMID: 14508603.
- Grahl N, Shepardson KM, Chung D, et al. Hypoxia and fungal pathogenesis: to air or not to air? Eukaryot Cell. 2012;11(5):560–570. Epub 2012/ 03/27.
- Brown JS, Aufauvre-Brown A, Brown J, et al. Signature-tagged and directed mutagenesis identify PABA synthetase as essential for Aspergillus fumigatus pathogenicity. Mol Microbiol. 2000;36(6): 1371–1380. PubMed PMID: 10931287.
- Yekutiel A, Shalit I, Shadkchan Y, et al. In vitro activity of caspofungin combined with sulfamethoxazole against clinical isolates of Aspergillus spp. Antimicrob Agents Chemother. 2004;48(9):3279–3283. PubMed PMID: 15328085; PubMed Central PMCID: PMCPMC514744.
- Bourne CR. Utility of the biosynthetic folate pathway for targets in antimicrobial discovery. Antibiotics (Basel). 2014;3(1):1–28. PubMed PMID: 27025730; PubMed Central PMCID: PMCPMC4790348.
- Balloy V, Huerre M, Latge JP, et al. Differences in patterns of infection and inflammation for corticosteroid treatment and chemotherapy in experimental invasive pulmonary aspergillosis. Infect Immun. 2005;73(1):494–503. PubMed PMID: 15618189; PubMed Central PMCID: PMCPMC538925.
- Dick T, Manjunatha U, Kappes B, et al. Vitamin B6 biosynthesis is essential for survival and virulence of Mycobacterium tuberculosis. Mol Microbiol. 2010;78(4):980–988. PubMed PMID: 20815826.
- Reidling JC, Lambrecht N, Kassir M, et al. Impaired intestinal vitamin B1 (thiamin) uptake in thiamin transporter-2-deficient mice. Gastroenterology. 2010;138(5):1802–1809. PubMed PMID: 19879271; PubMed Central PMCID: PMCPMC4916904.
- Purnell DM. The effects of specific auxotrophic mutations on the virulence of Aspergillus nidulans for mice. Mycopathol Mycol Appl. 1973;50(3): 195–203. PubMed PMID: 4580921.
- Gudipati V, Koch K, Lienhart WD, et al. The flavoproteome of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2014;1844(3):535–544. PubMed PMID: 24373875; PubMed Central PMCID: PMCPMC3991850.
- Lapp K, Vodisch M, Kroll K, et al. Characterization of the Aspergillus fumigatus detoxification systems for reactive nitrogen intermediates and their impact on virulence. Front Microbiol. 2014;5:469. PubMed PMID: 25309516; PubMed Central PMCID: PMCPMC4160965.
- Haas H. Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat Prod Rep. 2014;31(10):1266–1276. PubMed PMID: 25140791; PubMed Central PMCID: PMCPMC4162504.
- Leonardi R, Zhang YM, Rock CO, et al. Coenzyme A: back in action. Prog Lipid Res. 2005;44(2–3):125–153. PubMed PMID: 15893380.
- Long Q, Ji L, Wang H, et al. Riboflavin biosynthetic and regulatory factors as potential novel anti-infective drug targets. Chem Biol Drug Des. 2010;75(4):339–347. PubMed PMID: 20148904.
- Chen J, Illarionov B, Bacher A, et al. A high-throughput screen utilizing the fluorescence of riboflavin for identification of lumazine synthase inhibitors. Anal Biochem. 2005;338(1):124–130. PubMed PMID: 15707942.
- Zhao Y, Bacher A, Illarionov B, et al. Discovery and development of the covalent hydrates of trifluoromethylated pyrazoles as riboflavin synthase inhibitors with antibiotic activity against Mycobacterium tuberculosis. J Org Chem. 2009;74(15):5297–5303. PubMed PMID: 19545132.
- Spry C, Kirk K, Saliba KJ. Coenzyme A biosynthesis: an antimicrobial drug target. FEMS Microbiol Rev. 2008;32(1):56–106. PubMed PMID: 18173393.
- Sambandamurthy VK, Wang X, Chen B, et al. A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat Med. 2002;8(10):1171–1174. PubMed PMID: 12219086.
- Hung AW, Silvestre HL, Wen S, et al. Optimization of inhibitors of Mycobacterium tuberculosis pantothenate synthetase based on group efficiency analysis. ChemMedChem. 2016;11(1):38–42. PubMed PMID: 26486566; PubMed Central PMCID: PMCPMC4949533.
- Calder RB, Williams RS, Ramaswamy G, et al. Cloning and characterization of a eukaryotic pantothenate kinase gene (panK) from Aspergillus nidulans. J Biol Chem. 1999;274(4):2014–2020. PubMed PMID: 9890959.
- Ben Yaakov D, Shadkchan Y, Albert N, et al. The quinoline bromoquinol exhibits broad-spectrum antifungal activity and induces oxidative stress and apoptosis in Aspergillus fumigatus. J Antimicrob Chemother. 2017;72(8):2263–2272. PubMed PMID: 28475687.
