ABSTRACT
Background: Staphylococcus pseudintermedius is an opportunistic pathogen that is the major cause of pyoderma affecting dogs. Conventional antimicrobial treatment for infections caused by this organism have failed in recent years due to widespread resistance and alternative treatment strategies are a high priority. Protein A encoded in Staphylococcus aureus by spa protects the bacterium by binding IgG and acts as a superantigen. Staphylococcus pseudintermedius possess two genes orthologous to S. aureus spa, spsP, and spsQ. Methods: SpsQ and SpsQ-M, a non-toxigenic SpsQ, were cloned and expressed as recombinant proteins and their cytotoxic effect on canine B cells was measured. The neutralizing ability of antibody raised against them in clinically healthy dogs was evaluated. Results: S. pseudintermedius SpsQ induced apoptosis of canine B cells. Specific amino acid substitutions diminished SpsQ-M binding to immunoglobulin and its super-antigenic activity, while its antigenicity was maintained. This recombinant, non-toxigenic S. pseudintermedius SpsQ stimulated the production of antibodies in dogs that specifically reacted with SpsQ and greatly diminished its cytotoxic effect on canine B cells. Conclusions: The production of neutralizing antibody suggests that attenuated, non-toxic SpsQ produced in this study is a good candidate for inclusion in a vaccine for use in the treatment and prevention of S. pseudintermedius infections.
Abbreviations: SpA: Staphylococcus aureus protein A; SpsP: Staphylococcus pseudintermedius protein A; SpsQ: Staphylococcus pseudintermedius protein A; SpsQ-M: attenuated Staphylococcus pseudintermedius protein A; MRSP: methicillin resistant Staphylococcus pseudintermedius; IgA: immunoglobulin A; IgG: immunoglobulin G; IgM: immunoglobulin M; VH: variable region of immunoglobulin heavy chain; IgBD: immunoglobulin binding domains; MFI: mean fluorescent intensity; SEM: standard error of the mean; PBMC: Peripheral blood mononuclear cells; CD21: complement receptor type 2; ST: Sequence type; OD: Optical density; ORF: open reading frame; PBS: Phosphate buffered saline; Tween 20: Polyethylene glycol sorbitan monolaurate 20; HRP: horseradish peroxidase; TMB- 3,3',5,5'-Tetramethylbenzidine
Introduction
Staphylococcus pseudintermedius, a gram-positive, coagulase-positive bacterium, is commonly found on the normal skin and nasal flora of household pets [Citation1,2]. It is the major cause of canine skin and ear infections [Citation2,Citation3]. Furthermore, human infections are sporadically reported [Citation4,Citation5], suggesting that, although it is estimated to be low, there is a threat of zoonotic disease. Approximately 30–35% of the S. pseudintermedius isolates tested in our University of Tennessee College of Veterinary Medicine Clinical Bacteriology Laboratory are methicillin-resistant. Methicillin resistant S. pseudintermedius (MRSP) are resistant to most antimicrobial agents accessible at present and the number of multidrug resistant staphylococcal isolates is increasing [Citation6–Citation9].
Alternative approaches to antimicrobial therapy to treat staphylococcal infections, such as vaccines, have been difficult to develop. This is likely rooted in the ability of the bacteria to evade and/or destroy important components of their host defenses [Citation10,Citation11]. Protein A is anchored on the peptidoglycan cell wall and is also secreted during the exponential growth phase [Citation12–Citation15]. It is an important virulence factor, able to bind IgG via its Fc region [Citation16–Citation18] protecting staphylococci from opsonophagocytosis. It also binds to and crosslinks the VH3-family of immunoglobulin idiotypes (the Ig fragment responsible for antigen recognition), triggering apoptosis [Citation19,Citation20]. Consequently, protein A plays an important role in enhancing pathogenicity of staphylococci by impairing the development of the host immune response [Citation10,Citation21]. Recently, it has been demonstrated that immunization with nontoxigenic S. aureus protein A (SpAKKAA) (with amino acid substitutions in five repeated domains, E, D, A, B, and C) protected guinea pigs in experimental models of S. aureus infection [Citation22]. Also, S. aureus expressing the mutant variant was deficient in abscess formation, and laboratory animals infected with these mutant bacteria overcame the immunosuppressive effects of protein A and developed adaptive immune responses [Citation22]. Likewise, Pankey et al. [Citation23] investigated the use of S. aureus protein A (SpA) as a vaccine for bovine mastitis with promising results.
In vertebrates, the immunoglobulin variable region of the heavy chain (VH) is encoded by diverse genes and the VH differ in length and amino acid composition between species and each species has it is own repertoire [Citation24–Citation26]. In dogs, the VH1-family of immunoglobulin idiotypes represent the largest portion of VH genes in B cell populations [Citation25].
Previous studies on SpA [Citation27] have demonstrated that SpA binds to the Fab portion of VH3-type IgM on the surface of B cells resulting in cross-linking of B cell receptors that lead to B cell clonal expansion and subsequent apoptotic collapse, thereby limiting the host’s capability to produce antibody to other staphylococcal antigens [Citation21].
In 2011, Bannoehr et al. [Citation28] found that S. pseudintermedius possess two genes orthologous to S. aureus spa, spsP (encoding SpsP) and spsQ (encoding SpsQ, analogous to SpA). Balachandran et al. [Citation18] recently found that SpsP and SpsQ consist of approximately 377 and 462 amino acids, respectively. Both proteins have predicted N-terminal sequences of 33 amino acids, followed by a repeat region of three or four IgG-binding domains and a C terminal region that shares about 63% sequence similarity to the X-region of S. aureus. The Immunoglobulin-binding domains (IgBDs) are highly conserved between SpsP and SpsQ, with amino acid sequence identity ranging from 67 to 90%. Balachandran et al. [Citation18] found that spsQ was present in all clinical isolates of S. pseudintermedius tested within diverse multilocus sequence types, although they had different levels of expression, whereas spsP was less conserved and often occurs in degenerate form. They showed that S. pseudintermedius SpsQ is cell wall anchored and was detected in the bacterial secretome during log phase [Citation18]. SpsQ bound to canine IgG primarily via its Fc region [Citation18]. This interaction was inhibited by anti-SpA raised in chickens which made S. pseudintermedius more susceptible to phagocytosis [Citation18]. Grandolfo [Citation29] suggested that S. pseudintermedius SpsQ could be considered a possible vaccine target, however, information about SpsQ is scarce and it is necessary to determine if there are toxic effects of S. pseudintermedius protein A on canine B cells that can be neutralized with antibody.
The purpose of this study was to characterize the cytotoxic effect of recombinant S. pseudintermedius SpsQ on dog B cells, develop attenuated SpsQ (SpsQ-M), and evaluate antibody raised against SpsQ-M in clinically healthy dogs. Staphylococcus pseudintermedius SpsQ-M was tested for its antigenicity and B cell killing. The results from this study suggest that S. pseudintermedius SpsQ may serve as a key component in a vaccine or as part of an immunotherapeutic treatment.
Results
Bioinformatics analysis and S. pseudintermedius protein A characteristics
S. pseudintermedius ED99, representing isolate that expresses both SpsQ and SpsP, SpsQ contained four and SpsP had genes encoding a maximum of three Ig-binding domains (). This compares to five domains with AA identities of 74% in Staphylococcus aureus subsp. aureus strain Newman SpA (). Three out of four SpsQ domains share 84–88% identity with the C domain of S. aureus SpA with threonine at position 19, while only one domain is more similar to the B domain of S. aureus SpA ().
Figure 1. S. pseudintermedius SpsQ characteristics and basis for generation of recombinant nontoxigenic protein A (SpsQ-M). (a) Pairwise alignment between S. pseudintermedius ED99 SpsQ and SpsP. (b) SpA of S. aureus subsp. aureus strain Newman and SpsQ of S. pseudintermedius harbors an N-terminal signal peptide (orange boxes), Ig binding domains, 13 variable region X (grey boxes), LysM sequence (green boxes) and C-terminal sorting signal 14 (black boxes). (c) Amino acid sequences of the four Ig binding domains of SpsQ, as well as Ig 15 binding domain C of SpA, with the positions glutamine (Q, highlighted in blue) 5 and 6 and 16 aspartate (D, blue highlight) 32 and 33 as indicated. (d) SpsQ secondary structure composition. 17 SpsQ is composed of alpha helices with the positions of triple α-helical bundles shown for each 18 domain.

Multiple sequence alignment (MSA) between each domain of SpsQ and domain C of SpA, the most similar domain to SpsQ IgBDs, showed that each domain in SpsQ has binding sites for the IgG Fcγ (Glutamine (Q) 5 and Q 6), and Fab region (aspartate (D) 32 and D33) of surface membrane-associated variable heavy 1 (VH1)-encoded B cell antigen receptors (). Furthermore, as with SpA of S. aureus, the secondary structure of SpsQ consists only of alpha helices and each Ig binding domain consists of three helices ().
Cloning, expression, and purification of recombinant SpsQ and SpsQ-M
Recombinant polyhistidine tagged SpsQ and SpsQ-M were produced in E.coli and purified using HisPur Ni-NTA resin under native conditions and eluted using an imidazole gradient. The molecular weights of SpsQ and SpsQ-M determined in western blots were of the expected sizes (47.2 and 47.6 kDa, respectively) ().
Antigenicity of SpsQ and SpsQ-M and their reactivity with serum from a dog with pyoderma
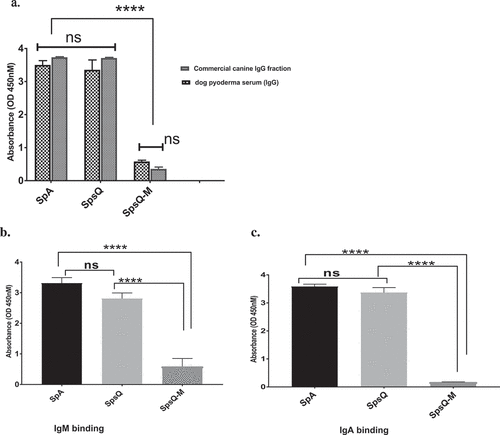
Chicken anti-protein A, chosen because protein A does not bind chicken immunoglobulin, had strong reactivity with affinity-purified SpsQ-M similar to that with S. aureus SpA and S. pseudintermedius SpsQ, indicating the attenuated protein retained its antigenicity (Figure 3). Sera from a dog with a history of chronic pyoderma showed significantly lower reactivity against SpsQ-M (Figure 4(a)) compared to recombinant wild-type SpsQ and SpA. SpsQ-M had significantly lower binding (p-value of 0.0001) to commercial dog IgG (), IgM (p-value of 0.0001) ()or IgA (p-value of 0.0001) compared to SpsQ and SpA. ().
Figure 2. Western blot of recombinant S. pseudintermedius wild and mutant protein A with HRP-conjugated anti-6xhis tag monoclonal antibody, SpsQ and SpsQ-M were expressed in E.coli with M.W. of 47.2 and 47.6 KDa, respectively.
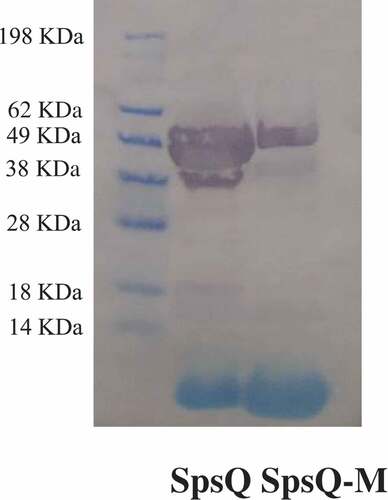
Figure 3. HRP-conjugated chicken anti-protein a recognizes and binds to recombinant SpsQ, SpsQ-M and commercial S. aureus SpA. The values represent averages from three independent experiments. (*P < 0.05 was considered significant). There was no statistically significant difference between any sample P > 0.05 (ns).
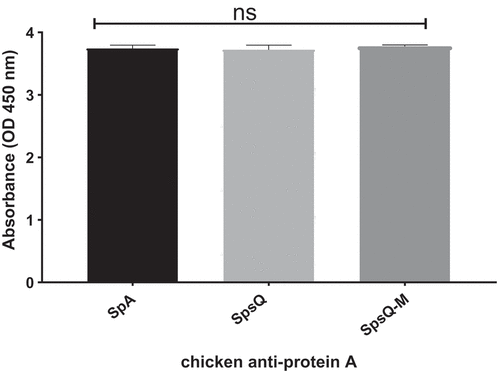
Figure 4. Natural infection with S. pseudintermedius did not elicit antibody specifically reactive with staphylococcal protein A Serum from a dog with a history of chronic pyoderma showed significantly lower reactivity with SpsQ-M compared to wild type SpsQ and SpA for (a), IgG, (b), IgM and (c), IgA (p-value of 0.0001 for each) ****. The values represent averages from three independent experiments. (*P < 0.05 was considered significant). ns – Not significant.
S. pseudintermedius protein A induced apoptosis of canine B cells
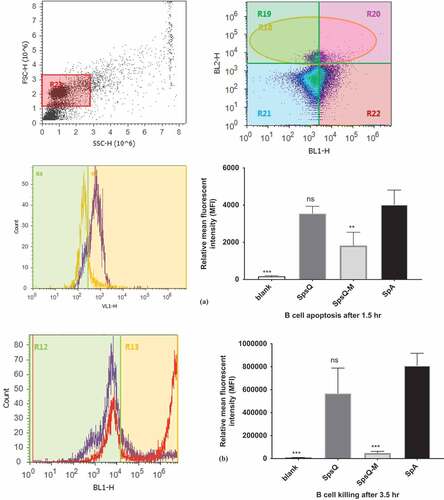
Affinity-purified SpsQ-M had a low apoptotic effect on B cells at 1.5 h in contrast to recombinant SpsQ at the same time point, with a mean ± SEM fluorescent intensity (MFI) difference of 2052 ± 285.2 between SpsQ and SpsQ-M (p-value of 0.0063) (Figure 5(a)). Incubation of the cells for 3.5 h with SpsQ-M resulted in a Sytox MFI reduction of 519,065 ± 128,292 SEM compared to that of SpsQ (p-value of 0.0003) ().
Figure 5. S. pseudintermedius recombinant SpsQ has a superantigenic effect on canine B cells. (a) Gating on canine peripheral blood mononuclear cells (PBMC) based on side and forward scatter (shown in dot plot) and on B cells using PE-anti-CD21 antibody (shown in density plot), SpsQ induced B cell apopstosis after 1.5 hr (violet peak on histogram) compared to SpsQ-M(yellow peak on histogram). The mean fluorescent intensity (MFI) of the blank, SpsQ and SpsQ-M relative to SpA was calculated based on average values from three independent experiments. (*P < 0.05 was considered significant). ns – Not significant. MFI of SpsQ-M was significantly lower (P = 0.0063) ** than that of SpsQ and SpA. (b) Gating on canine B cells using PE-anti-CD21 antibody, SpsQ incubated for 3.5 hr, killed B cells (red peak) compared to SpsQ-M (violet peak on histogram). The MFI% of the blank, SpsQ and SpsQ-M relative to SpA was calculated based on average values from three independent experiments (*P < 0.05 was considered significant). ns – Not significant. MFI of SpsQ-M was significantly lower (P = 0.0003***) than that of SpsQ and SpA.
SpsQ-M induces specific antibody responses in dogs that reduce the effect of SpsQ on B cells in vitro
ELISA analysis of sera obtained from dogs on days −7, 8, 15 and 29 (relative to injections) showed that antibodies against S. pseudintermedius SpsQ-M and SpsQ were detected on day 15 and reached the highest level on day 29 (P < 0.0001) compared to pre-injection control sera ().
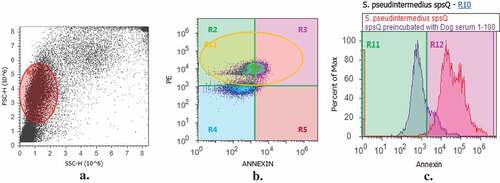
Pre-incubation of SpsQ, at a concentration of 100 µg/ml in PBS with dog anti-SpsQ-M resulted in 105,665 ± 900.3 SEM, n = 1 reduction in annexin mean fluorescent intensity (MFI) as compared with that of SpsQ treatment alone ().
Figure 6. Dogs injected with spsQ-M developed specific IgG reactive with recombinant wildtype and mutant protein A. Specific antibodies against S. pseudintermedius SpsQ-M (a) and SpsQ (b) were detected using an indirect ELISA after the second injection (on day 15) and were higher on day 29 (p < 0.0001) compared to pre-injection control sera.
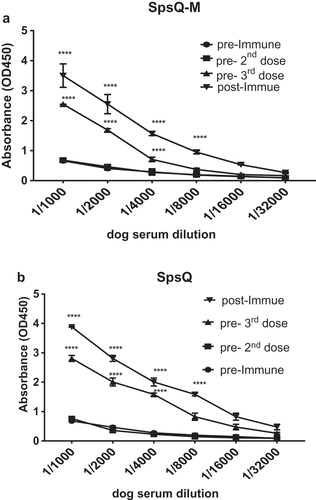
Figure 7. Dog anti- spsQ-M antibody protects canine B cells from the superantigenic effects of SpsQ. Pre-incubation of SpsQ with dog anti-SpsQ-M resulted in mean 105,665 ± 900.3 SEM, n = 1 reduction in mean fluorescent intensity (MFI) from SpsQ on canine B cells compared to that of SpsQ treatment alone. a, Gating on canine peripheral blood mononuclear cells (PBMC) based on side and forward scatter (shown in dot plot) and (b) on B cells using PE-anti-CD21 antibody (shown in density plot). c, SpsQ induced B cell apopstosis after 1.5 hr (red peak on histogram) compared to SpsQ preincubated with dog serum at dilution 1:100 (blue peak on histogram).
Discussion
Protective host immune responses against extracellular pathogens are typically antibody-mediated. Unfortunately, staphylococcal infection does not usually establish protective immunity [Citation24,Citation30–Citation32] and efficacious staphylococcal vaccines have proven difficult to develop. Most vaccine research has focused on S. aureus, which most commonly affects humans and is distinguished from S. pseudintermedius, which is primarily of veterinary concern.
Two forms of S. pseudintermedius protein A contain IgBDs that are similar to each other and likely to be antigenically cross-reactive. Because of their similarity and the presence of SpsQ in all tested isolates, SpsQ was used for this study. Understanding how S. pseudintermedius subverts the canine immune response is important for vaccine development and to facilitate natural host humoral immunity.
S. pseudintermedius SpsQ binding with the Fab region of surface membrane-associated VH1-encoded B-cell antigen receptors induces apoptosis in canine B cells. We developed a non-toxigenic SpsQ (SpsQ-M) by substitution of residues responsible for dual reactivity of each SpsQ domain with IgG Fcγ and Fab regions of surface membrane-associated VH1-encoded B-cell antigen receptors. Compared to staphylococcal protein A [Citation33], SpsQ-M had much lower reactivity with IgG. Moreover, IgG, IgM, and IgA in sera from a dog with a history of chronic pyoderma had little immune-mediated binding to SpsQ-M indicating a lack of natural production of antibody elicited by infection with S. pseudintermedius. Comparing purified recombinant wild-type and mutant SpsQ, we found SpsQ-M had a significantly lower toxic effect on canine B lymphocytes. These results are in agreement with the findings of Kim et al [Citation21]. with S. aureus SpA(KKAA) as determined using mouse B lymphocytes and human immunoglobulins.
It should be noted that specific antibody directed against native protein A cannot be measured due to the non-immune binding of protein A to immunoglobulin. After injecting a clinically healthy dog with SpsQ-M, we observed a high titer of SpsQ-specific antibodies in agreement with Kim et al’s observations with SpAKKAA injected in mice [Citation21]. These results indicate that SpsQ, similar to SpA, suppresses adaptive immune responses during staphylococcal infection which may explain why previous S. pseudintermedius infections are not associated with protective immunity against recurrent infection.
Dog anti-SpsQ-M abolished the superantigenic effect of SpsQ that triggers apoptotic cell death in canine B cells, consistent with the findings of others with SpA studied in non-canine systems [Citation21].
Collectively, the use of SpsQ-M will improve our understanding of the immune interactions between S. pseudintermedius and the dog. Data presented in our study suggest that blocking the Fcγ and Fab binding activities of SpsQ could be an alternative or adjunct to conventional antimicrobial treatments for infections caused by S. pseudintermedius including canine pyoderma [Citation11]. Canine anti-SpsQ-M may reduce SpsQ immune suppression in dogs, stimulate the production of specific antibodies against S. pseudintermedius and establish a protective immunity against recurrent infection. A SpsQ-M vaccine would likely contain additional S. pseudintermedius virulence factors.
Material and methods
Ethics Statement
Experimental protocols were reviewed and approved by the University of Tennessee Institutional Animal Care and Use Committee (IACUC) including obtaining blood samples of dog (2474–0716) and injecting dogs with recombinant protein for producing antibodies (2572–1217).
Bioinformatics analysis
MSA of spsQ from diverse isolates of S. pseudintermedius (n = 100), was performed using Geneious, version 9.1.3[Citation34]. The bacterial localization prediction tool, PSORTb version 3.0.2 (http://www.psort.org/psortb/) [Citation35], was used to determine the topology and domain structure of SpsQ and SpsP. SpsQ modeling and binding site prediction were performed using Protein Homology/analogY Recognition Engine V 2.0 (Phyre2) (http://www.sbg.bio.ic.ac.uk/phyre2) [Citation36], and the 3DLigandSite (http://www.sbg.bio.ic.ac.uk/3dligandsite/) [Citation37], using SpA as a basis to predict the IgBDs in each domain. A pairwise sequence alignment of SpsQ and SpsP was used to identify conserved amino acids critical for IgG Fc and B cell receptor binding.
Identification of IgBDs was guided by S. aureus SpA secondary structure [Citation38] and based on S. aureus SpA residues [Citation21] responsible for dual reactivity of each domain in SpA with Fcγ [Citation39] and Fab [Citation40] and shared by S. pseudintermedius SpsQ.
Geneious, version 9.1.3 was used to select the locations for amino acids to be substituted in each IgBD and to design a full-length, four domain (SpsQ-M) attenuated S. pseudintermedius protein A construct (SpsQ-M). Glutamine (Q) 5 and 6, as well as aspartate (D) 32 and 33 in each domain of SpsQ were selected as critical amino acids for the association of SpsQ with immunoglobulin. To test this, substitutions of Q5K (lysine), Q6K, D32A (alanine), and D33A were introduced into each IgBDs of SpsQ.
Bacterial strains and growth conditions
The S. pseudintermedius strain used in this study, strain 06–3228 [Citation41], was isolated at the University of Tennessee, College of Veterinary Medicine Bacteriology Laboratory. It represents the most common multilocus sequence type (ST) previously reported in the United States (ST68) [Citation6,Citation8,Citation42]. Bacterial colonies grown on blood agar plates were inoculated into 5ml of sterile trypticase soy broth (TSB) (BD Biosciences, San Jose, CA, USA Cat No. RS1-011–21) and incubated overnight at 37°C with shaking at 225 rpm (Excella E24 Incubator Shaker, New Brunswick Scientific). Fifty microliters of overnight culture were inoculated into 5ml of fresh, sterile TSB to initiate log-phase bacterial cultures. Bacteria were grown at 37°C with shaking at 225rpm until an optical density of OD 600 = 0.4–0.6 was reached [Citation43].
Cloning, expression, and purification of recombinant wild-type and non-toxigenic S. pseudintermedius SpsQ
Bacteria from a single colony of S. pseudintermedius strain 06–3228 obtained from blood agar plates were grown in TSB at 37°C with 225 rpm shaking. DNA was extracted using a MO BIO UltraClean® Microbial DNA Isolation Kit (QIAGEN Inc., USA Cat No.12224–50) according to the manufacturer’s instructions. Oligonucleotide primers (Integrated DNA Technology, Coralville, USA) () were designed using a PrimerQuest Tool (https://www.idtdna.com/Primerquest/Home/Index) based on the whole genomic sequence of S. pseudintermedius strain 06–3228 determined by Riley et al [Citation41].
Table 1. Primers used to amplify recombinant wild and attenuated protein A (SpsQ) from Staphylococcus pseudintermedius. NotI and BamHI restriction sites are underlined.
The spsQ open reading frame (ORF) without the regions encoding the predicted N-terminal signal peptide was amplified from S. pseudintermedius 06–3228 genomic DNA. The ORF of spsQ-M was amplified from a PMA-SpsQ-M plasmid () (Life Technologies Corp., Carlsbad, CA, USA), containing a synthetic spsQ-M gene. PCR was performed using taq polymerase (rTaq, Takara, USA Cat No. R004) and the following cycling conditions were performed: initial denaturation at 95°C for 90 seconds, 30 cycles of denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 1 minute followed by a final extension at 72°C for 5 minutes. All ORFs were amplified without a 6x histidine tag because pETBlue-2 () allowed T7lac promoter-based expression of target genes with C-terminal histidine •Tag® sequences. PCR products were Sanger sequenced at The University of Tennessee Genomics Core facility.
Table 2. Plasmids and competent cells used to clone and express recombinant wild and attenuated protein A (SpsQ) from Staphylococcus pseudintermedius.
To clone full length S. pseudintermedius spsQ and spsQ-M, the PCR products were digested with NotI and BamHI, then ligated into pETBlue-2, an expression vector with C-terminal HSV•Tag® and His•Tag® sequences (Novagen, USA Cat No .70674). The pETBlue-2 construct transformed into DH5-alpha E. coli chemically-competent cells () (New England BioLabs Inc., USA Cat No .C2987I) by heat shock and DH5-alpha bacteria were plated on LB agar plates with 100 µg/mL ampicillin. The plasmid constructs were transformed into Tuner ™ (DE3) pLacI E. coli chemically-competent cells () (Novagen, USA Cat No .70623) by heat shock and the bacteria were plated on LB agar containing 50µg/ml ampicillin and 20µg/ml chloramphenicol.
To express recombinant protein, a single colony of Tuner ™ (DE3) pLacI E. coli was inoculated into LB broth containing 50µg/ml ampicillin and 20µg/ml chloramphenicol and bacteria were grown overnight at 37ºC with 225 rpm shaking. LB broth containing 50µg/ml ampicillin and 20µg/ml chloramphenicol was inoculated with a 1:100 dilution of overnight culture and grown at 37ºC with 225 rpm shaking until a 600 nm optical density between 0.4 and 0.6 was reached. Protein expression was induced by addition of 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Teknova, USA Cat. No. I3431) and bacteria were grown for 4 h at 30ºC with shaking at 225 rpm. Bacterial cultures were centrifuged at 12,000 x g for 5 min in 5 ml of protein extraction reagent (BugBuster, Novagen, USA Cat No. 70,584) and 20 µl of 100X protease inhibitor (Cocktail Set III, EDTA-Free Calbiochem, USA Cat No. 539,134), and subsequently incubated for 30 min at 37ºC in a shaking incubator at 225 rpm. Bacteria were pelleted by centrifugation at 12,000 x g for 45 min at 4°C. Recombinant protein was purified from the supernatant using affinity purification (HisPur™ Ni-NTA Spin Purification Kit, Thermo Scientific, USA Cat No. 88,228). Protein concentrations were determined using a bicinchoninic acid (BCA) assay (Thermo Scientific, USA Cat No.23227).
Production of antibodies against recombinant proteins
Recombinant SpsQ-M at 100 µg/ 0.5 cc in phosphate buffered saline (PBS) (pH 7.2) was injected in the lateral thorax by the subcutaneous route, into three clinically normal dogs. Injections were given once every 7 days for a total of three injections with a control dog receiving PBS (pH 7.2) only. Blood (6 cc) was collected from a jugular vein using a 20 g needle and 12 cc syringe 4 times, on days −7, 8, 15 and 29. The collected blood was left undisturbed at room temperature for 30 min followed by centrifugation at 2,000 x g for 10 min in a refrigerated centrifuge.
SDS-PAGE and western blots
Protein samples were resolved by SDS-PAGE in 4–12% polyacrylamide gels (Invitrogen, USA Cat No. NP0322BOX) and electrophoretically transferred onto nitrocellulose membranes (Thermo Scientific, USA Cat No. 77,010). The blots were blocked overnight in 5% (w/v) nonfat dried milk powder dissolved in phosphate buffered saline containing 0.05% polyethylene glycol sorbitan monolaurate (Tween 20) (PBS-T) at 4°C. The blocked membranes were incubated with a 1:2,000 dilution of horseradish peroxidase (HRP)-conjugated anti-6xhis tag monoclonal antibody (Thermo Scientific, USA Cat No. MA1-21,315-HRP) in 0.05% PBS-T for 1 h with 225 rpm shaking at room temperature. After five washes with 0.05% PBS-T, bound antibodies were detected using 1-Step™ chloronaphthol substrate solution (Thermo Scientific, USA Cat No. 34,012).
Enzyme-linked immunosorbent assay
For measurement of the antigenicity of recombinant proteins, affinity-purified SpsQ and SpsQ-M were coated on ELISA plates (Corning, USA Cat No. 3590) at 2 µg/ml in PBS (pH 7.2). The plates were washed with PBS-T and incubated with HRP- chicken anti-protein A antibody (Gallus Immunotech, USA Cat No. APA) for 1 h at 37°C, then washed. For this and all subsequent ELISA assays, plates were washed three times with PBS-T between all incubations, bound antibodies were detected using TMB substrate (Thermo Scientific, USA Cat No. N301), reactions were stopped with 0.18 M sulphuric acid and optical density read at 450 nm on a plate reader (Bio TEK, USA Cat No. EL800). The experiment was repeated at least three times and a p-value of < 0.05 was considered significant, which was the same for all the experiments unless mentioned otherwise.
To test the reactivity of serum from a dog with chronic pyoderma, S. aureus SpA and S. pseudintermedius SpsQ and SpsQ-M were coated on ELISA plates (Corning, USA Cat No. 3590) at 2 µg/ml in PBS. They were incubated with serum at a dilution of 1:2000. Bound IgG, IgM, and IgA were detected by HRP- goat anti-dog IgG-heavy and light chain (Bethyl Laboratories, Inc., USA Cat No. A40-123–1), HRP-goat anti-dog IgM µ chain (Bethyl Laboratories, Inc., USA Cat No. A40-116–2), HRP-goat anti-dog IgA (Bethyl Laboratories, Inc., USA Cat No. A40-104P) at a dilution of 1:8000 in PBS-T. Dog whole IgG molecule (Rockland, USA Cat No. 004–0102-1000) was used for comparison to measure non-specific IgG binding.
To detect a specific antibody response against SpsQ-M in injected dogs, recombinant S. pseudintermedius proteins and commercial SpA were coated on ELISA plates as previously described and incubated with two-fold serially diluted serum from injected dogs (1/1000–1/32,000). Bound IgG was detected using HRP- goat anti-dog IgG-heavy and light chain (Bethyl Laboratories, Inc., USA Cat No. A40-123–1) with serum from uninjected dogs used as negative controls. The experiment was run in duplicate and a p-value of < 0.05 was considered significant.
The ability of S. pseudintermedius SpsQ to kill B cells and induce B cell apoptosis
A total of 100 µg of purified recombinant SpsQ or SpsQ-M was mixed with isolated peripheral blood mononuclear cells (PBMC) in 1 ml of RPMI medium supplemented with 10% fetal bovine serum and incubated for 1.5 h at 37ºC in a 5% CO2 incubator. To detect early phases of B cell apoptosis, phosphatidylserine was measured on the surface of cells using pacific blue- conjugated annexin (Thermo Scientific, USA Cat No. A35136). B cells were identified using phycoerythrin (PE) conjugated mouse anti-canine CD21 (clone: CA2.1D6) (BIO-RAD, USA Cat No. MCA1781PE) that recognizes canine CD21 (complement receptor type 2) on mature B lymphocytes. Stained B cells were analyzed using a flow cytometer (Attune acoustic focusing cytometer). B cells were also incubated with the same recombinant proteins as described above but for 3.5 h in order to detect B cell death. Cells were stained with Sytox green (Life Technologies, Inc., USA Cat No. 1,776,406) and PE-conjugated mouse anti-canine CD21. Gates were placed on cells positive for PE and these B cells were analyzed by flow cytometry.
To determine the protective effect of canine anti-SpsQ-M on B cells, recombinant S. pseudintermedius SpsQ was incubated for 30 minutes at 37ºC with serum from SpsQ-M injected dogs. The experiment was run in duplicate and a p-value of < 0.05 was considered significant.
For flow cytometry analysis the cut-off for apoptosis or cell death was established using leukocytes incubated without SpsQ. Mean fluorescent intensity was determined from all B cells.
Statistical analysis
A one-way ANOVA and Tukey–Kramer method were used to measure the significant differences between SpsQ, SpsQ-M, and SpA on inducing apoptosis and causing B cell death. However, two-way ANOVA and Tukey–Kramer methods were performed to test if there were significant differences in SpA, SpsQ or SpsQ-M binding with canine antibodies. All analyzes were conducted using the GraphPad Prism software (Version 7, GraphPad Software Inc.).
Supplemental Material
Download PDF (258.5 KB)Acknowledgments
This research was supported by Egyptian cultural and educational bureau, Washington DC and the University of Tennessee, Center of Excellence in Livestock Diseases and Human Health.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Rubin JE, Chirino-Trejo M. Prevalence, sites of colonization, and antimicrobial resistance among Staphylococcus pseudintermedius isolated from healthy dogs in Saskatoon, Canada. J Vet Diagn Invest. 2011 Mar;23(2):351–354. PubMed PMID: 21398462.
- Bannoehr J, Guardabassi L. Staphylococcus pseudintermedius in the dog: taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet Dermatol. 2012 Aug;23(4):253. PubMed PMID: 22515504.
- van Duijkeren E, Catry B, Greko C, et al. Review on methicillin-resistant Staphylococcus pseudintermedius. J Antimicrob Chemother. 2011 Dec;66(12):2705–2714. PubMed PMID: 21930571.
- Stegmann R, Burnens A, Maranta CA, et al. Human infection associated with methicillin-resistant Staphylococcus pseudintermedius ST71. J Antimicrob Chemother. 2010 Sep;65(9):2047–2048. PubMed PMID: 20601356.
- Riegel P, Jesel-Morel L, Laventie B, et al. Coagulase-positive Staphylococcus pseudintermedius from animals causing human endocarditis. Int J Med Microbiol. 2011 Mar;301(3):237–239. PubMed PMID: 21075051.
- Videla R, Solyman SM, Brahmbhatt A, et al. Clonal complexes and antimicrobial susceptibility profiles of Staphylococcus pseudintermedius isolates from dogs in the United States. Microb Drug Resist. 2017 May 15; PubMed PMID: 28504897. DOI:10.1089/mdr.2016.0250
- Moodley A, Damborg P, Nielsen SS. Antimicrobial resistance in methicillin susceptible and methicillin resistant Staphylococcus pseudintermedius of canine origin: literature review from 1980 to 2013. Vet Microbiol. 2014 Jul 16;171(3–4):337–341. PubMed PMID: 24613081.
- Solyman SM, Black CC, Duim B, et al. Multilocus sequence typing for characterization of Staphylococcus pseudintermedius. J Clin Microbiol. 2013 Jan;51(1):306–310. PubMed PMID: 23115265; PubMed Central PMCID: PMCPMC3536184.
- Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012 Mar 1;18(3):268–281.
- Thammavongsa V, Kim HK, Missiakas D, et al. Staphylococcal manipulation of host immune responses [Review article]. Nat Rev Microbiol. 2015;13:529–543. .
- Kim HK, Emolo C, DeDent AC, et al. Protein A-specific monoclonal antibodies and prevention of Staphylococcus aureus disease in mice. Infect Immun. 2012;80(10):3460–3470.
- Sjoquist J, Movitz J, Johansson IB, et al. Localization of protein A in the bacteria. Eur J Biochem. 1972 Oct 17;30(1):190–194. PubMed PMID: 5086604.
- Movitz J. Formation of extracellular protein A by Staphylococcus aureus. Eur J Biochem. 1976 Sep;68(1):291–299. PubMed PMID: 964266.
- Becker S, Frankel MB, Schneewind O, et al. Release of protein A from the cell wall of Staphylococcus aureus. Proc Natl Acad Sci U S A. 2014 Jan 28;111(4):1574–1579. PubMed PMID: 24434550; PubMed Central PMCID: PMCPMC3910568.
- Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992 Jul 24;70(2):267–281. PubMed PMID: 1638631.
- Lindmark R, Thorén-Tolling K, Sjöquist J. Binding of immunoglobulins to protein A and immunoglobulin levels in mammalian sera. J Immunol Methods. 1983 Jan 1;62(1):1–13.
- Jensen K. A normally occurring Staphylococcus antibody in human serum. APMIS. 2007 May;115(5):533; discussion 540-1. PubMed PMID: 17504410.
- Balachandran M, Bemis DA, Kania SA. Expression and function of protein A in Staphylococcus pseudintermedius. Virulence. 2018 Jan 1;9(1):390–401. PubMed PMID: 29157101.
- Boyle MDP. CHAPTER 1 - Introduction to bacterial immunoglobulin-binding proteins. In Boyle, MDP. editor, Bacterial Immunoglobulin-binding proteins. Cambridge Massachusetts: academic press. 1990. p. 1–21.
- Sasso EH, Silverman GJ, Mannik M. Human IgM molecules that bind staphylococcal protein A contain VHIII H chains. J Immunol. 1989;142(8):2778–2783.
- Kim HK, Cheng AG, Kim HY, et al. Nontoxigenic protein A vaccine for methicillin-resistant Staphylococcus aureus infections in mice. J Exp Med. 2010 Aug 30;207(9):1863–1870. PubMed PMID: 20713595; PubMed Central PMCID: PMCPMC2931167.
- Kim HK, Falugi F, Thomer L, et al. Protein A suppresses immune responses during Staphylococcus aureus bloodstream infection in guinea pigs. MBio. 2015 Jan 6;6(1):e02369-14. PubMed PMID: 25564466; PubMed Central PMCID: PMCPMC4313907.
- Pankey JW, Boddie NT, Watts JL, et al. Evaluation of protein A and a commercial bacterin as vaccines against Staphylococcus aureus mastitis by experimental challenge. J Dairy Sci. 1985 Mar 1;68(3):726–731.
- Dryla A, Prustomersky S, Gelbmann D, et al. Comparison of antibody repertoires against Staphylococcus aureus in healthy individuals and in acutely infected patients. Clin Diagn Lab Immunol. 2005;12(3):387–398.
- Bao Y, Guo Y, Xiao S, et al. Molecular characterization of the VH repertoire in Canis familiaris. Vet Immunol Immunopathol. 2010 Sep 15;137(1–2):64–75. PubMed PMID: 20483487.
- Pauli NT, Kim HK, Falugi F, et al. Staphylococcus aureus infection induces protein A-mediated immune evasion in humans. J Exp Med. 2014 Nov 17;211(12):2331–2339. PubMed PMID: 25348152; PubMed Central PMCID: PMCPMC4235641.
- Goodyear CS, Silverman GJ. Death by a B cell superantigen: in vivo VH-targeted apoptotic supraclonal B cell deletion by a Staphylococcal Toxin. J Exp Med. 2003 May 5;197(9):1125–1139. PubMed PMID: 12719481; PubMed Central PMCID: PMCPMC2193973.
- Bannoehr J, Ben Zakour NL, Reglinski M, et al. Genomic and surface proteomic analysis of the canine pathogen Staphylococcus pseudintermedius reveals proteins that mediate adherence to the extracellular matrix. Infect Immun. 2011 Aug;79(8):3074–3086. PubMed PMID: 21576333; PubMed Central PMCID: PMCPMC3147560.
- Grandolfo E, Looking through Staphylococcus pseudintermedius infections: could SpA be considered a possible vaccine target? Virulence. 2018 Feb 19;1–10. PubMed PMID: 29457988. DOI:10.1080/21505594.2018.1426964
- Falugi F, Kim HK, Missiakas DM, et al. Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. MBio. 2013 Aug 27;4(5):e00575–13. PubMed PMID: 23982075; PubMed Central PMCID: PMCPMC3760252.
- Gjertsson I, Hultgren OH, Stenson M, et al. Are B lymphocytes of importance in severe Staphylococcus aureus infections? Infect Immun. 2000;68(5):2431–2434.
- Hermos CR, Yoong P, Pier GB. High levels of antibody to Panton-Valentine leukocidin are not associated with resistance to Staphylococcus aureus—associated skin and soft-tissue infection. Clin Infect Dis. 2010;51(10):1138–1146.
- Forsgren A, Nordström K. Protein A from Staphylococcus aureus: the biological significance of its reaction with IgG. Ann N Y Acad Sci. 1974;236:252–266. 19750106 DCOM- 19750106(0077-8923 (Print)). eng.
- Kearse M, Moir R, Wilson A, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012 Jun 15;28(12):1647–1649. PubMed PMID: 22543367; PubMed Central PMCID: PMCPMC3371832.
- Yu NY, Wagner JR, Laird MR, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010 Jul 1;26(13):1608–1615. PubMed PMID: 20472543; PubMed Central PMCID: PMCPMC2887053.
- Kelley LA, Mezulis S, Yates CM, et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845.
- Wass MN, Kelley LA, Sternberg MJE. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010 Jul;38:W469–W473. PubMed PMID: WOS:000284148900076; English.
- Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry. 1981 Apr 28;20(9):2361–2370. PubMed PMID: 7236608.
- Gouda H, Shiraishi M, Takahashi H, et al. NMR study of the interaction between the B domain of staphylococcal protein A and the Fc portion of immunoglobulin G. Biochemistry. 1998 Jan 6;37(1):129–136. PubMed PMID: 9425032.
- Graille M, Stura EA, Corper AL, et al. Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity. Proc Natl Acad Sci U S A. 2000 May 9;97(10):5399–5404. PubMed PMID: 10805799; PubMed Central PMCID: PMCPMC25840.
- Riley MC, Perreten V, Bemis DA, et al. Complete genome sequences of three important methicillin-resistant clinical isolates of Staphylococcus pseudintermedius. Genome Announc. 2016 Oct 20;4(5). PubMed PMID: 27795289; PubMed Central PMCID: PMCPMC5073276. DOI:10.1128/genomeA.01194-16
- Perreten V, Kadlec K, Schwarz S, et al. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and north America: an international multicentre study. J Antimicrob Chemother. 2010 Jun;65(6):1145–1154. PubMed PMID: 20348087.
- Kobayashi A, Hirakawa H, Hirata T, et al. Growth phase-dependent expression of drug exporters in Escherichia coli and its contribution to drug tolerance. J Bacteriol. 2006 Feb 9;188(16):5693–5703. /received 05/16/accepted. PubMed PMID: PMC1540079.
