ABSTRACT
The Gram-negative enteropathogen Yersinia pseudotuberculosis possesses a number of regulatory systems that detect cell envelope damage caused by noxious extracytoplasmic stresses. The CpxA sensor kinase and CpxR response regulator two-component regulatory system is one such pathway. Active Cpx signalling upregulates various factors designed to repair and restore cell envelope integrity. Concomitantly, this pathway also down-regulates key determinants of virulence. In Yersinia, cpxA deletion accumulates high levels of phosphorylated CpxR (CpxR~P). Accumulated CpxR~P directly repressed rovA expression and this limited expression of virulence-associated processes. A second transcriptional regulator, RovM, also negatively regulates rovA expression in response to nutrient stress. Hence, this study aimed to determine if CpxR~P can influence rovA expression through control of RovM levels. We determined that the active CpxR~P isoform bound to the promoter of rovM and directly induced its expression, which naturally associated with a concurrent reduction in rovA expression. Site-directed mutagenesis of the CpxR~P binding sequence in the rovM promoter region desensitised rovM expression to CpxR~P. These data suggest that accumulated CpxR~P inversely manipulates the levels of two global transcriptional regulators, RovA and RovM, and this would be expected to have considerable influence on Yersinia pathophysiology and metabolism.
Introduction
All bacteria contain a cell envelope or cell wall, and its preservation is essential for cell viability. External environmental conditions that threaten bacterial envelope integrity are referred to as extracytoplasmic stresses (ECSs). In order to survive in the presence of ECSs, bacteria must respond rapidly with the activation of quality control systems that have the purpose to maintain an intact bacterial envelope and to ensure the continued delivery of functional proteins throughout the bacterial envelope. Responding to ECSs is performed by a number of sentry regulatory systems partly located in the bacterial envelope, such as the CpxA-CpxR (CpxAR) two-component regulatory pathway and the extracytoplasmic function sigma factor RpoE [Citation1–Citation7].
CpxA is both a sensor kinase and phosphatase to the cognate CpxR response regulator. Upon sensing ECSs, CpxA is first an autokinase and then a phosphoryl donor to CpxR. Based upon available genome-wide transcriptome data, phosphorylated CpxR (CpxR~P) then acts as a transcription factor to activate or repress ~100 gene targets in bacteria [Citation8–Citation11]. Among these are a number of small regulatory RNAs [Citation9], including the newly described regulatory RNA, CpxQ, which works together with the Hfq protein to repress mRNAs of envelope proteins [Citation12,Citation13].
In fact, it is generally accepted that the primary function of the Cpx pathway is to preserve the integrity of cell envelope when bacteria encounter ECSs [Citation14,Citation15]. In this regard, a major target of the Cpx response is believed to be processes that ensure correct biogenesis and function of certain inner membrane (IM) respiratory complexes, with disturbances in these processes serving to activate the Cpx response [Citation16]. The Cpx response counteracts the effects of ECSs by the CpxR~P-dependent production of selected periplasmic protein folding and degradation factors, either directly or through the action of CpxQ [Citation12–Citation15]. An active Cpx response is also responsible for up-regulation of the lipopolysaccharide and phospholipid biosynthesis-transport operons [Citation8,Citation17]. This is telling given how lipopolysaccharide and phospholipid products serve a critical purpose in membrane biosynthesis and barrier function, and are in particular demand by bacteria exposed to noxious stresses.
Intriguingly, it is now also apparent that an intact Cpx signalling pathway is essential for full virulence of diverse clinically- and agriculturally-relevant bacteria most probably because CpxR~P can influence the levels of virulence gene expression [Citation11,Citation18–Citation25]. These findings are being exploited with the purpose of identifying small molecule modulators of the Cpx response that could be used as novel virulence blockers [Citation26]. Given the widespread genetic conservation of the CpxA and CpxR signalling components within the Gram-negative bacterial community [Citation27], these results may mean that a broad spectrum virulence blocker could be identified that targets this two-component regulatory pathway.
In the absence of ECSs, CpxA phosphatase activity dominates over its kinase activity to limit levels of CpxR~P through a feedback inhibitory mechanism [Citation28] that acts in concert with the inhibitory function of the periplasmic regulatory protein, CpxP [Citation14,Citation29,Citation30]. This means that bacterial strains deficient in CpxA phosphatase activity, or a cpxA deletion mutant lacking both phosphatase and kinase activities, are prone to accumulated active phosphorylated CpxR [Citation28, Citation31–Citation35]. Initially, it was not intuitively obvious how CpxR is phosphorylated without CpxA, but now it is clear that this can occur through the indiscriminate action of low molecular weight high-energy phosphodonors such as Acetyl phosphate, which are typically by-products of metabolism [Citation28,Citation36–Citation40].
The Yersinia genus possesses three clinically relevant species – Y. pestis, Y. pseudotuberculosis and Y. enterocolitica. The most dreaded is Y. pestis, the causative agent of plague that can have high mortality rates especially in the absence of prompt pharmacological intervention [Citation41]. On the other hand, enteropathogenic Y. pseudotuberculosis and Y. enterocolitica are both usually associated with mild, self-limiting, and diverse, food-borne infections that are collectively referred to as yersiniosis [Citation42]. Nevertheless, many of the disease manifestations brought about by pathogenic Yersinia are an initial consequence of their decisive contact with host eukaryotic cells. For example, enteropathogenic Yersinia utilise invasin, a dominant surface-located adhesin that plays significant roles in the initial steps of the infection process [Citation43,Citation44]. The characteristic thermal regulation of invasin expression requires the global transcription regulator RovA [Citation45–Citation51].
RovA belongs to SlyA/Hor/Rap family of MarR-type dimeric winged-helix DNA-binding proteins [Citation52]. Its production in the cell is tightly controlled, and this is consistent with its role as a master regulator of several physiological properties of pathogenic Yersinia, including the production of some virulence factors [Citation53,Citation54]. Transcription of rovA occurs from two distinct promoters that are positively and negatively autoregulated in a temperature-dependent manner [Citation47,Citation55]. Depending on the Yersinia background and the prevailing environmental growth conditions, activity from one or both of these promoters is also directly or indirectly negatively influenced by a collection of other DNA binding elements – such as RovM, H-NS, YmoA, PhoP, LeuO, UvrY, cAMP receptor protein (Crp) and the carbon storage regulatory system (CsrABC) [Citation46,Citation47,Citation55–Citation59]. Furthermore, our recent studies in Y. pseudotuberculosis showed that artificially accumulated CpxR~P can suppress rovA expression through direct binding to its promoter [Citation45,Citation60]. In addition, post-translational modification of RovA controls its activity in response to temperature, i.e. structural changes in a RovA homodimer specifically limits target DNA binding [Citation50]. Thermoregulatory control of RovA availability operates as a molecular switch that plays a key role in adapting bacterial populations to the ever-changing conditions encountered during the infectious cycle within a host [Citation61].
A central negative regulator of rovA is the LysR-type transcription regulator, RovM [Citation56]. This means that the amount of accumulated RovA in a bacterial cell is indirectly proportional to the amount of accumulated RovM. Additionally, RovM is a pleiotropic regulator of Y. pseudotuberculosis pathophysiology on the basis that rovM deletion leads to a hyper-virulent bacterial state, contrasting with the effect of RovM over-expression that attenuates bacterial virulence, despite an enhancement of flagella-mediated motility [Citation56]. Expression of RovM is tightly coupled to the prevailing growth environments, with enhanced expression occurring upon growing Y. pseudotuberculosis in nutrient-deprived media, and this effect is mediated by a complex regulatory cascade involving communication between RovM, Crp and the Csr system [Citation57,Citation59,Citation62,Citation63]. Control of rovM expression and the CsrA-Crp-RovM-RovA regulatory cascade has clear implications for the lifestyle choices made by Yersinia spp. in terms of planktonic versus sessile growth and survivability in mildly acidic environments or in the flea gut [Citation64–Citation67].
We have earlier shown that active phosphorylated CpxR suppressed rovA expression through direct binding to its promoter [Citation45,Citation60]. Stemming from that finding, this study explores another possible route of rovA regulation by CpxR~P, which is through direct CpxR~P activation of rovM transcription. Indeed we found that accumulated CpxR~P in Y. pseudotuberculosis can directly bind to the rovM regulatory region to enhance its transcriptional output leading to accumulated RovM levels, which in turn limits RovA accumulation. Hence, the CpxR regulator can influence the CsrA-Crp-RovM-RovA regulatory hierarchy at both rovM and rovA transcription. A consequence of this input is to connect nutritional and ECS sensing pathways to enhance the fine-tuning of virulence gene expression in pathogenic Yersinia.
Results
Elevated RovM levels in a CpxA phosphatase defective mutant
Activation of the ECS responsive Cpx pathway ultimately leads to the phosphorylation of the response regulator, CpxR. Active CpxR~P then positively influences the level of transcription from genes that are involved in envelope biogenesis and negatively regulates genes involved in virulence [Citation4,Citation5,Citation68–Citation70]. In Y. pseudotuberculosis, a full-length ΔcpxA deletion mutant accumulates CpxR~P when grown in LB media [Citation60,Citation71]. This phosphorylation is mediated by small phospho-donors (e.g.: acetyl~P) that are intermediates of metabolic processes in the bacterial cytoplasm [Citation60,Citation71]. We have previously shown that in Y. pseudotuberculosis ∆cpxA, accumulated CpxR~P binds to the promoter regions of rovA leading to a lowered transcriptional output [Citation60]. As rovA expression is also suppressed by the RovM transcriptional regulator [Citation56], we wondered whether accumulated CpxR~P influences the amount of RovM in the cell. Bacteria were grown at 26°C in LB media until late stationary phase. Total cell lysates were generated and analysed by western blot for changes in RovM levels. Lysates derived from ∆cpxA had enhanced levels of RovM and this enhanced RovM disappeared on trans complementing with pcpxA+ in the ∆cpxA mutant ()). On the other hand, RovM production was only just detectable in control bacteria that did not accumulate CpxR~P, such as the parental strain and bacteria lacking cpxR and this ∆cpxR mutant complemented with pcpxR+ ()).
Figure 1. Cpx signalling influences the expression of RovM in Y. pseudotuberculosis. Steady state levels of accumulated RovM was analysed in protein pools sampled from bacteria grown in LB (A) or RPMI (B) media at 26°C until late stationary phase. Protein samples were separated on a 12% acrylamide SDS-PAGE followed by western immunoblot and detection with polyclonal rabbit antiserum raised against RovM. As a protein loading control, samples were also probed with antiserum specific for the cytoplasmic molecular chaperone DnaJ. The indicated immunoblots stem from one independent experiment. The molecular weights shown in the parenthesis are deduced from primary sequence. Strains: parent, YPIII/pIB102; parent/empty vector, YPIII/pIB102, pWKS30; rovM null mutant, YPIII171/pIB102; cpxA null mutant, YPIII07/pIB102; cpxA null mutant/empty vector, YPIII07/pIB102, pWKS30; cpxA null mutant/pcpxA+, YPIII07/pIB102, pJF067; cpxR null mutant, YPIII08/pIB102; cpxR null mutant/empty vector, YPIII08/pIB102, pWKS30; cpxR null mutant/pcpxR+, YPIII08/pIB102, pJF068. ImageJ software was used to quantify from five independent experiments the levels of RovM relative to the levels of DnaJ. Results from this analysis are represented in a scatter plot with each dot indicating data derived from a single independent experiment. The mean value from all independent experiments is indicated by a red line. Differences with a P value of < 0.01 or < 0.001 were considered significantly different from parent and are indicated by a blue-coloured double (**) or triple (***) asterisk situated immediately above the respective data points on the scatter plot.
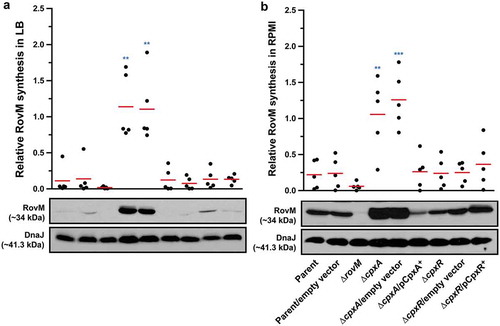
RovM levels are affected by the amount of nutrients available for bacterial growth, with production being most prominent following bacterial growth in RPMI media [Citation56]. In actuality, by our hands RovM production was ~3.5 fold higher in parental bacteria grown in RPMI compared to LB (Supplementary Fig. S1). Hence, we continued to analyse if RovM expression was still influenced by CpxR during growth of bacteria in RPMI media. Lysates for western blotting were prepared from the various strains grown at 26°C in RPMI media until late stationary phase. While all bacteria except the rovM null mutant produced detectable amounts of RovM, once again RovM accumulated to a greater extent in the ∆cpxA null mutant ()). Moreover, complementing ∆cpxA mutant with pcpxA+ reduced RovM levels to below that of the parent strain ()). Interestingly, RovM production was only ~1.3 fold higher in the ∆cpxA null mutant grown in RPMI compared to LB (Supplementary Fig. S1), indicating that maximum RovM levels are reached in this background independent of growth media. Taken together, these data indicate that steady state RovM production by Y. pseudotuberculosis is affected by Cpx signalling independent of the growth medium used to culture the bacteria.
Active CpxR isoform affects positively RovM accumulation
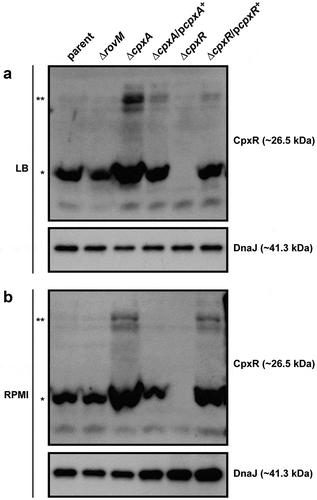
Active CpxR exists as a phosphorylated isoform (CpxR~P), whereas inactive CpxR is considered to be non-phosphorylated. We have successfully used Phos-tag acrylamide technology to distinguish these two isoforms [Citation60,Citation71]. Hence, to observe the impact of active CpxR isoform on levels of accumulated RovM, lysates derived from bacteria grown to late stationary phase at 26°C in either LB or RPMI media were fractionated on a Phos-tag gel, and the CpxR level was detected by western blot. Regardless of growth media, both the parent and ∆rovM null mutant consistently produced similar levels of total CpxR, and with little visible active CpxR~P isoform (). This suggested that RovM per se does not impact on Cpx signalling. On the other hand, the total pools of detectable CpxR were higher in the ∆cpxA null mutant and the ∆cpxR null mutant complemented with pcpxR+ (). Moreover, active CpxR isoform accumulated to higher levels in these two strains. Significantly, complementation of the ΔcpxA null mutant with pcpxA+ lowered the total pools and restored the phosphorylation status of CpxR back to parental levels (). Notably, bacterial strains that favoured accumulation of active CpxR~P () concomitantly favoured RovM production ().
Figure 2. Accumulation of CpxR~P in the cytoplasm of Yersinia. The Phos-tag acrylamide system was used to measure accumulated CpxR~P in vivo. Bacteria were cultured at 26°C until late stationary phase in LB (A) or RPMI (B) media. After harvesting by centrifugation, bacteria were lysed with formic acid and samples immediately fractionated on Phos-tag acrylamide, immunoblotted, and detected with anti-CpxR antiserum. The cytoplasmic molecular chaperone DnaJ served as a loading control. Strains: parent, YPIII/pIB102; rovM null mutant, YPIII171/pIB102; cpxA null mutant, YPIII07/pIB102; cpxA null mutant/pcpxA+, YPIII07/pIB102, pJF067; cpxR null mutant, YPIII08/pIB102; cpxR null mutant/pcpxR+, YPIII08/pIB102, pJF068. The double asterisk (**) reflects the active phosphorylated CpxR isoform accumulated in the Yersinia cytoplasm, while the single asterisk (*) indicates the accumulated inactive non-phosphorylated CpxR isoform.
If CpxR~P does affect positively the amounts of accumulated RovM, then reducing CpxR~P accumulation should lessen this effect. We know that the non-specific low molecular weight phosphodonor acetyl~P derived from the phosphotransacetylase (Pta)–acetate kinase (AckA) pathway can amplify CpxR~P levels in Y. pseudotuberculosis lacking CpxA phosphatase activity [Citation60,Citation71]. Hence, to investigate if this phosphodonor impacts on the CpxR-dependent regulation of RovM, we utilised a full-length ΔackA, pta null mutation in parental Yersinia, as well as in the isogenic mutant harbouring the full-length ΔcpxA null mutation [Citation60]. Significantly, this second-site mutation in the ΔcpxA null mutant totally suppressed the production of RovM following growth in LB broth (Supplementary Fig. S2A), while reducing RovM production following growth in RPMI media (Supplementary Fig. S2B). To confirm that a reduction in RovM production in these strains occurred because of active CpxR~P also diminished, we analysed lysates with Phos-tag acrylamide technology. Indeed, the introduction of a ΔackA, pta mutation into the ΔcpxA null mutant diminished levels of detectable CpxR~P isoform, especially following growth in LB broth (Supplementary Fig. S3A) compared to growth in RPMI media (Supplementary Fig. S3B). The fact that reduction of CpxR~P was far more evident in the ΔcpxA, ΔackA, pta double mutant grown in LB explains why RovM was most efficiently repressed in this mutant when grown in LB (see Supplementary Fig. S2A). The different phenotypes observed following growth in LB versus RPMI, correlates with growth media dependent accumulation of phosphodonor intermediates other than acetyl~P [Citation40], which likely can then contribute to modulating CpxR~P levels.
In the past, we have also worked with the CpxAT253P deficient phosphatase mutant [Citation60,Citation71], which in E. coli is designated as the cpxA101* allele [Citation28,Citation31]. We know that the CpxAT253P deficient phosphatase mutant of Y. pseudotuberculosis accumulates active phosphorylated CpxR, and in the absence of acetyl~P the level of phosphorylated CpxR is reduced [Citation60,Citation71]. Thus, we examined RovM production in the CpxAT253P deficient phosphatase mutant, and in this mutant lacking both ackA and pta. As expected, far greater production of RovM was observed in the CpxAT253P deficient phosphatase mutant strain, and these levels were comparable to the ΔcpxA null mutant (Supplementary Fig. S4). On the other hand, RovM levels in the CpxAT253P deficient phosphatase mutant also lacking ackA and pta barely produced detectable RovM (Supplementary Fig. S4).
As further evidence that CpxR~P accumulation enhances RovM production, we utilised two cpxR mutants with a disruption in the auto-amplification loop controlling expression of the cpxAR operon [Citation29,Citation34,Citation72]. The first mutant produces CpxRD51A with reduced capacity for phosphorylation, while the second mutant produces CpxRM199A with reduced capacity for DNA binding. As anticipated, these two mutants produced lower levels of total CpxR pools compared to the parent irrespective of the growth media used (Supplementary Fig. S3A and Fig. S3B). It follows that these low levels of accumulated CpxR restricted the production RovM (Supplementary Fig. S2A and Fig. S2B).
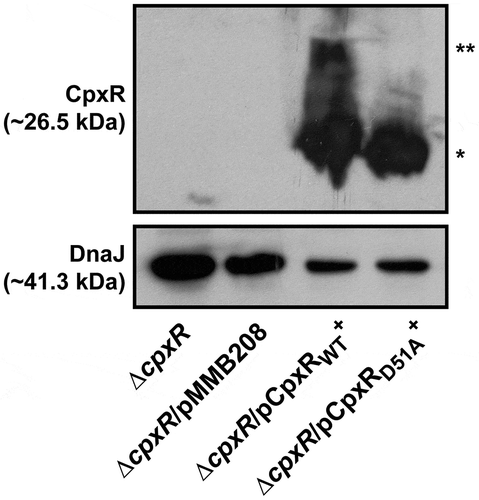
Even so, we appreciate that replacing the wild type cpxR allele in the chromosome with the cpxRD51A or cpxRM199A allele under the control of the cpxR native promoter destroys the auto-amplification loop. The consequence is much lower total pools of CpxRD51A or CpxRM199A in these strains (Supplementary Fig. S3). Indeed, lower total pools of CpxR occur also in the double mutant backgrounds of ∆cpxA, ∆ackA, pta (Supplementary Fig. S3) and cpxAT253P, ∆ackA, pta [Citation60,Citation71]. Since the total CpxR level is lower in these four strains compared to the ∆cpxA strain (Supplementary Fig. S3), it is difficult to say that the lower production of RovM in these strains (Supplementary Fig. S2) is really due to a lower amount of CpxR~P in the bacterial cells. We attempted to address this by analysing RovM production in a previously published background where cpxR or cpxRD51A expression occurs in trans from the plasmid pMMB208 and under the control of a leaky IPTG inducible promoter. In this case, the endogenous cpxR auto-amplification loop will no longer exist, and this should allow a constant production of CpxR or its mutant CpxRD51A. From past experience, we know that IPTG induction of cpxR expression severely affects the growth of Yersinia bacteria [Citation73]. However, levels adequate to complement the ΔcpxR null mutant can be achieved by trans-expression of cpxR from the leaky inducible promoter on pMMB208 without IPTG induction, and this avoids any measurable bacterial growth defect [Citation73]. Hence, to directly see the impact of CpxR’s phosphorylation state on RovM expression in vivo, we analysed the effect of cpxR or cpxRD51A expression from pMMB208 in the ΔcpxR background following bacterial growth in LB media without the addition of IPTG. A significant level (*, p = 0.0396) of RovM accumulation was restored in the ΔcpxR null mutant expressing cpxR in trans, although not to the degree of RovM accumulation achieved in the ΔcpxA null mutant (). On the other hand, RovM accumulation was not observed in the ΔcpxR null mutant expressing in trans cpxRD51A (). Significantly, we observed equivalent levels of total CpxR produced by the ΔcpxR mutant harbouring either cpxR or cpxRD51A on pMMB208, but only phosphorylated form could be detected for the wild type CpxR variant (). Hence, it is the phosphorylation state of CpxR that impacts on accumulated levels of RovM in vivo. All these data are therefore consistent with the idea that accumulated active CpxR~P isoform acts as a transcriptional activator to enhance RovM production.
Figure 3. RovM accumulation in the cytoplasm of Yersinia in the absence of the endogenous cpxR auto-amplification loop. Steady state levels of accumulated RovM was analysed in protein pools sampled from bacteria harbouring plasmid-borne cpxR variants under the control of an IPTG inducible promoter and grown in LB media at 26°C until late stationary phase. Protein samples were separated on a 12% acrylamide SDS-PAGE followed by western immunoblot and detection with polyclonal rabbit antiserum raised against RovM (arrowhead). A lower molecular weight unidentified protein that cross-reacted with the anti-RovM antibodies was used as a convenient protein loading control (arrow). The indicated immunoblots stem from one independent experiment. The molecular weight shown in the parenthesis is deduced from primary sequence. Strains: parent, YPIII/pIB102; parent/pMMB208 (empty vector), YPIII/pIB102, pMMB208; cpxA null mutant, YPIII07/pIB102; cpxR null mutant, YPIII08/pIB102; cpxR null mutant/pMMB208 (empty vector), YPIII08/pIB102, pMMB208; cpxR null mutant/pCpxRWT+, YPIII08/pIB102, pKEC021; cpxR null mutant/pCpxRD51A+, YPIII08/pIB102, pJF015. ImageJ software was used to quantify from three independent experiments the levels of RovM relative to the levels of the lower molecular weight band cross-reacting with anti-RovM antibodies. Results from this analysis are represented in a scatter plot with each dot indicating data derived from a single independent experiment. The mean value from all independent experiments is indicated by a red line. Differences with a P value of < 0.05 or < 0.001 were considered significantly different from parent and are indicated by a blue-coloured double (*) or triple (***) asterisk situated immediately above the respective data points on the scatter plot.
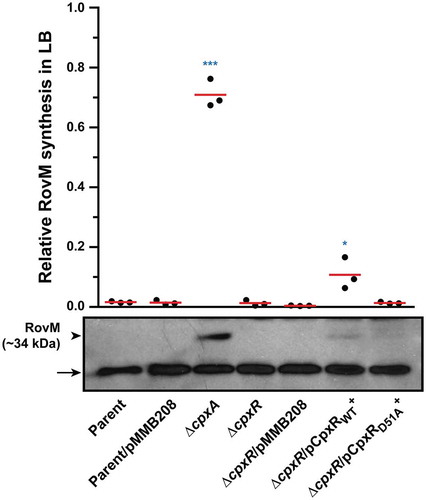
Figure 4. Accumulation of CpxR~P in the cytoplasm of Yersinia in the absence of the endogenous cpxR auto-amplification loop. The Phos-tag acrylamide system was used to measure accumulated CpxR~P in vivo. Bacteria harbouring plasmid-borne cpxR variants under the control of an IPTG inducible promoter were cultured at 26°C until late stationary phase in LB media. After harvesting by centrifugation, bacteria were lysed with formic acid and samples immediately fractionated on Phos-tag acrylamide, immunoblotted, and detected with anti-CpxR antiserum. The cytoplasmic molecular chaperone DnaJ served as a loading control. Strains: cpxR null mutant, YPIII08/pIB102; cpxR null mutant/pMMB208 (empty vector), YPIII08/pIB102, pMMB208; cpxR null mutant/pCpxRWT+, YPIII08/pIB102, pKEC021; cpxR null mutant/pCpxRD51A+, YPIII08/pIB102, pJF015. The double asterisk (**) reflects the active phosphorylated CpxR isoform accumulated in the Yersinia cytoplasm, while the single asterisk (*) indicates the accumulated inactive non-phosphorylated CpxR isoform.
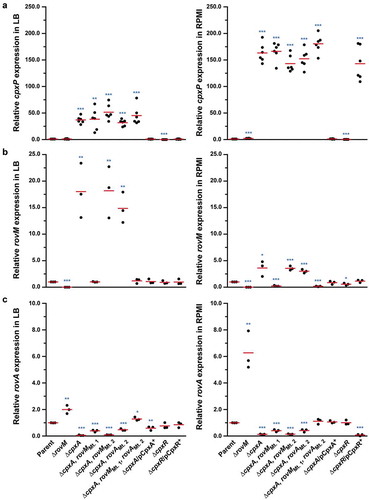
Accumulated CpxR~P induces transcription from the rovM promoter
Increased RovM production was observed in Y. pseudotuberculosis strains that accumulated CpxR~P. As active CpxR~P can act as a transcriptional regulator, a likely scenario was that CpxR~P is a transcriptional activator of rovM transcription. To investigate this, we utilised quantitative RT-PCR on mRNA isolated from strains that accumulated CpxR~P and in control strains in which the cpxR allele was removed. We first examined the amount of cpxP transcript, since the cpxP promoter is known to be highly inducible upon Cpx signalling activation [Citation39]. As anticipated, cpxP expression levels were radically induced in the ΔcpxA mutant that accumulated CpxR~P, with the most pronounced induction relative to parent occurring following growth in RPMI media (~163-fold) compared to growth in LB (~37-fold) ()). By comparison, cpxP expression was reduced to basal levels in the ΔcpxR mutant ()). As these data verified our experimental system, we next focused on examining rovM transcript levels. In the ΔcpxA mutant where active CpxR~P accumulates, a ~ 18-fold and ~4-fold increase in rovM transcript amounts were observed relative to parent bacteria following growth in LB and RPMI, respectively ()). On the other hand, rovM transcript levels relative to parent did not increase in the ΔcpxR mutant devoid of active CpxR~P ()). In parallel, we examined the levels of rovA transcripts given that RovM represses rovA transcription [Citation56,Citation62]. Consistent with this, only minimal amounts of rovA mRNA transcripts were detected in the ΔcpxA mutant where rovM transcription was elevated ()). In contrast, rovA transcripts increased markedly in the ΔrovM mutant ()). Hence, these data indicate that active CpxR~P affects RovM levels primarily by activating transcription from the rovM promoter.
Figure 5. Transcription of rovM and rovA in Y. pseudotuberculosis. For transcriptional analysis of cpxP (A), rovM (B) and rovA (C), quantitative RT-PCR was performed on mRNA isolated from Y. pseudotuberculosis strains cultured at 26°C until late stationary phase in LB and RPMI media. Each sample was normalised against the mean cycle threshold of rpoA for that sample. Results are represented in a scatter plot with each dot indicating data derived from a single independent experiment. The mean value from all independent experiments is indicated by a red line. Differences with a P value of <0.05, < 0.01 or < 0.001 were considered significantly different from parent and are indicated by a blue-coloured single (*), double (**) or triple (***) asterisk situated immediately above the respective data points on the scatter plot. Strains: parent, YPIII/pIB102; rovM null mutant, YPIII171/pIB102; cpxA null mutant, YPIII07/pIB102; cpxA null mutant, rovM(Mt. 1), YPIII177/pIB102; cpxA null mutant, rovM(Mt. 2), YPIII179/pIB102; cpxA null mutant, rovA(Mt. 2), YPIII183/pIB102; cpxA null mutant, rovM(Mt. 1), rovA(Mt. 2), YPIII181/pIB102; cpxA null mutant/pcpxA+, YPIII07/pIB102/pJF067; cpxR null mutant, YPIII08/pIB102; cpxR null mutant/pcpxR+, YPIII08/pIB102/pJF068.
Direct CpxR~P interaction with regulatory sequences within the rovM promoter
As a negative transcriptional regulator, RovM plays a significant role in growth media-dependent fine-tuning of rovA expression [Citation56,Citation57,Citation62], which in turn is needed for the production of colonisation factors such as invasin [Citation45–Citation49]. Therefore, this regulatory cascade is a vital contributor for coordination of the initial phase of Y. pseudotuberculosis infection [Citation49,Citation56]. It follows that knowledge of how accumulated CpxR~P regulates rovM gene expression will benefit our general understanding of how the RovM-RovA-invasin cascade adapts Y. pseudotuberculosis pathophysiology to the prevailing environmental conditions. Our data so far can be explained by a direct interaction of active CpxR~P with regulatory DNA sequences within the rovM promoter. To assay for this, recombinant CpxRHis was purified and in vitro phosphorylated with acetyl~P and then used in a nuclease protection (“foot-printing”) assay of six short PCR amplified DNA fragments (labelled FP-A to FP-F in )) that together spanned the entire rovM promoter region. Two protected regions were identified within the promoter area of rovM. The most prominent protection was observed in the amplified DNA segment FP-D () – left panel) as well as the overlapping segments lying upstream (FP-C) and downstream (FP-E and FP-F) of FP-D () and Supplementary Fig. S5). The second protected region was detected in segment FP-B () – left panel) and the overlapping downstream segment FP-C () and Supplementary Fig. S5). The protected regions mapped between −87 to −117 bp and −318 to −352 bp upstream of the rovM transcriptional start site (indicated by ‘+1ʹ in )).
Figure 6. Mapping the CpxR~P DNA binding site upstream of rovM by nuclease protection (foot-printing) analysis. The promoter region of rovM was divided into six shorter amplified DNA segments; FP-A (−618 to −364), FP-B (−471 to −222), FP-C (−348 to −92), FP-D (−256 to −6), FP-E (−194 to +51) and FP-F (−108 to +146) (shown in ). Numbers in parentheses reflect the amount of nucleotides upstream (-) and downstream (+) of the +1 transcriptional start site. DNase I foot-printing assays were performed to investigate the binding of CpxR~P to a region within these DNA fragments of the rovM promoter. The 32P labelled FP-D (B) and FP-B (C) sense strands of parent and mutated DNA fragments were incubated with CpxR~P at the following final concentrations: 0 nM, lane a and b (indicated by “–”); 100 nM, lane c; 200 nM, lane d; 400 nM, lane e; 600 nM, lane f, 800 nM, lane g. Reactions were resolved by denaturing PAGE and analysed with a Molecular Dynamics PhosphorImager. Labelled pBR322 DNA digested with MspI (New England Biolabs) was used as a size marker (lane a). An estimation of the protected sequence is given on the right hand side of the panels. Based upon the E. coli consensus sequence of 5ʹ-GTAAA(N)4-8GTAAA-3ʹ, a putative CpxR~P consensus binding sites, box 1 and box 2, are highlighted in a grey box with the mutagenized sequence immediately to the right and labelled as Mt. 1 (for CpxR~P binding box 1 mutation) and Mt. 2 (box 2 mutation) respectively.
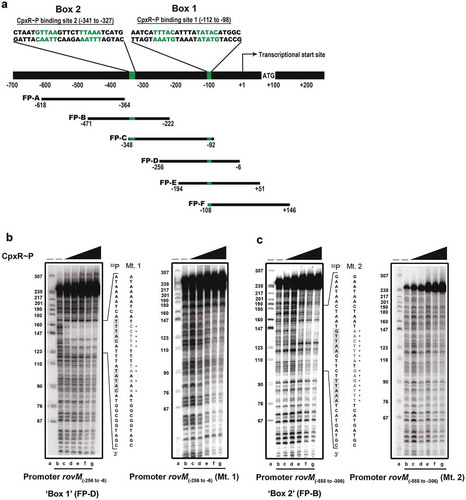
In E. coli, the consensus CpxR~P binding site is considered to be 5´-GTAAA(N)4-8GTAAA-3´ [Citation74,Citation75]. Our earlier work with Y. pseudotuberculosis verified variants of this consensus sequence in the promoter regions of cpxP-cpxR, ppiA and rovA [Citation60]. Here, we manually inspected the two protected regions in the rovM promoter for the presence of a similar 5´-GTAAA(N)4-8GTAAA-3´ sequence. Indeed, two conserved potential CpxR~P binding sites were observed in the rovM promoter region. The one in the first protected region was present on the non-coding strand at position −98 to −112 bp and having the sequence 5´-GTATA(N)5GTAAA-3´ (i.e.: 9 of 10 positions represent consensus) (Box 1; ) – left panel). The second was in the other protected region present on the coding strand at position −327 to −341 bp and having the sequence 5´-GTTAA(N)5TTAAA-3´ (i.e.: 8 of 10 positions represent consensus) (Box 2; ) – left panel). Critically, this subtle difference in consensus is relevant, since a concentration of 100 nM CpxR~P was sufficient to protect the binding region incorporating the residues −98 to −112 bp () – left panel), whereas four times this concentration of CpxR~P (400 nM) was required to protect the binding region encompassing the residues from −327 to −341 () – left panel). Collectively, these data demonstrate that CpxR~P binds to two specific elements in the rovM promoter region.
CpxR~P binding is required for endogenous rovM transcription
We have identified CpxR~P binding sites in the regulatory region of rovM. Next, we sought to verify that CpxR~P binding to the rovM promoter region is biologically relevant i.e.: accumulated CpxR~P activates rovM transcription in vivo via direct engagement with the rovM promoter. To do so, we first used site-directed mutagenesis to shuffle the order of nucleotides in the predicted binding regions of CpxR~P in the rovM promoter. A mutation created in each of Box 1 and Box 2 were respectively defined as Mt. 1 ()) and Mt. 2 ()). Using these mutants in foot-printing assays clearly revealed reduced binding of CpxR~P, since no protected region was observed on the Mt. 1 template () – right panel) and the Mt. 2 template () – right panel). These data confirm that Box 1 and Box 2 in the regulatory region of the rovM promoter are sites for CpxR~P binding.
To address the direct influence of CpxR~P binding box 1 and box 2 on rovM transcription in vivo, the Mt 1 and Mt 2 were introduced in cis by homologous recombination into the ∆cpxA mutant of Y. pseudotuberculosis that is known to accumulate CpxR~P in the cytoplasm. Mutant bacteria were then grown at 26°C until late stationary phase in both LB broth and RPMI media. Total cell lysates were prepared and the level of RovM was analysed by western immunoblot. RovM levels decreased markedly in the ∆cpxA mutant that also contain the promoter mutant Mt. 1 (∆cpxA, rovMMt. 1) following growth in LB broth () – left panel) and RPMI media () – left panel). On the other hand, the ∆cpxA, rovMMt. 2 double mutant retained RovM levels comparable to that produced by the ∆cpxA strain (). The most likely explanation for this is that the ∆cpxA, rovMMt. 2 strain still contains an intact higher affinity CpxR~P binding site in the rovM promoter region (i.e.: box 1) (see ). These data corroborated quantitative RT-PCR analysis that identified a decrease in expression of rovM in the ∆cpxA, rovMMt. 1 strain, but not in the ∆cpxA, rovMMt. 2 strain ()). Hence, taken together this data indicates that the regulatory region designated Box 1 that lies between −98 to −112 bp upstream of the rovM transcriptional site (see ) is primarily responsible for CpxR~P dependent de novo expression of RovM. A second regulatory region designated Box 2 and lying at position −327 to −341 bp upstream of the rovM transcriptional site (see )) contributes a lesser role in CpxR~P-dependent control of endogenous rovM expression.
Figure 7. Mutation of the box 1 binding site effects rovM promoter responsiveness to active CpxR~P. Protein levels were analysed from lysed bacterial samples previously grown in LB (A) or RPMI (B) media at 26°C until late stationary phase. Protein samples were separated on a 12% (RovM) or 15% (RovA) acrylamide SDS-PAGE and specific proteins were identified using a western immunoblot and detection analysis with polyclonal rabbit antiserum raised against RovM and RovA. DnaJ served as a protein loading control. The indicated immunoblots stem from one independent experiment. The molecular weights shown in the parenthesis are deduced from primary sequence. Strains: parent, YPIII/pIB102; rovM null mutant, YPIII171/pIB102; cpxA null mutant, YPIII07/pIB102; cpxA null mutant, rovM(Mt. 1), YPIII177/pIB102; cpxA null mutant, rovM(Mt. 2), YPIII179/pIB102. ImageJ software was used to quantify from at least four independent experiments the levels of RovM and RovA relative to the levels of DnaJ. Results from this analysis are represented in a scatter plot with each dot indicating data derived from a single independent experiment. The mean value from all independent experiments is indicated by a red line. Differences with a P value of <0.05, < 0.01 or < 0.001 were considered significantly different from parent and are indicated by a blue-coloured single (*), double (**) or triple (***) asterisk situated immediately above the respective data points on the scatter plot.
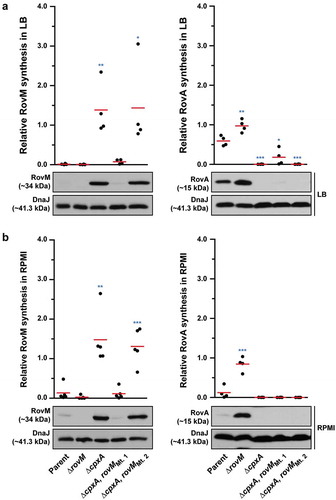
We also examined to what extent uncoupling rovM expression from the influence of CpxR~P would have on accumulated levels of RovA. Minor amounts of RovA could be visualised, but only when RovM was no longer accumulated in the ∆cpxA, rovMMt. 1 strain during growth in LB () – right panel). These RovA levels were still well below the levels seen in the parental strain () – right panel), corroborating low rovA transcription levels detectable in the ∆cpxA, rovMMt. 1 strain, which were higher than in the ∆cpxA mutant but lower than the parental strain ()). The low recovery of RovA in the ∆cpxA, rovMMt. 1 strain, despite the absence of elevated RovM, can be easily explained by the direct repression of rovA expression mediated by active CpxR~P [Citation60]. On the other hand, RovA was not detectable in the ∆cpxA, rovMMt. 2 strain irrespective of growth media () – right panels), and this also reflected in a very low level of rovA transcription that was the equivalent to the levels observed in the full length ∆cpxA mutant ()). In this case, the inability to detect RovA in either condition is simply due to the fact that levels of RovM still remain high in this strain.
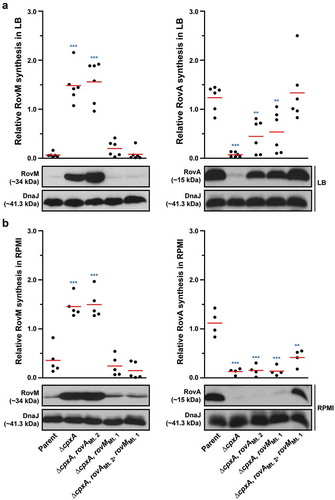
RovM and CpxR~P both contribute towards transcriptional silencing of rovA
In Y. pseudotuberculosis, rovA transcription is repressed by both RovM and CpxR~P. The rovA regulatory region contains two promoters termed P1 and P2, whereby RovM engages near the P1 promoter [Citation56] and CpxR~P near the P2 promoter [Citation60]. In light of this, we wondered if RovM and CpxR~P act independently in mediating repression of rovA transcription. We approached this by looking at steady-state RovA levels, analysed in total lysates derived from bacteria that had been grown in both LB and RPMI media at 26°C until late stationary phase. A full length rovM deletion (∆rovM) was introduced into a strain that either accumulates active CpxR~P (i.e.: isogenic ∆cpxA null mutant) or lacks an ability to produce CpxR (i.e.: isogenic ∆cpxR null mutant). Irrespective of whether cpxR was present or absent, we observed a decrease in detectable levels of steady state RovA when bacteria were grown in RPMI compared to LB media (compare ) with 8A – right panels), and this reflected a general elevation in RovM pools when bacteria were grown in RPMI compared to LB media (compare ) with 8A – left panels). Moreover, the ∆cpxA, rovM double mutant that still accumulated active CpxR~P maintained RovA production at high levels comparable to the ∆rovM mutant () with 8B – right panels). Additionally, RovA production was fully restored in the ∆cpxR null mutant that also lacks the rovM gene () with 8B – right panels). These results indicate that RovM has a greater capacity to mediate repression of rovA compared to the magnitude to which CpxR~P can mediate repression of rovA.
Figure 8. Contribution of RovM and CpxR~P to the steady state accumulation of RovA. Protein levels were analysed from lysed bacterial samples previously grown in LB (A) or RPMI (B) media at 26°C until late stationary phase. Protein samples were separated on a 12% (RovM) or 15% (RovA) acrylamide SDS-PAGE and specific proteins identified using western immunoblot and detection analysis with polyclonal rabbit antiserum raised against RovM (left panels) and RovA (right panels). DnaJ served as a protein loading control. The indicated immunoblots stem from one independent experiment. The molecular weights shown in the parenthesis are deduced from primary sequence. Strains: parent, YPIII/pIB102; rovM null mutant, YPIII171/pIB102; cpxA null mutant, YPIII07/pIB102; cpxA null mutant, rovM null mutant, YPIII165/pIB102; cpxR null mutant, YPIII08/pIB102; cpxR null mutant, rovM null mutant, YPIII173/pIB102. ImageJ software was used to quantify from three independent experiments the levels of RovM and RovA relative to the levels of DnaJ. Results from this analysis are represented in a scatter plot with each dot indicating data derived from a single independent experiment. The mean value from all independent experiments is indicated by a red line. Differences with a P value of <0.05, < 0.01 or < 0.001 were considered significantly different from parent and are indicated by a blue-coloured single (*), double (**) or triple (***) asterisk situated immediately above the respective data points on the scatter plot.
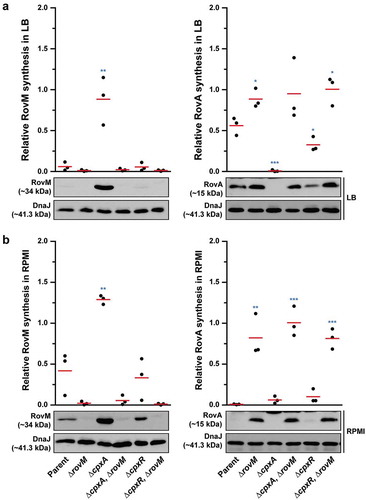
Nevertheless, data in shows that RovA levels were still restricted in the ∆cpxA, rovMMt. 1 mutant even though RovM production was also low, which suggests a situation exists when CpxR~P mediated repression of rovA has a regulatory role. To demonstrate this in another way, into both the single ∆cpxA mutant, and the double ∆cpxA, rovMMt. 1 mutant (where RovM production is blind to elevated CpxR~P), we introduced an in cis mutation within the rovA promoter that rendered RovA production blind to elevated CpxR~P (mutation is termed rovAMt. 2 from our earlier work [Citation60]). Compared to the single ∆cpxA mutant, the ∆cpxA, rovAMt. 2 double mutant grown in LB broth restored a portion of RovA production corroborating earlier findings [Citation60], () – right panel), and this reflected the extent of CpxR~P-dependent inhibition of rovA expression given that it occurs despite the abundance of RovM produced by this strain () – left panel). The accumulative effect of RovM and CpxR~P repression on RovA levels can be observed in the ∆cpxA, rovAMt 2,rovMMt. 1 triple mutant, which encodes variants of the rovM and rovA promoters that are no longer recognised by the abundantly produced CpxR~P in this strain. Here, RovA levels are fully restored to levels observed in parental bacteria () – right panel). The difference between RovA amounts in the ∆cpxA, rovAMt 2,rovMMt. 1 triple mutant versus ∆cpxA, rovAMt. 2 double mutant reflects the extent to which RovM has influence over RovA production. On the other hand, a portion of RovA production was recovered in the ∆cpxA, rovMMt. 1 double mutant () – right panel). Given that this strain produces very little RovM (because the mutated rovM promoter does not respond to accumulated CpxR~P) () – left panel), the difference between RovA amounts in the parent versus the ∆cpxA, rovMMt. 1 double mutant reflects the extent to which CpxR~P can influence RovA production. Interestingly, RovA production is low in the two double mutants when bacteria are grown in RPMI media () – right panel), which correlates to the greater amounts of RovM () – left panel) and active CpxR~P (see Supplementary Fig. S3) that accumulates in these growth conditions. Most likely the amounts of these two repressors combine to keep RovA production low. The one exception is the ∆cpxA, rovAMt 2,rovMMt. 1 triple mutant that produces elevated RovA levels albeit not to the same degree as produced by parent bacteria () – right panel). In this case, partial recovery of RovA production must be due entirely to the rovA promoter being blind to the action of active CpxR~P, given that similar amounts of RovM are produced in this triple mutant compared to the ∆cpxA, rovMMt. 1 double mutant () – left panel) where RovA production is greatly restricted () – right panel). As anticipated, these effects occur at the level of transcription since with quantitative RT-PCR we could demonstrate partial restoration of rovA expression in ∆cpxA, rovAMt. 2 and ∆cpxA, rovMMt. 1 double mutants and full restoration of rovA expression in ∆cpxA, rovAMt. 2, rovMMt. 1 triple mutant when compared to parent bacteria ()). Hence, these data demonstrate a direct involvement of both RovM and CpxR~P in controlling negative rovA expression, although the relative contributions can be influenced by the prevailing growth conditions.
Figure 9. RovM and CpxR cooperate to establish negative control on rovA transcription. A mutation of the box 1 CpxR~P-binding site in the rovM promoter (rovM(Mt. 1)) was combined with a mutation in the CpxR~P-binding box of the rovA promoter (rovA(Mt. 2)). To analyse for RovM and RovA production, protein pools were isolated from bacteria grown in LB (A) or RPMI (B) media at 26°C until late stationary phase. Protein samples were separated on a 12% (RovM) or 15% (RovA) acrylamide SDS-PAGE and specific proteins identified using western immunoblot and detection analysis with polyclonal rabbit antiserum raised against RovM (left panels) and RovA (right panels). DnaJ served as a protein loading control. The indicated immunoblots stem from one independent experiment. The molecular weights shown in the parenthesis are deduced from primary sequence. Strains: parent, YPIII/pIB102; cpxA null mutant, YPIII07/pIB102; cpxA null mutant, rovA(Mt. 2), YPIII183/pIB102; cpxA null mutant, rovM(Mt. 1), YPIII177/pIB102; cpxA null mutant, rovA(Mt. 2), rovM(Mt. 1), YPIII181/pIB102. ImageJ software was used to quantify from at least four independent experiments the levels of RovM and RovA relative to the levels of DnaJ. Results from this analysis are represented in a scatter plot with each dot indicating data derived from a single independent experiment. The mean value from all independent experiments is indicated by a red line. Differences with a P value of < 0.01 or < 0.001 were considered significantly different from parent and are indicated by a blue-coloured double (**) or triple (***) asterisk situated immediately above the respective data points on the scatter plot.
Discussion
Our data contributes to ongoing progress in defining determinants of rovM regulation and demonstrates that transcription is induced by a positive auto-activation loop Citation56] and by CpxR~P (this study). In response to nutrient limitation, an auto-activation loop is engaged whereby RovM activates its own transcription albeit via an indirect mechanism that involves as yet unknown regulators () [Citation56]. To this end, CRP-cAMP and the carbon storage regulatory (Csr) system cooperate to repress rovM transcription in “feast” conditions and enable transcription in “famine” conditions, although the molecular mechanism for this control at the rovM promoter remains obscure [Citation57,Citation62]. In response to nutrient levels, Crp activates csrC transcription and represses csrB transcription [Citation57]. This is further refined through a response of the PhoQ-PhoP two-component system to certain diverse stimuli that induces transcription of the CsrC regulatory RNA molecule [Citation59]. In parallel, active BarA-UvrY two-component system signalling enhances levels of the CsrB regulatory RNA molecule [Citation62]. Together, this has direct implications for the sequestration and inactivation of CsrA, and this is considered to diminish rovM transcription. Our findings add to this complexity by defining the regulatory role played by active Cpx signalling (). The precise physiological cues that activate Cpx signalling to promote rovM transcription are not yet defined. However, these cues are expected to negatively influence the maintenance of the bacterial envelope, including the ability to ensure correct protein folding in the periplasm. Thus, Y. pseudotuberculosis dedicates significant resources to incorporate two seemingly very diverse cues – internal nutrient limitation and external noxious stresses – for the purpose of coordinating the RovM-RovA regulatory cascade; a clear indication that this pathway is central to controlling multiple aspects of Yersinia pathophysiology. We propose that signal integration stemming from these diverse cues is somehow coordinated through a coupling of Cpx signalling to the nutritional biosensor pathway encoded by csrA and crp ().
Figure 10. A scheme depicting CpxR~P influence on the CsrA-Crp-RovM-RovA regulatory cascade. The RovM-RovA regulatory pathway leading to the controlled production of several virulence determinants – including the Y. pseudotuberculosis adhesin invasin – is strictly controlled by cascade regulation at both the transcriptional and post-transcriptional levels in response to multiple environmental cues. The strongest influence on RovA production is through two opposing pathways. The first is an auto-amplification loop, which in turn is controlled by thermo-regulated proteolysis via the action of ClpXP and Lon proteases. The second is repression via RovM acting in concert with H-NS. RovM levels are principally controlled by an uncharacterised auto-activation loop and the prominent Csr and Crp pathways responsive to carbon and glucose availability. Additionally, we now show that the CpxR~P pathway responsive to extracytoplasmic (periplasmic) noxious stresses that is another critical player in this cascade regulation. Active CpxR~P isoform directly induces production of the RovM repressor, and also directly represses production of both RovA and invasin. By an uncharacterised mechanism active CpxR~P also negatively influences Csr output. Thus the net effect of CpxR~P is a significant downturn in RovA and invasin production, as well as other prominent virulence factors such as the Ysc-Yop T3SS (not shown here). In this context, it is interesting that the FliA sigma factor plays a role in the coordination of inverse regulation of invasin and the Ysc-Yop system (not shown). On this basis, a prediction would be that the regulatory events of CpxR~P and FliA are somehow connected. In the diagram, direct (filled line) or indirect (dotted line) induction events are indicated by a green arrow, while direct (filled line) or indirect (dotted line) repression events are indicated by a blunted red line.
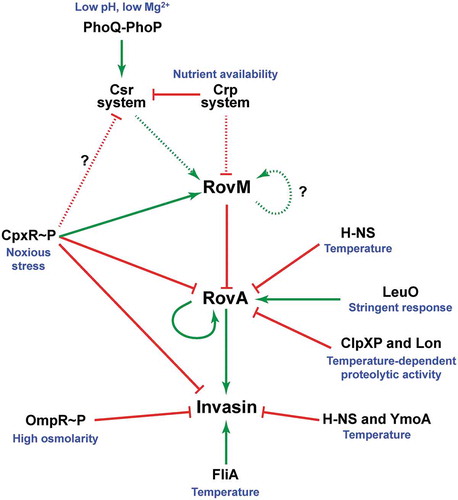
Interestingly, the Crp-Csr-RovM-RovA regulatory cascade of Y. pseudotuberculosis influences a gross lifestyle switch from acute infection to chronic persistent infection in an in vivo mouse model [Citation76]. Presumably, a drive towards persistence enables the bacteria to establish a protected haven within a host from where it can survive long-term despite an ongoing robust immune response. This may even provide a bacterial reservoir from where a new round of acute-phase infection and/or transmission to a new host can be initiated. Evident during this phenotypic transition is a clear downturn in expression of prominent virulence factors such as the Ysc-Yop type III secretion system (T3SS) and an upturn in stress survival factors [Citation76]. Pointedly, close parallels exist between this in vivo phenomenon and the in vitro phenotypic effects observed when Y. pseudotuberculosis accumulate active CpxR~P. We have shown that accumulated CpxR~P forces in vitro grown bacteria to undergo a pronounced phenotypic switch that represses many prominent virulence factors, such as various adhesins including invasin and also the Ysc-Yop T3SS, at that same time that key periplasmic protein folding and degradation factors are induced, such as DegP, DsbA and PpiA [Citation45,Citation60,Citation71,Citation73]. Determination of the environmental conditions that drive this dominant Cpx signalling event promises to benefit greatly our understanding of Y. pseudotuberculosis survival in diverse environments both inside and outside of a host.
Notably, Yersinia virulence determinants such as invasin and the Ysc-Yop T3SS are inversely thermo-regulated to a large degree by the FliA sigma factor () [Citation77]. Presumably, this permits Y. pseudotuberculosis to initiate internalisation by the gut epithelium and then to survive induced pro-inflammatory immune responses. Hence, to understand the overarching molecular mechanisms of the Cpx signalling life style switch, it would be prudent to explore the existence of a connection between Cpx and FliA regulatory outputs. This information would help to shed light on some of the phenomena concerning cross-regulation between virulence determinants in response to different stages of Y. pseudotuberculosis transit within an infected host.
CpxR is a member of the large class of OmpR/PhoB response regulators [Citation78,Citation79]. Members of this class have been the subject of extensive experimentation with the purpose to uncover their mechanisms of activation and promoter binding. For the most part, activation by phosphorylation (e.g.: by the cognate sensor kinase) prompts homodimerisation and subsequent DNA binding at the target promoter [Citation80]. Deviations of this theme exist however, since it is possible for DNA binding to stimulate the phosphorylation and subsequent homodimerisation of two monomers or for phosphorylation to promote DNA binding prior to homodimerisation [Citation80]. With active CpxR~P capable of both activating and repressing gene transcription, a goal for the future is to determine the mechanism(s) of CpxR activation and promoter binding. In this study we observed that the choice of bacterial growth medium impacted on the overall amount of CpxR production as well as the ratio between active (phosphorylated) and inactive (non-phosphorylated) isomers. This is significant as it raises the possibility that high levels of the inactive CpxR isoform could compete with active CpxR~P isoform in homodimerisation and/or DNA binding with ramifications for promoter output. Our preliminary mobility shift analysis suggested that non-phosphorylated CpxR could not bind DNA under the conditions used [Citation60]. However, more sensitive competition assays are required to analyse whether non-phosphorylated CpxR alters the manner or efficiency in which active CpxR~P homodimerises and/or binds to target DNA.
Our nuclease protection assay further showed that CpxR~P can bind at two distinct regions in rovM promoter region, which we termed CpxR~P binding box 1 and box 2. Our data indicates that box 1 is a higher affinity target site for CpxR~P, and this binding is biologically significant with respect to rovM transcriptional output. This might be related to its positioning in the reverse orientation to transcription, although in E. coli there is no obvious correlation between binding site orientation and promoter responsiveness to CpxR~P [Citation17]. Actually, more important might be the positioning of box 1 around ~100 nucleotides upstream of the transcriptional start, since this is predicative of a promoter strongly responsive to active CpxR~P [Citation17]. We are yet to define if box 2 is biologically relevant for rovM transcriptional control. However, there is a precedent for promoters with dual binding sites for transcription factors. Specifically, within the E. coli CpxR regulon exists a number of promoters that engage CpxR at more than one site [Citation17,Citation81]. Thus, the fact that we identified two binding sites with disparate affinity for CpxR~P indicates that several CpxR molecules may bind cooperatively to regulatory regions within the rovM promoter.
Finally, transcription of rovA encoding for a major transcription factor in Yersinia is repressed by dual mechanisms; the first involving cooperative binding between H-NS and RovM [Citation47,Citation56], and the second involving active CpxR~P () [Citation45,Citation60]. From in vitro studies it is evident that both RovM [Citation62] and CpxR~P (this study) can bind to rovA regulatory regions independent of each other. Moreover, the relative effects of this binding on RovA steady state levels is further influenced by the prevailing growth conditions. The rovA regulatory region possesses two active promoters designated ‘P1ʹ and ‘P2ʹ [Citation47]. It is reasonable to assume that CpxR~P inhibits the activity of ‘P2ʹ, for its binding site overlaps with the −35 region [Citation60]. In fact, a mutation specifically disrupting this −35 region did prevent rovA transcription under the conditions tested [Citation60], so it’s inhibition by CpxR~P does serve to influence in vivo levels of RovA. Crucially, the CpxR~P binding site dissects that of H-NS and RovM; a regulatory complex thought to alter DNA curvature of the rovA regulatory sequences [Citation62]. It is possible therefore that under growth conditions exposing bacteria to ECSs, in vivo CpxR~P binding specifically fine-tunes rovA expression from the P2 promoter. However, this could still require cooperative binding of the H-NS/RovM regulatory complex to the rovM promoter. Perhaps this binding facilitates a change in DNA topology that maximises CpxR access to the DNA target, and this might be a prerequisite for stimulating activation and/or homodimerisation of this response regulator.
In summary, this study expands the role of Cpx signalling in regulation of the RovA global regulator in Y. pseudotuberculosis. New-found roles focus on the activation of RovM production, which is a known repressor of rovA transcription. The molecular mechanism for RovM induction via Cpx signalling involved direct engagement of active CpxR~P at the rovM promoter that presumably serves to recruit RNA polymerase to increase rovM transcription. Hence, Cpx signalling is another component of an expanding regulatory cascade that has potential to link nutritional status to the control of virulence gene expression in the enteropathogen Y. pseudotuberculosis.
Materials and methods
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids used in this study can be viewed in Supplementary Table S1. Y. pseudotuberculosis YPIII/pIB102 (serotype III) is designated as the parental strain, where the plasmid pIB102 encodes for the Ysc-Yop T3SS. Significantly, this plasmid is marked by a kanamycin resistance cartridge inserted into the yadA gene that does not attenuate virulence in mouse models [Citation82]. This strain is also defined by a defective phoP allele that limits survival inside phagocytic immune cells [Citation83]. The cpxA (pJF067) and cpxR (pJF068) expression plasmids in pWKS30 used for mutant complementation are based on synthetic genes consigned by GenScript USA Inc. (Piscataway, New Jersey, USA). Unless otherwise mentioned, bacteria were normally cultivated in Luria-Bertani (LB) agar or broth at either 26°C (Y. pseudotuberculosis) or 37°C (E. coli) with aeration. For comparisons, Y. pseudotuberculosis were also grown in Roswell Park Memorial Institute (RPMI) 1640 medium (ThermoFisher Scientific; Cat no. 51,800,035). This medium was initially prepared as a 2x stock, which was further supplemented with 0.2% (w/v) Glucose, 0.2% (w/v) Casamino acids and 1mM MgSO4, prior to being diluted to 1x medium with sterilised non-distilled water. Importantly, phenotypic analyses were routinely conducted on bacteria grown to late stationary phase of growth. This choice was motivated by several telling reasons. First, late stationary phase allows for the cpxA deletion to accumulate phosphorylated CpxR [Citation11]. Second, transcriptome analyses of CpxR-regulated genes have consistently shown maximal activation of the Cpx pathway in bacteria grown to late stationary phase [Citation11,Citation34,Citation84]. Third, maximal rovM expression in Yersinia occurs at late stationary phase when nutrients needed for growth have been exhausted [Citation56,Citation62,Citation64,Citation65]. Where required, antibiotics were added at the final concentrations of carbenicillin (Cb; 100 μg/ml), kanamycin (Km; 50 μg/ml), Trimethoprim (Tp; 10 μg/ml) and chloramphenicol (Cm; 25 μg/ml).
Mutant construction
To construct site-directed and deletion mutants, we applied the overlap PCR technique [Citation85] using the relevant primer combinations listed in Supplementary Table S2. The primers were synthesised by Sigma-Aldrich Sweden AB (Stockholm, Sweden). The amplified fragments were cloned into the sequencing vector pTZ57R using the InsTAclone PCR cloning kit (Thermo Scientific) and mutations were confirmed by sequence analysis (Eurofins MWG Operon, Ebersberg, Germany). Confirmed mutated fragments were cloned into the suicide plasmid, pDM4, following XbaI-XhoI restriction enzyme digestion. Plasmid DNA was maintained in E. coli SY327λpir, while S17-1λpir was the donor strain of choice for conjugal mating with Yersinia recipients. Mutated alleles were introduced into the Y. pseudotuberculosis genome by a double cross-over homologous recombination event and the desired genotype was recovered by sacB-dependent sucrose sensitivity [Citation86]. The presence of desired mutations in the genome of Y. pseudotuberculosis was verified by PCR and sequence analysis of the amplified regions flanking the mutation.
RNA isolation and real-time quantitative reverse transcription-PCR
Y. pseudotuberculosis variants were grown to late stationary phase at 26°C with aeration in either LB or RPMI medium. RNAprotect bacteria reagent (QIAGEN) was added immediately to the bacterial culture to stabilise RNA transcripts. Total RNA was then isolated by the Nucleospin RNA II method (Macherey Nagel) followed by on column DNase treatment. To remove contaminating DNA from each sample, the TurboDNAfree kit (Ambion) was used according to the manufacturer’s directions. Total RNA (1 µg) was reverse transcribed to synthesise cDNA using RevertAid Reverse Transcriptase (ThermoFisher Scientific). The qRT-PCR was performed in an iCycler iQ5 real-time PCR detection system (Bio-Rad) using KAPA SYBR FAST Bio-Rad iCycler qPCR kit (KAPA Biosystems). Internal primer combinations specific for rovM, rovA, cpxP, csrA, crp and rpoA were used in qRT-PCR (Supplementary Table S2). The 20 µl total reaction mixture consists of cDNA that is less than 20 ng, 10 µl of KAPA SYBR FAST qPCR master mix and 200 nM of forward and reverse primers. At least four independent samples were tested in duplicate.
Protein production and western blot analysis
Y. pseudotuberculosis strains were grown until late stationary phase at 26°C with aeration in LB broth or RPMI media. Bacterial suspensions were lysed directly with 4× loading buffer (250 mM Tris-HCl pH 6.8, 8% SDS, 40% glycerol, 4% β-Mercoptoethanol, and 0.08% Bromophenol Blue) and heat denatured at 95°C for 10 min. Total protein was fractionated by SDS-PAGE with 12% acrylamide (for RovM and DnaJ) and 15% acrylamide (for RovA), and then subjected to western immunoblotting most often using a Trans Blot® semi-dry transfer system (BioRad) to transfer proteins onto Polyvinylidene difluoride (PDVF) membranes. Specific proteins of interest were bound with specific rabbit polyclonal antibodies that were then detected with an anti-rabbit monoclonal antibody conjugated with horse radish peroxidase (GE Healthcare) and a homemade chemiluminescent solution.
Relative protein levels were quantified from protein bands on scanned western blot X-ray films using the gel analysis tool ImageJ [Citation87]. In every case, the lane profile plot area of each protein band of interest was normalised to the corresponding protein band appearing in the same lane in the loading control blot.
Visualisation of in vivo accumulated CpxR~P
Our approach for in vivo visualisation of phosphorylated and non-phosphorylated isoforms of CpxR produced by various Y. pseudotuberculosis utilised Phos-tag™ Acrylamide AAL-107 essentially according to the manufacturer’s directions (Wako Nard Institute, Japan). Equal amounts of harvested Y. pseudotuberculosis strains grown to late stationary phase at 26°C with aeration in either LB or RPMI media were mixed vigorously with 33 μl of 1.2 M Formic acid. These suspensions were subsequently mixed with 13 μl of 1x SDS-PAGE Loading buffer (250 mM Tris-HCl; pH 6.8, 8% SDS, 40% Glycerol, 4% β-mercaptoethanol and 0.08% Bromophenol blue), and following a brief incubation on ice, a volume of 6 μl of 5M NaOH was added to neutralise the pH to ~7.0. Following heat denaturation at 95°C for 5 min and clarification by brief centrifugation, volumes of 5 μl cleared supernatants were immediately fractionated on a freshly prepared Phos-tagTM gel at constant current (30 mA; ~70 volts) for 2.5 h at room temperature. Specifically, the running gel was prepared with 2 ml 30% (w/v) Acrylamide Solution, 1.875 ml 1 M Tris-HCl, pH 8.8, 50 μl 5.0 mM Phos-TagTM AAL Solution, 50 μl 10 mM Manganese(II) chloride, 50 μl 10% (w/v) SDS, 5 μl Tetramethylethylenediamine (TEMED), 925 μl Milli-Q® H2O, 20 μl 10% Ammonium persulfate, while the stacking gel was prepared with 375 μl 30% Acrylamide Solution, 312.5 μl 1 M Tris-HCl, pH 6.8, 25 μl 10% (w/v) SDS, 2.5 μl TEMED, 1.772 ml Milli-Q® H2O, 12.5 μl 10% Ammonium persulfate. In preparation for blotting, the gel was washed for 10 min with 30 ml of 1x Transfer buffer containing 1 mM EDTA, and then for 20 min with 1x transfer buffer (without EDTA). Following wet electrotransfer onto PVDF membrane (50 Volts for 2 h at 4°C), the two CpxR isoforms were bound with rabbit polyclonal anti-CpxR antibody, followed by anti-rabbit-HRP, and then detected with PierceTM ECL Plus Western blotting system according to the manufacturer’s instructions.
DNA foot-printing
The DNA sequences of all primers used in this foot-printing analysis are detailed in Supplementary Table S2. The primers pFP-A-rovMFw, pFP-B-rovMFw, pFP-C-rovMFw, pFP-D-rovMFw, pFP-E-rovMFw and pFP-F-rovMFw were all radioactively labelled with 32P using γ32P-ATP (Perkin Elmer) by T4 polynucleotide kinase (Thermo Scientific). The 32P-labelled primers were paired with unlabelled pFP-A-rovMRev, pFP-B-rovMRev, pFP-C-rovMRev, pFP-D-rovMRev, pFP-E-rovMRev and pFP-F-rovMRev, respectively, and used to PCR amplify overlapping segments of the rovM promoter (see text for details). As a control the labelled pcpxRfor was paired with unlabelled pcpxRrev for the PCR amplification of an internal region of cpxR. Reaction mixtures contained within a volume of 40 µl consisted of 1.5 nM of individual amplified DNA fragments and 0, 100, 200, 400, 600 and 800 nM of CpxRwt::His6 ~ P (phosphorylated in vitro by acetyl~P) along with 25 mM Hepes (pH 8), 100 mM potassium glutamate, and 0.5 mg/ml BSA. To analyse samples by electrophoresis, we followed previously described methods [Citation60].
Statistical analysis
Mean ± standard deviation were calculated for at least three biological replicates. Significance from the parental control was determined using the nonparametric, unpaired, two-tailed student t test. Analysis was performed using GraphPad Prism, version 5.00, for Windows (GraphPad Software, Inc. La Jolla, CA, USA). Differences with a P value of <0.05 were considered significant.
Author Contributions
EJT, DKG and MSF designed the study and wrote the manuscript including the supplementary material; cloning and mutant construction were performed by EJT, DKG, JL and IRO; contributions to immunoblotting or RT-qPCR was achieved by EJT, DKG, JMG, MFS, KR; critical reagents or technical and conceptual support was provided by MKF, KMT, SC, and PD. All authors discussed the results and implications and commented on the manuscript at all stages, including the supplementary material.
Supplemental Material
Download MS Word (25.7 MB)Acknowledgments
This work was performed within the virtual framework of the Umeå Centre for Microbial Research-Linnaeus Program (UCMR-LP). We express gratitude to Emeritus Prof. Hans Wolf-Watz (Department of Molecular Biology, Umeå University) for gifting antiserum to DnaJ, and thank international masters in Molecular Biology student Abebe Aseffa Negeri for his contributions in making mutant strains.
Disclosure statement
No potential conflicts of interest were disclosed.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Raffa RG, Raivio TL. A third envelope stress signal transduction pathway in Escherichia coli. Mol Microbiol. 2002;45:1599–1611.
- Clarke DJ. The Rcs phosphorelay: more than just a two-component pathway. Future Microbiol. 2010;5:1173–1184.
- Darwin AJ. Stress relief during host infection: the phage shock protein response supports bacterial virulence in various ways. PLoS Pathog. 2013;9:e1003388.
- Merdanovic M, Clausen T, Kaiser M, et al. Protein quality control in the bacterial periplasm. Annu Rev Microbiol. 2011;65:149–168.
- Rowley G, Spector M, Kormanec J, et al. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol. 2006;4:383–394.
- Raivio TL. Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim Biophys Acta. 2014;1843:1529–1541.
- Hunke S, Keller R, Muller VS. Signal integration by the Cpx-envelope stress system. FEMS Microbiol Lett. 2012;326:12–22.
- Bury-Mone S, Nomane Y, Reymond N, et al. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 2009;5:e1000651.
- Vogt SL, Evans AD, Guest RL, et al. The Cpx envelope stress response regulates and is regulated by small noncoding RNAs. J Bacteriol. 2014;196:4229–4238.
- Labandeira-Rey M, Brautigam CA, Hansen EJ. Characterization of the CpxRA regulon in Haemophilus ducreyi. Infect Immun. 2010;78:4779–4791.
- Dbeibo L, van Rensburg JJ, Smith SN, et al. Evaluation of CpxRA as a therapeutic target for uropathogenic Escherichia coli infections. Infect Immun. 2018;86(3). pii:e00798–17.
- Chao Y, Vogel J. A 3ʹ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol Cell. 2016;61:352–363.
- Grabowicz M, Koren D, Silhavy TJ. The CpxQ sRNA negatively regulates skp to prevent mistargeting of beta-barrel outer membrane proteins into the cytoplasmic membrane. MBio. 2016;7:e00312–16.
- Surmann K, Cudic E, Hammer E, et al. Molecular and proteome analyses highlight the importance of the Cpx envelope stress system for acid stress and cell wall stability in Escherichia coli. Microbiologyopen. 2016;5:582–596.
- Delhaye A, Collet JF, Laloux G. Fine-tuning of the Cpx envelope stress response is required for cell wall homeostasis in Escherichia coli. MBio. 2016;7:e00047–16.
- Guest RL, Wang J, Wong JL, et al. A bacterial stress response regulates respiratory protein complexes to control envelope stress adaptation. J Bacteriol. 2017;199(20). pii: e00153–17.
- Price NL, Raivio TL. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J Bacteriol. 2009;191:1798–1815.
- Humphreys S, Rowley G, Stevenson A, et al. Role of the two-component regulator CpxAR in the virulence of Salmonella enterica serotype Typhimurium. Infect Immun. 2004;72:4654–4661.
- Debnath I, Norton JP, Barber AE, et al. The Cpx stress response system potentiates the fitness and virulence of uropathogenic Escherichia coli. Infect Immun. 2013;81:1450–1459.
- Bontemps-Gallo S, Madec E, Lacroix JM. The two-component system CpxAR is essential for virulence in the phytopathogen bacteria Dickeya dadantii EC3937. Environ Microbiol. 2015;17(11):4415–4428.
- Thomassin JL, Giannakopoulou N, Zhu L, et al. The CpxRA two-component system is essential for Citrobacter rodentium virulence. Infect Immun. 2015;83:1919–1928.
- Tanner JR, Li L, Faucher SP, et al. The CpxRA two-component system contributes to Legionella pneumophila virulence. Mol Microbiol. 2016;100:1017–1038.
- Nakayama S, Watanabe H. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J Bacteriol. 1995;177:5062–5069.
- Gangaiah D, Raterman EL, Wu H, et al. Both MisR (CpxR) and MisS (CpxA) are required for Neisseria gonorrhoeae infection in a murine model of lower genital tract infection. Infect Immun. 2017;85(9). pii:e00307–17.
- Li H, Liu F, Peng W, et al. The CpxA/CpxR two-component system affects biofilm formation and virulence in Actinobacillus pleuropneumoniae. Front Cell Infect Microbiol. 2018;8:72.
- van Rensburg JJ, Fortney KR, Chen L, et al. Development and validation of a high-throughput cell-based screen to identify activators of a bacterial two-component signal transduction system. Antimicrob Agents Chemother. 2015;59:3789–3799.
- De Wulf P, Akerley BJ, Lin EC. Presence of the Cpx system in bacteria. Microbiology. 2000;146(Pt 2):247–248.
- Raivio TL, Silhavy TJ. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol. 1997;179:7724–7733.
- Raivio TL, Popkin DL, Silhavy TJ. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J Bacteriol. 1999;181:5263–5272.
- Zhou X, Keller R, Volkmer R, et al. Structural basis for two-component system inhibition and pilus sensing by the auxiliary CpxP protein. J Biol Chem. 2011;286:9805–9814.
- Cosma CL, Danese PN, Carlson JH, et al. Mutational activation of the Cpx signal transduction pathway of Escherichia coli suppresses the toxicity conferred by certain envelope-associated stresses. Mol Microbiol. 1995;18:491–505.
- Pogliano J, Dong JM, De Wulf P, et al. Aberrant cell division and random ftsz ring positioning in escherichia coli cpxA* mutants. J Bacteriol. 1998;180:3486–3490.
- De Wulf P, Lin EC. Cpx two-component signal transduction in Escherichia coli: excessive CpxR-P levels underlie CpxA* phenotypes. J Bacteriol. 2000;182:1423–1426.
- De Wulf P, Kwon O, Lin EC. The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J Bacteriol. 1999;181:6772–6778.
- Danese PN, Snyder WB, Cosma CL, et al. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 1995;9:387–398.
- Danese PN, Silhavy TJ. CpxP, a stress-combative member of the Cpx regulon. J Bacteriol. 1998;180:831–839.
- Danese PN, Murphy CK, Silhavy TJ. Multicopy suppression of cold-sensitive sec mutations in Escherichia coli. J Bacteriol. 1995;177:4969–4973.
- Pogliano J, Lynch AS, Belin D, et al. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182.
- Lima BP, Lennon CW, Ross W, et al. In vitro evidence that RNA polymerase acetylation and acetyl phosphate-dependent CpxR phosphorylation affect cpxP transcription regulation. FEMS Microbiol Lett. 2016;363:fnw011.
- Wolfe AJ. Physiologically relevant small phosphodonors link metabolism to signal transduction. Curr Opin Microbiol. 2010;13:204–209.
- Zietz BP, Dunkelberg H. The history of the plague and the research on the causative agent Yersinia pestis. Int J Hyg Environ Health. 2004;207:165–178.
- Galindo CL, Rosenzweig JA, Kirtley ML, et al. Pathogenesis of Y. enterocolitica and Y. pseudotuberculosis in human yersiniosis. J Pathog. 2011;2011:182051.
- Mikula KM, Kolodziejczyk R, Goldman A. Yersinia infection tools-characterization of structure and function of adhesins. Front Cell Infect Microbiol. 2013;2:169.
- Grassl GA, Bohn E, Muller Y, et al. Interaction of Yersinia enterocolitica with epithelial cells: invasin beyond invasion. Int J Med Microbiol. 2003;293:41–54.
- Carlsson KE, Liu J, Edqvist PJ, et al. Influence of the Cpx extracytoplasmic-stress-responsive pathway on Yersinia sp.-eukaryotic cell contact. Infect Immun. 2007;75:4386–4399.
- Ellison DW, Miller VL. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J Bacteriol. 2006;188:5101–5112.
- Heroven AK, Nagel G, Tran HJ, et al. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol Microbiol. 2004;53:871–888.
- Nagel G, Lahrz A, Dersch P. Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol Microbiol. 2001;41:1249–1269.
- Revell PA, Miller VL. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol Microbiol. 2000;35:677–685.
- Quade N, Mendonca C, Herbst K, et al. Structural basis for intrinsic thermosensing by the master virulence regulator RovA of Yersinia. J Biol Chem. 2012;287:35796–35803.
- Herbst K, Bujara M, Heroven AK, et al. Intrinsic thermal sensing controls proteolysis of Yersinia virulence regulator RovA. PLoS Pathog. 2009;5:e1000435.
- Ellison DW, Miller VL. Regulation of virulence by members of the MarR/SlyA family. Curr Opin Microbiol. 2006;9:153–159.
- Cathelyn JS, Ellison DW, Hinchliffe SJ, et al. The RovA regulons of Yersinia enterocolitica and Yersinia pestis are distinct: evidence that many RovA-regulated genes were acquired more recently than the core genome. Mol Microbiol. 2007;66:189–205.
- Yang F, Ke Y, Tan Y, et al. Cell membrane is impaired, accompanied by enhanced type III secretion system expression in Yersinia pestis deficient in RovA regulator. PLoS One. 2010;5:e12840.
- Zhang Y, Gao H, Wang L, et al. Molecular characterization of transcriptional regulation of rovA by PhoP and RovA in Yersinia pestis. PLoS One. 2011;6:e25484.
- Heroven AK, Dersch P. RovM, a novel LysR-type regulator of the virulence activator gene rovA, controls cell invasion, virulence and motility of Yersinia pseudotuberculosis. Mol Microbiol. 2006;62:1469–1483.
- Heroven AK, Sest M, Pisano F, et al. Crp induces switching of the CsrB and CsrC RNAs in Yersinia pseudotuberculosis and links nutritional status to virulence. Front Cell Infect Microbiol. 2012;2:158.
- Lawrenz MB, Miller VL. Comparative analysis of the regulation of rovA from the pathogenic yersiniae. J Bacteriol. 2007;189:5963–5975.
- Nuss AM, Schuster F, Kathrin Heroven A, et al. A direct link between the global regulator PhoP and the Csr regulon in Y. pseudotuberculosis through the small regulatory RNA CsrC. RNA Biol. 2014;11:580–593.
- Liu J, Obi IR, Thanikkal EJ, et al. Phosphorylated CpxR restricts production of the RovA global regulator in Yersinia pseudotuberculosis. PLoS One. 2011;6:e23314.
- Nuss AM, Schuster F, Roselius L, et al. A precise temperature-responsive bistable switch controlling Yersinia virulence. PLoS Pathog. 2016;12:e1006091.
- Heroven AK, Bohme K, Rohde M, et al. A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol Microbiol. 2008;68:1179–1195.
- Dai Q, Xu L, Xiao L, et al. RovM and CsrA negatively regulate urease expression in Yersinia pseudotuberculosis. Front Microbiol. 2018;9:348.
- Zhao R, Song Y, Dai Q, et al. A starvation-induced regulator, RovM, acts as a switch for planktonic/biofilm state transition in Yersinia pseudotuberculosis. Sci Rep. 2017;7:639.
- Song Y, Xiao X, Li C, et al. The dual transcriptional regulator RovM regulates the expression of AR3- and T6SS4-dependent acid survival systems in response to nutritional status in Yersinia pseudotuberculosis. Environ Microbiol. 2015;17(11):4631–4645.
- Vadyvaloo V, Hinz AK. A LysR-Type transcriptional regulator, RovM, Senses nutritional cues suggesting that it is involved in metabolic adaptation of Yersinia pestis to the flea gut. PLoS One. 2015;10:e0137508.
- Liu L, Fang H, Yang H, et al. Reciprocal regulation of Yersinia pestis biofilm formation and virulence by RovM and RovA. Open Biol. 2016;6.
- Raivio TL. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol. 2005;56:1119–1128.
- Dorel C, Lejeune P, Rodrigue A. The Cpx system of Escherichia coli, a strategic signaling pathway for confronting adverse conditions and for settling biofilm communities? Res Microbiol. 2006;157:306–314.
- Flores-Kim J, Darwin AJ. Links between type III secretion and extracytoplasmic stress responses in Yersinia. Front Cell Infect Microbiol. 2012;2:125.
- Liu J, Thanikkal EJ, Obi IR, et al. Elevated CpxR~P levels repress the Ysc-Yop type III secretion system of Yersinia pseudotuberculosis. Res Microbiol. 2012;163:518–530.
- DiGiuseppe PA, Silhavy TJ. Signal detection and target gene induction by the CpxRA two-component system. J Bacteriol. 2003;185:2432–2440.
- Carlsson KE, Liu J, Edqvist PJ, et al. Extracytoplasmic-stress-responsive pathways modulate type III secretion in Yersinia pseudotuberculosis. Infect Immun. 2007;75:3913–3924.
- Yamamoto K, Ishihama A. Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci Biotechnol Biochem. 2006;70:1688–1695.
- De Wulf P, McGuire AM, Liu X, et al. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J Biol Chem. 2002;277:26652–26661.
- Avican K, Fahlgren A, Huss M, et al. Reprogramming of Yersinia from virulent to persistent mode revealed by complex in vivo RNA-seq analysis. PLoS Pathog. 2015;11:e1004600.
- Horne SM, Pruss BM. Global gene regulation in Yersinia enterocolitica: effect of FliA on the expression levels of flagellar and plasmid-encoded virulence genes. Arch Microbiol. 2006;185:115–126.
- Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993;32:11741–11753.
- Galperin MY. Diversity of structure and function of response regulator output domains. Curr Opin Microbiol. 2010;13:150–159.
- Barbieri CM, Wu T, Stock AM. Comprehensive analysis of OmpR phosphorylation, dimerization, and DNA binding supports a canonical model for activation. J Mol Biol. 2013;425:1612–1626.
- Jubelin G, Vianney A, Beloin C, et al. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol. 2005;187:2038–2049.
- Bölin I, Wolf-Watz H. Molecular cloning of the temperature-inducible outer membrane protein 1 of Yersinia pseudotuberculosis. Infect Immun. 1984;43:72–78.
- Grabenstein JP, Marceau M, Pujol C, et al. The response regulator PhoP of Yersinia pseudotuberculosis is important for replication in macrophages and for virulence. Infect Immun. 2004;72:4973–4984.
- Wolfe AJ, Parikh N, Lima BP, et al. Signal integration by the two-component signal transduction response regulator CpxR. J Bacteriol. 2008;190:2314–2322.
- Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367.
- Francis MS, Amer AA, Milton DL, et al. Site-directed mutagenesis and its application in studying the interactions of T3S components. Methods Mol Biol. 2017;1531:11–31.
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675.
- Pogliano J, Dong JM, De Wulf P, et al. Aberrant cell division and random FtsZ ring positioning in Escherichia coli cpxA* mutants. J Bacteriol. 1998;180:3486–3490.
